Abstract
The T-cell leukemia/lymphoma 1 (TCL1) oncogene is a target of chromosomal translocations and inversions at 14q31.2, and its rearrangement in T cells causes T-cell prolymphocytic leukemias. TCL1 dysregulation in B cells is responsible for the development of an aggressive form of chronic lymphocytic leukemia (CLL), the most common human leukemia. We have investigated the mechanisms underlying the oncogenic functions of Tcl1 protein using a mass spectrometry approach and have identified Atm (ataxia-telangiectasia mutated) as a candidate Tcl1-interacting protein. The Tcl1-Atm complex formation was validated by coimmunoprecipitation experiments. Importantly, we show that the association of Atm with Tcl1 leads to enhanced IκBα phosphorylation and ubiquitination and subsequent activation of the NF-κB pathway. Our findings reveal functional cross-talk between Atm and Tcl1 and provide evidence for a novel pathway that could be targeted in leukemias and lymphomas.
Introduction
The T-cell leukemia/lymphoma 1 (TCL1) gene maps to chromosome 14q31.2 and is involved in chromosomal translocations and inversions in T-cell prolymphocytic leukemia (T-PLL). It is also dysregulated in several B-cell malignancies, including B-cell chronic lymphocytic leukemia (B-CLL) and in germ cell tumors, such as seminoma and dysgerminoma.1,2 Normally, Tcl1 protein is expressed in early embryos and fetal tissues, germ cells, and early T and B cells.3-5 We previously showed that aggressive human CLLs overexpress Tcl16 and that transgenic mice expressing TCL1 in B cells developed an aggressive form of CLL.7,8 These previous studies demonstrated that Tcl1 up-regulation is critical in the pathogenesis of the aggressive form of human CLL. Tcl1 is also known to be a coactivator of the Akt oncoprotein9 and of the NF-κB transcription pathway.10 Despite efforts to clarify the molecular mechanisms of Tcl1 oncogenic function, the identification of specific pathways through which Tcl1 modulates oncogenic functions has been elusive. This was mainly because Tcl1 interacting proteins could not be identified by conventional methods aimed to isolate protein-protein complexes, with the exception of Akt, a protein unlikely to play a significant role in B-cell malignancies.10,11 The ATM (ataxia-telangiectasia mutated) gene, located at chromosome 11q22.3,12 is responsible for a rare disorder called ataxia-telangiectasia (AT), an autosomal recessive disease characterized by progressive cerebellar degeneration, variable immunodeficiency, genomic instability, and susceptibility to cancer, especially lymphoid malignancies.12-14 ATM is thought to be a tumor suppressor gene because DNA double-strand breaks caused by ionizing radiation or chemicals result in rapid Atm autophosphorylation and activation, leading to checkpoint activation and phosphorylation of substrates that regulate cell-cycle progression, DNA repair, recombination, transcription, and cell death.12,15
Atm is a Ser/Thr kinase responsible for the phosphorylation of many substrates, including p53, Nbs1, Chk2, H2A.X, Mdm2, 4E-BP1, and IκBα. IκBα is an inhibitor of NF-κB, a transcription factor involved in a variety of hematologic and solid tumor malignancies.16 IκBα17-21 phosphorylation leads to its degradation followed by activation of NF-κB. Atm has a carboxy-terminal sequence with significant homology to the catalytic domain of phosphatidylinositol-3-OH kinase (PI3K).15 AKT is a downstream target of PI3K and interacts directly with Tcl1.22 ATM biallelic loss in AT leads to chromosomal translocations and inversions at 14q31.2, resulting in Tcl1 overexpression in AT patients. Preleukemic and leukemic T cells from AT patients with 14q31.2 chromosomal rearrangements have been found to overexpress TCL1.12,23,24
We assumed that Tcl1 interacts with some proteins involved in important pathways up-regulated in hematologic malignancies. The aim of this work was to identify new partners of Tcl1 and uncover its role in oncogenesis using a mass spectrometry approach.
Methods
DNA constructs
Human TCL1 full-length was cloned into a pCMV5 vector to obtain both pCMV5-TCL1 WT and pCMV5-TCL1 WT HA-tagged vectors. The pcDNA 3.1 ATM-wt HIS6-FLAG-tagged and pcDNA 3.1 ATM-KD HIS6-FLAG constructs were obtained from Dr M. B. Kastan (Department of Oncology, St Jude Children's Research Hospital, Memphis, TN).
A glutathione S-transferase (GST)–encoding DNA fragment was cloned between the Omni-tag and TCL1 cDNA sequence of the Omni-TCL1 construct to generate the Omni-GST-TCL1 construct. IκBα cDNA was cloned into a pcDNA3.1-V5-His6 tag vector in frame with the tags. The Dual-luciferase Reporter Assay System and Renilla luciferase reporter vector pRL-TK were purchased from Promega. pNF-κB-Luc and the construct encoding the kinase domain of MEKK1 under the transcriptional control of the CMV promoter, pFC-MEKK, were purchased from Stratagene.
Cells, transfections, and antisera
HEK-293 cells were grown in DMEM with 10% FBS and 100 μg/L gentamicin at 37°C. FuGene 6 transfection reagent and protease inhibitor mixture tablets were purchased by Roche Diagnostics. Daudi lymphoma cells were grown in RPMI 1640 medium with 10% FBS and 100 μg/L gentamicin at 37°C. Daudi cells and CLL samples were transfected with nucleofector following AMAXA guidelines for cells in suspension.
Immunoblots were developed using denville HyGlo ECL. Antibodies used were: anti-Tcl1 (Santa Cruz Biotechnology; sc-32331), for Western blots and immunoprecipitation, anti-Atm (Santa Cruz Biotechnology; sc-23921), resin conjugated with Atm from Bethyl (for IPs), anti-Egr1 (Santa Cruz Biotechnology, sc-189) and anti-IκBα (Cell Signaling), resin-conjugated HA (Bethyl for mass spectometry experiments and IPs), anti–HA-HRP (Roche Diagnostics), and anti-V5 (Invitrogen for ubiquitination experiments).
In vitro cell growth assessment
Daudi cells were seeded at 2 × 104 cells per 60-mm diameter dish; cells were either untreated or treated with TCL1 siRNA, Kudos, or combinations of both. Cells were monitored and counted at 24-hour intervals.
Western blotting, immunoprecipitation analysis, and cell fractionation
Coimmunoprecipitation experiments were performed by incubating 1 mg of total lysates with resin-conjugated HA (Bethyl) or with anti-Tcl1 antibody conjugated with Sepharose beads, or with Atm-resin conjugated overnight at 4°C; after washing, beads were boiled in 1× SDS sample buffer and proteins separated on 4% to 20% polyacrylamide gels (Bio-Rad). A kit from Biovision was used to perform protein fractionation of cytosol and nucleus. GST pull-down was done by incubating 1 mg of whole-cell extracts with GST beads with rocking overnight at 4°C. Dithiobis (succinimidylpropionate) crosslinker was from Pierce Chemical.
RNA isolation and quantitative RT-PCR
Purification of total RNA was performed using TRIzol reagent (Invitrogen). A total of 500 ng of total RNA was used for the retrotranscription step. The 2−ΔCt method was used to calculate the relative abundance of EGR1 compared with GAPDH expression. To generate complete cDNAs for expression of proteins, 500 ng of total RNA was retrotranscribed using a high-capacity cDNA reverse transcription kit (Applied Biosystems) with random primers, according to the manufacturer's protocol. Real-time PCR analysis of the samples was carried out with SYBR Green PCR master mix (Applied Biosystems). EGR1 and GAPDH primers were bought from Real-Time Primers LLC.
Mass spectrometry studies
Protein pellets were solubilized and digested by trypsin. Protein constituents were identified by liquid chromatography tandem mass spectrometry (LC-MS/MS). Inspection of LC-MS/MS data was undertaken to assess exclusive presence of mass peaks belonging to candidate partner proteins in samples from cells transfected with tagged TCL1-HA.
Digestion and MALDI analysis
Immunoprecipitated protein complexes were digested with sequencing grade trypsin from Promega using the Multiscreen Solvinert Filter Plates from Millipore. Briefly, complexes were incubated with a dithiothreitol solution (25mM in 100mM ammonium bicarbonate) for 30 minutes before the addition of 55mM iodoacetamide in 100mM ammonium bicarbonate solution. Iodoacetamide was incubated with protein complexes in the dark for 30 minutes before removal. Enzymatic digestion was carried out with trypsin (12.5 ng/μL) for 18 hours at 37°C. The digestion was stopped with addition of 0.5% trifluoroacetic acid. The MS analysis was immediately performed to ensure high-quality tryptic peptides with minimal nonspecific peptides.
Mass spectrometry, LTQ
Capillary-liquid chromatography-nanospray tandem mass spectrometry (LC/MS/MS) was performed on a Thermo Finnigan LTQ mass spectrometer equipped with a nanospray source operated in positive ion mode. The LC system was an UltiMate 3000 system from Dionex. The solvent A was water containing 50mM acetic acid, and the solvent B was acetonitrile. A total of 5 μL of each sample was first injected onto the m-Precolumn Cartridge (Dionex) and washed with 50mM acetic acid. The injector port was switched to inject, and the peptides were eluted off the trap onto the column. A 5-cm, 75-mm ID ProteoPep II C18 column (New Objective) packed directly in the nanospray tip was used for chromatographic separations. Peptides were eluted directly off the column into the LTQ system using a gradient of 2% to 80% B over 45 minutes, with a flow rate of 300 nL/min. The total run time was 65 minutes. The MS/MS was acquired according to standard conditions established in the laboratory. Briefly, a nanospray source operated with a spray voltage of 3 kV and a capillary temperature of 200°C. The scan sequence of the mass spectrometer was based on the TopTen method; the analysis was programmed for a full scan recorded between 350 and 2000 Da and an MS/MS scan to generate product ion spectra to determine amino acid sequence in consecutive instrument scans of the 10 most abundant peaks in the spectrum. The CID fragmentation energy was set to 35%. Dynamic exclusion was enabled with a repeat count of 2 within 10 seconds, a mass list size of 200, an exclusion duration 350 seconds, the low mass width was 0.5, and the high mass width was 1.5.
Protein identification
The RAW data files collected on the mass spectrometer were converted to mzXML and MGF files by use of MassMatrix data conversion tools (Version 1.3; http://www.massmatrix.net/download). For low mass accuracy data, tandem MS spectra that were not derived from singly charged precursor ions were considered as both doubly and triply charged precursors. The resulting MGF files were searched using Mascot Daemon by Matrix Science Version 2.2.2, and the database searched against the full SwissProt Version 57.5 database (471 472 sequences; 167 326 533 residues) or NCBI database Version 20091013 (9 873 339 sequences; 3 367 482 728 residues). The mass accuracy of the precursor ions was set to 2.0 Da, given that the data were acquired on an ion trap mass analyzer and the fragment mass accuracy was set to 0.5 Da. Considered modifications (variable) were methionine oxidation and carbamidomethyl cysteine. Two missed cleavages for the enzyme were permitted. A decoy database was searched to determine the false discovery rate, and peptides were filtered according to the false discovery rate and proteins identified required bold red peptides. Protein identifications were checked manually, and proteins with a Mascot score of 50 or higher with a minimum of 2 unique peptides from 1 protein having a -b or -y ion sequence tag of 5 residues or better were accepted.
Ubiquitination experiments
Total protein lysates (1 mg) were incubated overnight with V5 antibody (Invitrogen) and AG/agarose beads and then run on 4% to 20% polyacrylamide gels. The nitrocellulose membrane was immunoblotted with anti–HA-HRP to detect Ub-HA (ubiquitin), anti-IκBα, anti-Atm, and anti-Tcl1A.
Luciferase assay
HEK-293 cells were transfected with the indicated constructs. Firefly and Renilla luciferase activities were assayed with the Dual Luciferase Assay System (Promega), and firefly luciferase activity was normalized to Renilla luciferase activity, as suggested by the manufacturer. All experiments were carried out in triplicate and repeated 3 times with consistent results.
Isolation of B cells
CD19+ B cells were isolated from mouse spleen following the protocol and using reagents from Miltenyi Biotec (CD19 microbeads, mouse).
Patients
B-CLL samples were obtained after informed consent in accordance with the Declaration of Helsinki from patients diagnosed with B-CLL from the CLL Research Consortium. Research was performed with the approval of the Institutional Review Board of The Ohio State University. Briefly, blood was obtained from CLL patients, and lymphocytes were isolated through Ficoll/Hypaque gradient centrifugation (GE Healthcare) and processed for protein extraction.
Animal studies
Mice were maintained and animal experiments conducted under institutional guidelines established for the Animal Facility at The Ohio State University; nu/ν mice were obtained from The Jackson Laboratory. Tumors were established by injecting Daudi lymphoma cells (200 μL PBS, 7.5 × 107 cells/mouse) into the right flanks of female nude mice at 6 weeks of age (Charles River Breeding Laboratories). Tumor size was measured daily until tumors reached 50 mm3. Then, 5 μg of synthetic siRNA for Tcl1 or scrambled siRNA (si-Scr) diluted in Lipofectamine and w/wo Kudos 55933 (Atm inhibitor) in combination (50 μL total volume) were injected directly into the tumors and after 3, 7, and 10 days. Tumors were measured on the day of the injections and 4 days after the last injection. At that time, the mice were killed and tumors were weighed. Tumor volumes were calculated using the equation V (in mm3) = A × B2/2, where A is the largest diameter and B is the perpendicular diameter.
Statistics
All graph values represent the mean ± SEM from 3 independent experiments with each measured in triplicate. The differences between 2 groups were analyzed with unpaired 2-tailed Student t test. P < .05 was considered statistically significant and indicated with asterisks as described in the figure legends.
Results
Tcl1 interacts with Atm and both affect IκBα expression
To identify Tcl1-interacting proteins, we used a TCL1 cDNA modified at its 3′ end with a sequence encoding a HA tag (TCL1-HA). A549 lung cancer–derived cells, used as recipient, were transfected either either with wild-type TCL1 (used as a control) or TCL1-HA and treated with dithiobis (succinimidylpropionate), a cross-linker that fixes protein complexes in vivo. We used A549 cells in our experiment because no CLL cell lines are currently available; furthermore, these cells can be easily transfected at a very high efficiency, an important requirement for such an experiment.
Cells were lysed; and Tcl1-HA protein, along with candidate protein partners, was isolated using a HA-conjugated resin. Purified proteins were treated with dithiothreitol to cleave dithiobis (succinimidylpropionate) and dissociate complexes, and digested by trypsin; protein constituents were identified by LC-MS/MS (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Among the identified candidate interactors, we focused on Atm because its role in leukemogenesis has been already assessed.13
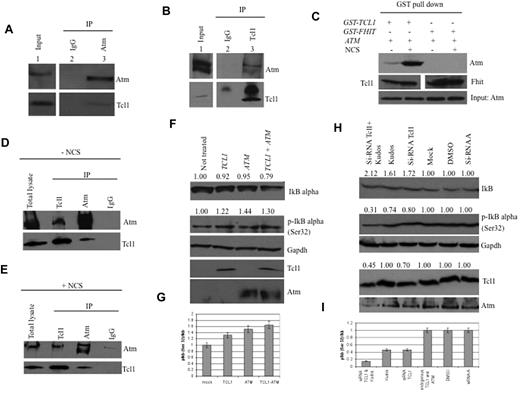
The potential interaction between Tcl1 and Atm proteins was investigated using coimmunoprecipitation experiments. HEK293 cells were stably transfected with a FLAG-ATM construct and infected with a recombinant Adeno-TCL1. Infected cell lysates were immunoprecipitated with Atm antisera, followed by immunoblotting with both Tcl1 and Atm antisera. As shown in Figure 1A (bottom panel lane 3), Tcl1 interacts with Atm; no Tcl1 protein was detected in anti-IgG immunoprecipitates (Figure 1A lane 2). The same complexes were detected in the reverse experiment, where cell lysates were immunoprecipitated with anti-Tcl1 and probed with both Atm and Tcl1 antisera (Figure 1B).
Tcl1 interacts with Atm, and both affect IκBα expression. (A) HEK293 cells were stably transfected with expression plasmid encoding FLAG-ATM and then infected with Ad-TCL1 wild-type (MOI 100). Forty-eight hours after infection, whole-cell lysates were immunoprecipitated with anti-Atm (resin conjugated to Atm). The immunoprecipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. Input: lysate expression of Atm (top) and Tcl1 (bottom). (B) HEK293 cells were transiently cotransfected with expression plasmids encoding mammalian FLAG-ATM (8 μg) and wild-type TCL1 (6 μg). Forty-eight hours after transfection, cell lysates were immunoprecipitated with anti-TCL1. The immunoprecipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. Input: lysate expression of Atm (top) and Tcl1 (bottom). (C) HEK293 cells were transiently cotransfected with expression plasmids encoding mammalian Omni-GST-TCL1 (6 μg) or Omni-GST-FHIT (6 μg) and FLAG-HIS-ATM (8 μg). Forty-eight hours after transfection, cells were treated with neocarzinostatin (NCS) 200 ng/mL for 2 hours. Cell lysates were GST-pulled down and immunoblotted with anti-His6, anti-Tcl1, and anti-Fhit. Input: lysate expression of Atm. (D-E) Daudi cells were untreated or treated with hydroxyurea, 50mM for 3 hours, then lysed and immunoprecipitated, performed with anti-Atm, anti-Tcl1, or IgG. The precipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. (F) Tcl1 and Atm coexpression is associated with decreased IκBα total protein. HEK293 cells were mock-transfected or transfected with TCL1 or ATM plasmids separately or together (4 μg for each plasmid, including empty vector). Forty-eight hours later, cells were treated with hydroxyurea 50mM for 3 hours and then analyzed by Western blot with the indicated antibodies. (G) Tcl1 and Atm coexpression is associated with increased IκBα (Ser32) phosphorylation as shown in the graph. (H) Daudi cells were mock-transfected or transfected with si-TCL1 or si-Scr, and then untreated or treated with Kudos 55933 or DMSO. The level of IκBα was measured by Western blot. (I) IκBα (Ser 32) levels are shown in the graph. Data are representative of 3 independent experiments.
Tcl1 interacts with Atm, and both affect IκBα expression. (A) HEK293 cells were stably transfected with expression plasmid encoding FLAG-ATM and then infected with Ad-TCL1 wild-type (MOI 100). Forty-eight hours after infection, whole-cell lysates were immunoprecipitated with anti-Atm (resin conjugated to Atm). The immunoprecipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. Input: lysate expression of Atm (top) and Tcl1 (bottom). (B) HEK293 cells were transiently cotransfected with expression plasmids encoding mammalian FLAG-ATM (8 μg) and wild-type TCL1 (6 μg). Forty-eight hours after transfection, cell lysates were immunoprecipitated with anti-TCL1. The immunoprecipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. Input: lysate expression of Atm (top) and Tcl1 (bottom). (C) HEK293 cells were transiently cotransfected with expression plasmids encoding mammalian Omni-GST-TCL1 (6 μg) or Omni-GST-FHIT (6 μg) and FLAG-HIS-ATM (8 μg). Forty-eight hours after transfection, cells were treated with neocarzinostatin (NCS) 200 ng/mL for 2 hours. Cell lysates were GST-pulled down and immunoblotted with anti-His6, anti-Tcl1, and anti-Fhit. Input: lysate expression of Atm. (D-E) Daudi cells were untreated or treated with hydroxyurea, 50mM for 3 hours, then lysed and immunoprecipitated, performed with anti-Atm, anti-Tcl1, or IgG. The precipitates were analyzed by Western blot with anti-Atm or anti-Tcl1. (F) Tcl1 and Atm coexpression is associated with decreased IκBα total protein. HEK293 cells were mock-transfected or transfected with TCL1 or ATM plasmids separately or together (4 μg for each plasmid, including empty vector). Forty-eight hours later, cells were treated with hydroxyurea 50mM for 3 hours and then analyzed by Western blot with the indicated antibodies. (G) Tcl1 and Atm coexpression is associated with increased IκBα (Ser32) phosphorylation as shown in the graph. (H) Daudi cells were mock-transfected or transfected with si-TCL1 or si-Scr, and then untreated or treated with Kudos 55933 or DMSO. The level of IκBα was measured by Western blot. (I) IκBα (Ser 32) levels are shown in the graph. Data are representative of 3 independent experiments.
Because DNA-damaging agents cause Atm phosphorylation, we investigated whether such treatment could affect the Tcl1-Atm interaction. HEK293 cells were cotransfected with FLAG-ATM and Omni-GST-TCL1 plasmids and treated with neocarzinostatin, a small protein that induces sequence-specific DNA single- and double-strand breaks.25 Tcl1 binds Atm with higher affinity after treatment with the DNA-damaging agent, whereas protein complexes with Omni-GST-FHIT protein, used as a negative control, were not detected (Figure 1C). The interaction was also validated with endogenous proteins. The results also indicated (Figure 1C) that Tcl1 has a higher affinity for activated Atm. The structural basis for this phenomenon will require further investigation. Tcl1 and Atm were immunoprecipitated and immunoblotted in Daudi lymphoma cells with or without the DNA-damaging agent (Figure 1D-E). To understand the functional relevance of the Tcl1-Atm interaction, we analyzed the phosphorylation status of several Atm substrates in the presence of Tcl1. Interestingly, we detected an increase of Atm-mediated IκBα phosphorylation in the presence of Tcl1 (Figure 1F). Because Atm-mediated phosphorylation of IκBα enhances its degradation, we examined the expression level of IkB α after Tcl1 and Atm coexpression. DNA double-strand breaks induced by hydroxyurea are responsible for Atm activation.26 HEK293 cells were mock-transfected or transfected with TCL1 and ATM constructs, separately or together (Figure 1F-G). Forty-eight hours later, cells were treated with hydroxyurea for 3 hours and cell lysates were analyzed by Western blotting. We found that Tcl1 and Atm coexpression was associated with increased levels of the phosphorylated form of IκBα and reduced levels of total IκBα (Figure 1F). Figure 1G shows an increased ratio of phosphorylated IκBα (Ser 32) relative to total IκBα. Daudi lymphoma cells were treated with siRNA for Tcl1, Kudos 55933 (an Atm inhibitor), and a combination of both; both IκBα expression and its phosphorylation were measured (Figure 1H-I). Taken together, these data indicate that the interaction of endogenous Tcl1 and Atm is responsible for an increase in phosphorylation and degradation of IκBα and in activation of NF-κB, whereas inhibition of Tcl1 and Atm resulted in decreased activity of the NF-κB pathway.
Expression of Tcl1 and Atm activates the NF-κB pathway
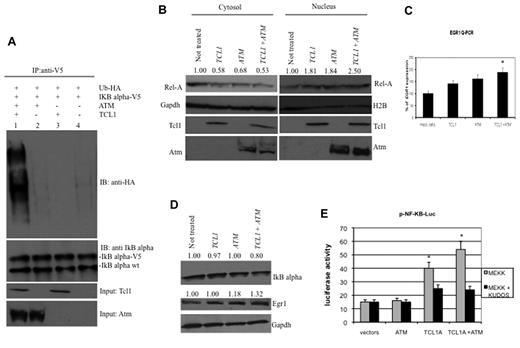
IκBα is phosphorylated by Atm, leading to its ubiquitination and degradation.17,19,27 To determine whether Tcl1 expression affects IκBα regulation through Atm, we carried out a ubiquitination assay. HEK293 cells were transfected with TCL1, ATM, Ub-HA, and IκBα-V5 expression vectors (Figure 2A). Forty-eight hours after transfection, cells were treated with MG132, a proteasome inhibitor, 10μM for additional 3 hours, then lysed and protein immunoprecipitated with anti-V5 followed by immunoblotting with anti-HA. Coexpression of Tcl1 and Atm resulted in IκBα ubiquitination (Figure 2A lane 1), whereas expression of Atm or Tcl1 alone did not (Figure 2A lanes 2 and 3). These results suggest that Tcl1 expression enhances Atm-mediated phosphorylation and ubiquitination of IκBα. Because ubiquitination of IκBα is associated with NF-κB activation and translocation from the cytosol to nucleus,20,21,28 we investigated whether active p65 NF-κB (Rel-A) translocates from the cytosol to the nucleus when TCL1 and ATM are coexpressed (Figure 2B). To this end, HEK293 cells were transfected with ATM and TCL1 constructs, separately or together; then, cell lysates were fractionated to obtain nuclear and cytosolic fractions and the fractions run on gels. The level of nuclear Rel-A increased when TCL1 and ATM were coexpressed (Figure 2B). Expression of Rel-A was compared with Gapdh and H2A (histone 2A) expression, as markers for cytosol and nucleus, respectively. These results suggest that Tcl1 and Atm coexpression is associated with Rel-A translocation into the nucleus. After its translocation into the nucleus, Rel-A enhances the transcription of several target genes, including EGR1 (early growth response 1); indeed, Rel-A binds EGR1 promoter sequences and enhances EGR1 mRNA and protein expression.29 Thus, we determined whether TCL1 expression could modulate this Rel-A function. HEK293 cells were transfected with TCL1 and ATM separately or together and treated with hydroxyurea for 3 hours and protein lysates fractionated on gels. Egr1 protein levels were increased when Tcl1 and Atm were coexpressed (Figure 2D middle panel). EGR1 mRNA expression measured by quantitative RT-PCR was also increased during coexpression of TCL1 and ATM (Figure 2C). To determine whether Tcl1 expression directly affects the transactivating function of NF-κB, we used a commercial system based on the ability of mitogen-activated protein kinase kinase 1 (MEKK1) to activate an NF-κB reporter construct, pNF-κB-Luc expressing luciferase under the control of an NF-κB–responsive element. Expression of ATM alone did not affect the reporter activity. Interestingly, Tcl1 by itself increased reporter activity by 2.5-fold (Figure 2E). When coexpressed, the Tcl1-Atm complex enhanced the NF-κB transactivating function approximately 4-fold (Figure 2E). To confirm that Tcl1 and Atm are equally important in NF-κB activation, we treated cells with Kudos 55933, an inhibitor of endogenous and exogenous Atm. Kudos inhibited NF-κB activation by Tcl1 or by Tcl1 plus Atm (Figure 2E right), suggesting that this function of Tcl1 is enhanced by endogenous as well as exogenous Atm. In summary, these results suggest that Tcl1 and Atm cooperate to induce NF-κB activation.
Expression of Tcl1 and Atm activates the NF-κB pathway. (A) HEK293 cells were transfected with indicated plasmids. Forty-eight hours later, cells were treated with MG132, 10μM for 3 hours, and then lysates immunoprecipitated with anti-V5. The precipitates were analyzed by Western blot with anti-HA or anti-IκBα. Input: lysate expression of Tcl1 and Atm. (B) HEK293 cells were mock-transfected or transfected with TCL1 and ATM plasmids separately or together (4 μg each plasmid). Forty-eight hours later, cells were treated with hydroxyurea for 3 hours, fractionated into cytosolic and nuclear fractions, and analyzed by Western blot with the indicated antibodies. (C) mRNA levels of EGR1 were measured by quantitative RT-PCR. HEK293 cells were mock-transfected or transfected with TCL1 and ATM plasmids, separately or together for 48 hours, and then treated with hydroxyurea, 50mM for 3 hours). Fold changes of EGR1 were calculated using the 2−ΔCt method. GAPDH mRNA levels were used as an internal normalization control. Samples transfected with TCL1 and ATM have been normalized to mock-transfected sample. (D) HEK293 cells were transfected with TCL1 and ATM (4 μg for each plasmid), treated with hydroxyurea for 3 hours and with MG132, 10μM 6 hours, and protein lysates run on gel. (E) Tcl1 activates NF-κB–dependent transcription synergistically with Atm. HEK-293 cells were cotransfected with 50 ng of pNF-κB-Luc reporter and 50 ng of pRL-TK Renilla reporter constructs. In addition, 0.75 μg of CMV5-empty vector, 0.75 μg of pcDNA 3.1 empty vector, 0.75 μg of CMV5-TCL1 WT, and 0.75 μg of pcDNA 3.1 empty vector constructs were used; 5 ng of pFC-MEKK was added where indicated. Cells were treated with 10μM Kudos for 5 hours, where indicated. Data are representative of 3 independent experiments. (C,E) Data are mean ± SEM of 3 independent experiments, and each is measured in triplicate. *P < .05.
Expression of Tcl1 and Atm activates the NF-κB pathway. (A) HEK293 cells were transfected with indicated plasmids. Forty-eight hours later, cells were treated with MG132, 10μM for 3 hours, and then lysates immunoprecipitated with anti-V5. The precipitates were analyzed by Western blot with anti-HA or anti-IκBα. Input: lysate expression of Tcl1 and Atm. (B) HEK293 cells were mock-transfected or transfected with TCL1 and ATM plasmids separately or together (4 μg each plasmid). Forty-eight hours later, cells were treated with hydroxyurea for 3 hours, fractionated into cytosolic and nuclear fractions, and analyzed by Western blot with the indicated antibodies. (C) mRNA levels of EGR1 were measured by quantitative RT-PCR. HEK293 cells were mock-transfected or transfected with TCL1 and ATM plasmids, separately or together for 48 hours, and then treated with hydroxyurea, 50mM for 3 hours). Fold changes of EGR1 were calculated using the 2−ΔCt method. GAPDH mRNA levels were used as an internal normalization control. Samples transfected with TCL1 and ATM have been normalized to mock-transfected sample. (D) HEK293 cells were transfected with TCL1 and ATM (4 μg for each plasmid), treated with hydroxyurea for 3 hours and with MG132, 10μM 6 hours, and protein lysates run on gel. (E) Tcl1 activates NF-κB–dependent transcription synergistically with Atm. HEK-293 cells were cotransfected with 50 ng of pNF-κB-Luc reporter and 50 ng of pRL-TK Renilla reporter constructs. In addition, 0.75 μg of CMV5-empty vector, 0.75 μg of pcDNA 3.1 empty vector, 0.75 μg of CMV5-TCL1 WT, and 0.75 μg of pcDNA 3.1 empty vector constructs were used; 5 ng of pFC-MEKK was added where indicated. Cells were treated with 10μM Kudos for 5 hours, where indicated. Data are representative of 3 independent experiments. (C,E) Data are mean ± SEM of 3 independent experiments, and each is measured in triplicate. *P < .05.
Tcl1 and Atm are crucial for proliferation in vitro and in vivo
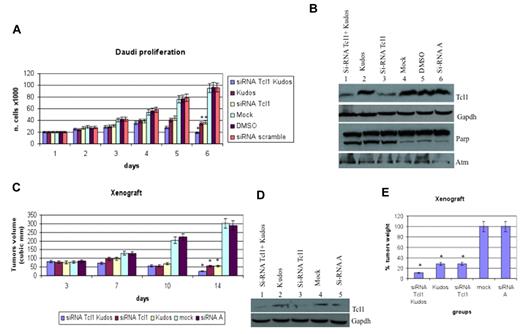
We investigated the role of Tcl1 depletion on the proliferation of Daudi lymphoma cells, which express both Tcl1 and Atm. The combination of Kudos 55933 treatment with Tcl1 depletion had an additive effect on cell proliferation (Figure 3A). Effects on proliferation were not observed after transfection with the carrier alone or scrambled siRNAs. The data reported represent the mean (SD) values of 3 different experiments (Figure 3A). Tcl1 protein expression was silenced with siRNAs. In Figure 3B, the effects of specific Tcl1-siRNA on Tcl1 protein expression levels are shown. Daudi cells were transfected with Tcl1-siRNAs in the absence or presence of Kudos 55933; in lanes 1 and 3, Tcl1 protein expression is down-regulated by Tcl1 siRNA. No effects on Tcl1 expression level were detected after scrambled siRNA transfection (lane 6). Total proteins extracted from Daudi cells, 5 days after transfection, were analyzed by immunoblotting (Figure 3B). The expression of Tcl1 was affected by Tcl1 siRNA; and more importantly, PARP was cleaved in the condition of depleted Tcl1 and Kudos inhibition of Atm. Finally, we investigated the effects of Tcl1 depletion by siTcl1 and Kudos 55933 treatment in vivo in a preclinical model of lymphoma; 5 groups of 4 mice each were injected with Daudi cells (Figure 3C). Xenograft tumors treated with Kudos 55933 or Tcl1 siRNA separately showed a reduction in volume compared with controls. A dramatic effect on inhibition of tumor growth was observed in the group of mice treated with Kudos 55933 and Tcl1 siRNA in combination. Total proteins extracted from tumors were analyzed by Western blotting, and Tcl1 expression was measured (Figure 3D). At the end of the experiment, tumors were excised and weighed (Figure 3E). These in vivo data confirmed the in vitro results and illustrated the effect of Atm and Tcl1 cross-talk.
Tcl1 and Atm are crucial for growth and proliferation of lymphoma cells in vitro and in vivo. (A) Daudi cells were mock-transfected or transfected with si-TCL1 or si-Scr and then untreated or treated with Kudos 55933 or DMSO. Cells were counted at 24-hour intervals. Results represent the average of 3 independent experiments. (B) Total lysates collected were analyzed by Western blot and tested with indicated antibodies. (C) Graph representing tumor volumes at indicated days during the experiment for the 5 groups indicated (4 mice/group). Tumor size was measured daily until the tumor reached 50 mm3. Then, 5 μg of synthetic si-TCL1or si-Scr diluted in Lipofectamine and with or without Kudos (50 μL total volume) were injected directly into the tumors and at 3, 7, and 10 days. Tumors were measured on the day of the injections and 4 days after the last injection. (D) Total lysates from tumors were analyzed by Western blot and tested with indicated antibodies. (E) Xenograft tumors were weighed. Data are mean ± SEM of 3 independent experiments, and each is measured in triplicate. *P < .05.
Tcl1 and Atm are crucial for growth and proliferation of lymphoma cells in vitro and in vivo. (A) Daudi cells were mock-transfected or transfected with si-TCL1 or si-Scr and then untreated or treated with Kudos 55933 or DMSO. Cells were counted at 24-hour intervals. Results represent the average of 3 independent experiments. (B) Total lysates collected were analyzed by Western blot and tested with indicated antibodies. (C) Graph representing tumor volumes at indicated days during the experiment for the 5 groups indicated (4 mice/group). Tumor size was measured daily until the tumor reached 50 mm3. Then, 5 μg of synthetic si-TCL1or si-Scr diluted in Lipofectamine and with or without Kudos (50 μL total volume) were injected directly into the tumors and at 3, 7, and 10 days. Tumors were measured on the day of the injections and 4 days after the last injection. (D) Total lysates from tumors were analyzed by Western blot and tested with indicated antibodies. (E) Xenograft tumors were weighed. Data are mean ± SEM of 3 independent experiments, and each is measured in triplicate. *P < .05.
Tcl1 and Atm affect the NF-κB pathway in CLL samples and transgenic Tcl1 mice
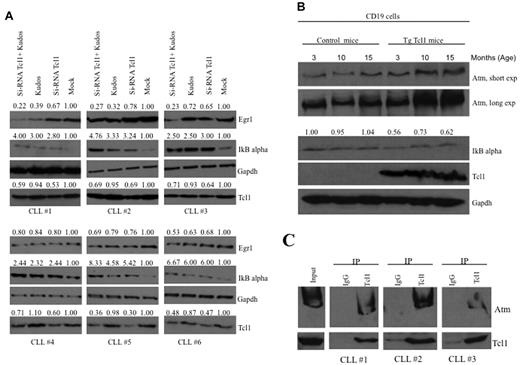
To investigate whether our results are relevant to human CLL, we studied the expression of IκBα and Egr1 in human CLL samples (Figure 4A). CLL samples (supplemental Table 2) were transfected with Tcl1 siRNA for 36 hours and/or treated with Kudos (10μM, 12 hours; Figure 4A). Mock-treated samples showed low expression of IκBα and high expression of Egr1. CLL samples treated either with Tcl1 siRNA or Kudos or both showed increased IκBα expression and decreased expression of Egr1 compared with the mock-treated control. These data confirmed cooperation of Tcl1 and Atm in NF-κB activation in CLL. In addition,we were able to confirm the interaction between endogenous Tcl1 and Atm in some B-CLL patient samples. B-CLLs were lysates and immunoprecipitated with anti-Tcl1 antibody; the immunoblotting with anti-Atm antibody confirmed the interaction (Figure 4C).
Tcl1 and Atm affect the NF-κB pathway in CLL patient samples and Tcl1 transgenic mice. (A) B-CLL samples were transfected with si-TCL1 for 36 hours or treated with Kudos (10μM, 12 hours) or both and in all cases treated with hydroxyurea, 50mM 3 hours. IκBα and Egr1 expression was detected by Western blot. (B) B-cell CD19 isolated from mouse spleen was lysed and analyzed by Western blot with indicated antibodies. Spleens were excised from wild-type and Tcl1 transgenic mice at 3, 10, and 15 months of age. Data are representative of 3 independent experiments. (C) Whole B-CLL cell lysates were immunoprecipitated with anti-Tcl1 antibody. The immunoprecipitates were analyzed by immunoblotting (IB) with anti-Atm and anti-Tcl1 antibodies. Input: total lysate.
Tcl1 and Atm affect the NF-κB pathway in CLL patient samples and Tcl1 transgenic mice. (A) B-CLL samples were transfected with si-TCL1 for 36 hours or treated with Kudos (10μM, 12 hours) or both and in all cases treated with hydroxyurea, 50mM 3 hours. IκBα and Egr1 expression was detected by Western blot. (B) B-cell CD19 isolated from mouse spleen was lysed and analyzed by Western blot with indicated antibodies. Spleens were excised from wild-type and Tcl1 transgenic mice at 3, 10, and 15 months of age. Data are representative of 3 independent experiments. (C) Whole B-CLL cell lysates were immunoprecipitated with anti-Tcl1 antibody. The immunoprecipitates were analyzed by immunoblotting (IB) with anti-Atm and anti-Tcl1 antibodies. Input: total lysate.
We also show evidence of NF-κB pathway activation in transgenic Tcl1 mice. A transgenic mouse model of CLL was generated by introducing the human TCL1 gene under control of a B cell–specific IgEμ enhancer.7,30 Overexpression of Tcl1 in these mice resulted in up-regulation of Atm and down-regulation of IκBα (Figure 4B). TCL1 transgenic mice develop monoclonal B-cell leukemia that is very similar to the aggressive form of human CLL. We isolated CD19+ B cells from spleens of Tcl1 transgenic and wild-type mice. We found that Tcl1 expression is associated with overexpression of Atm and down-regulation of IκBα in Tcl1 transgenic CD19+ B cells compared with wild-type CD19+ B cells used as a control (Figure 4B).
Discussion
Dysregulation of TCL1 in T and B cells causes T-cell prolymphocytic leukemias3,31 and the aggressive form of CLL,7,8 respectively. To clarify the function of Tcl1 in leukemia, we studied Tcl1 protein complexes using a proteomic approach. Atm was identified as a candidate Tcl1 partner, and the interaction was confirmed by coimmunoprecipitation experiments. Atm may act as an upstream kinase that mediates the constitutive NF-κB activation in marrow blasts in both high-risk myelodysplastic syndrome and acute myeloid leukemia.18 The analysis of the phosphorylation status of Atm substrate proteins directed our experiments toward the study of the NF-κB pathway. We previously demonstrated that Tcl1 is responsible for the activation of NF-κB,10 although that study did not show the mechanisms of such an activation. Here we propose the following mechanism based on the previous paragraph: through interacting with Atm, Tcl1 expression causes an increase in Atm kinase activity, an increase in phosphorylation of IκBα, and translocation Rel-A to the nucleus; Tcl1 and Atm have a common target: IκBα, a physiologic NF-κB inhibitor. Indeed, Tcl1 interacts with IκBα,32 whereas Atm phosphorylates it,17,18 which is subsequently degradated by ubiquitination.28
Because Tcl1 interacts with both IκBα and Atm, we detected an additive effect on IκBα ubiquitination when Tcl1 and Atm are coexpressed. As a result of degradation of IκBα, p65 NF-κB active (Rel-A) translocates from the cytosol to the nucleus. Rel-A interacts with the EGR1 promoter sequence and causes overexpression of Egr1 protein,29 as detected by Western blot and quantitative PCR. To confirm that our findings are relevant to human CLL, B-CLL samples were treated with TCL1 siRNA, Kudos, or both to reduce the expression/activity of Tcl1, Atm, or both proteins. Mock-treated samples, with endogenous expression of Atm and Tcl1, showed significant down-regulation of IκBα and overexpression of Egr1, confirming in B cells the data obtained with transfected HEK293 cells. The NF-κB pathway is down-regulated in Daudi Burkitt lymphoma cells when endogenous Tcl1 and Atm are targeted with si-TCL1 and Kudos 55933, both in vitro (proliferation) and in vivo (xenograft experiment). We also provided evidence of NF-κB pathway activation in Tcl1 transgenic mice. Overexpression of Tcl1 in these mice resulted in up-regulation of Atm and down-regulation of IκBα. Thus, we have shown cooperation of Tcl1 and Atm in NF-κB activation. Our results provide additional evidence of the role of the NF-κB pathway in CLL. Because Atm inhibitors are able to kill malignant cells by inhibiting upstream kinases that account for constitutive activation of the NF-κB pathway,17 our data suggest that members of the NF-κB pathway, as well as Tcl1 and Atm interaction, could be considered targets for novel therapies for CLL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr M. B. Kastan for the pcDNA 3.1 ATM-wt HIS6-FLAG tagged and pcDNA 3.1 ATM-KD HIS6-FLAG constructs, Dr Kari Green-Church (Mass Spectrometry and Protomics Facility, The Ohio State University) for the mass spectrometry analysis, Grace A. Hill (Molecular Virology, Immunology and Medical Genetics, The Ohio State University Medical Center) for the assistance with animal experiments, and Dr K. Huebner (Molecular Virology, Immunology, and Medical Genetics, The Ohio State University Medical Center) for the critical editing of the manuscript.
This work was supported by the American Cancer Society (Research Scholar Grant, Y.P.) and National Institutes of Health (grant PO1-CA81534 of the CLL Research Consortium, L.Z.R., T.J.K., and C.M.C.).
This work is dedicated to the memory of Pietro Gaudio, Geometra, who died on March 11, 1996.
National Institutes of Health
Authorship
Contribution: E.G., F.T., and C.M.C. designed the experiments; E.G., R.S., Z.L., F.P., M.K., A.S.L., N.Z., A.B., and S.C. performed research experiments; A.E., A.P., T.N., L.Z.R., and T.J.K. contributed new reagents; E.G., R.S., Y.P., R.I.A., F.T., and C.M.C. wrote the paper; and all authors critically reviewed the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, Department of Molecular Virology, Immunology, and Medical Genetics and Comprehensive Cancer Center, The Ohio State University, 1082 Biomedical Research Tower, 460 West 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal