Abstract
Angiogenic homeostasis is maintained by a balance between vascular endothelial growth factor (VEGF) and Notch signaling in endothelial cells (ECs). We screened for molecules that might mediate the coupling of VEGF signal transduction with down-regulation of Notch signaling, and identified B-cell chronic lymphocytic leukemia/lymphoma6-associated zinc finger protein (BAZF). BAZF was induced by VEGF-A in ECs to bind to the Notch signaling factor C-promoter binding factor 1 (CBF1), and to promote the degradation of CBF1 through polyubiquitination in a CBF1-cullin3 (CUL3) E3 ligase complex. BAZF disruption in vivo decreased endothelial tip cell number and filopodia protrusion, and markedly abrogated vascular plexus formation in the mouse retina, overlapping the retinal phenotype seen in response to Notch activation. Further, impaired angiogenesis and capillary remodeling were observed in skin-wounded BAZF−/− mice. We therefore propose that BAZF supports angiogenic sprouting via BAZF-CUL3-based polyubiquitination-dependent degradation of CBF1 to down-regulate Notch signaling.
Introduction
Vascular endothelial growth factor-A (VEGF-A) strongly stimulates endothelial cells (ECs) of preexisting blood vessels to break of their stable position in the vessel wall and jointly coordinate sprouting, branching, and new lumenized network formation. The tips of the sprouts are formed by specialized ECs called tip cells. Tip cells are migratory and extend numerous filopodia along gradients of VEGF-A. Following behind the tip cells, other ECs become stalk cells that proliferate and form the trunk of the new blood vessel.1 Growing evidence during the past few years has established that the Notch signaling pathway plays a key role in coordinating multiple aspects of endothelial cell behavior in angiogenesis.1 Recent genetic and pharmacologic studies in several systems demonstrate that the specification of ECs into tip and stalk cells is determined by a VEGF-Dll4/Notch cooperative negative or positive-feedback loop.2-15 Interestingly, ECs in preexisting vessels express both Dll4 and Notch, and their signaling are thought to be balanced between adjacent ECs. VEGF stimulation somehow breaks this balance. Consequently Dll4 expression is up-regulated in presumptive tip cells, and induced Notch signaling in adjacent cells to become stalk cells.16 However, the precise molecular mechanism to make such signal waves in adjacent cells is still unclear.
Recently, our studies of growth factors give a suggestive insight that a growth factor might signal not only to activate a growth promotion cascade, but also to unlock a growth suppression cascade via modulation of Zn-finger (ZnF)–type transcriptional repressors to progress cell growth coordinately.17 A concept we felt might be applicable to the VEGF-Notch signaling in ECs at the tip cell position of angiogenic sprouts. Based on the idea, we also hypothesized that one or more of the numerous ZnF transcriptional repressors, most of whose functions still remain unknown, might be key players in this signaling.
To understand the “tug-of-war” balance between VEGFR and Notch signaling, it seems to be essential to perform more intimate analysis for VEGF-A–targeting genes. Therefore, we screened for molecules that are up-regulated in response to VEGF-A stimulation, and also impact on Notch signaling based on a C2H2-type ZnF cDNA micro array. Then, we intensively characterized the biochemical and biologic properties of candidate factors involved in the VEGFR-Notch cross signaling. These efforts resulted in the identification of B-cell chronic lymphocytic leukemia/lymphoima6 (BCL6)–associated zinc finger protein (BAZF) as a crucial factor in angiogenesis, linking the VEGFR and Notch signal transduction pathways by targeting C-promoter binding factor 1 (CBF1).
Methods
Constructs
cDNAs encoding BAZF, CBF1, and CUL3 were PCR-amplified and subcloned into the expression vector pME18S. Human HEY1 promoter (3.9 kb)/pGL3 and Myc-N1ICD/pEFBOS were kindly provided by Drs Jiri Zavadil (NYU Langone Medical Center), Urban Lendahl (Karolinska Institute), and Tetsuya Taga (Tokyo Medical and Dental University), respectively.18,19 Adenoviral vectors were constructed by Adenovirus Expression Vector Kit (Takara).
Cell culture and network formation
Human umbilical vein endothelial cells (HUVECs) were obtained from Cell Systems. HUVECs (6.5 × 103 cells/cm2) were seeded onto solidified Matrigel (BD Biosciences). Before a treatment of 50 ng/mL VEGF-A (recombinant human VEGF165 protein; R&D systems), HUVEC were serum-starved for 12 hours. The images were analyzed by ImagePro Plus 4.5.1 (Media Cybernetics). In some experiments, DAPT (Calbiochem), cycloheximide (Wako), or MG132 (Sigma-Aldrich) were used as described in the figure legends. Time-lapse imaging analysis was performed using Eclipse TE300 microscope (Nikon) controlled by Simple PCI 6.60 software (Compix). HUVEC were infected with AdBAZF or control AdGFP at MOI of 50-250 and incubated for 18 to 24 hours before assays.
Microarray analysis
To identify VEGF-A response transcriptional regulators in HUVEC, 5 μg of total RNA was hybridized with a C2H2-type ZnF custom array (DNA Chip Research) and processed by CRBIOIIe/DNASIS Array ImageScanner (HitachiSoft) according to the manufacturer's protocol. All microarray data are available on the Gene Expression Omnibus (GEO) under accession number GSE35171.
Quantitative RT-PCR analysis
Total RNA (1 μg) was converted to cDNA using SuperScript III (Invitrogen) with oligo (dT)20 primer. For real-time polymerase chain reaction (PCR) amplification, the primer sequences are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Messenger RNA was quantified using SYBR green PCR master mix (Roche) or TaqMan gene expression assay by ABI Prism 7300 Fast Real-Time PCR system. The evaluation of relative differences of PCR product amounts among the treatment groups was carried out by the comparative cycle threshold method. The experiments were independently repeated at least 3 times, each performed in triplicate.
Gene silencing with small interfering RNA
HUVEC were transfected with small interfering RNA (siRNA) against human BAZF (15nM), CBF1 (100nM), Notch1 (100nM), or CUL3 (100nM) by CodeBreaker (Promega) or RNAiMAX (Invitrogen) siRNA transfection regent. The siRNAs were purchased from Qiagen [human BAZF siRNA and control siRNA (1027098)] or Thermo Fisher Scientific [human CBF1 (L-007772-00), Notch1 (L-007771-00), and CUL3 (L-010224-00)]. Thirty-six hours after the transfection, HUVEC were serum-starved for 12 hours, and then treated with 50 ng/mL VEGF-A.
Immunoprecipitation and Western blotting
Immunoprecipitation was performed with anti-CBF1 (Abcam) or anti-FLAG (clone: M2, Sigma-Aldrich) antibody. The samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose membranes (Whatman) or Immun-Blot PVDF Membrane (Bio-Rad), and probed with an appropriate antibody. Antibodies are anti–human BAZF, biotinylated anti–human BAZF (Immuno-Biologic Laboratories), anti-CBF1, antiubiquitin (clone: P4D1, Cell Signaling Technology), anti-Notch1 (Abcam), anti–Cleaved Notch1 (Val 1744; Cell Signaling Technology), anti-FLAG, anti-V5 (R960-25, Invitrogen), anti-Myc (clone: 9E10, Calbiochem), anti–β-actin (clone: AC-15, Sigma-Ardrich), anti–β-tubulin (clone: JDR.3B8, Sigma-Aldrich), antilamin A/C (clone: 636, Santa Cruz), or anti-histone H3 (Upstate) antibody. The proteins were detected by ECL Plus Western Blotting Detection System (GE Healthcare). See supplemental Methods for further information.
HUVEC fibrin gel bead assay with adenovirus infection
HUVEC fibrin gel bead assay was carried out according to the method described by Nakatsu et al.20 Briefly, 500 HUVEC were coated onto Cytodex-3 microcarrier (GE healthcare). HUVEC-coated beads were embedded in a fibrin clot in one well of a 12-well tissue culture plate. Human fibroblasts were plated on top of the clot. HUVEC were cultured for 3 days, and images were taken with an Olympus IX70 microscope (Olympus).
GST pull-down assay
GST, GST-CBF1, and GST-ZnF1-5 were prepared according to the protocol in the previous report.21 The extracts from HT1080 cells expressing various V5-CBF1 or BAZF mutants were incubated with 5 μg of recombinant GST, GST-CBF1, or GST-ZnF1-5 immobilized on glutathione-Sepharose beads, and the bound proteins were analyzed by SDS-PAGE, followed by Western blotting using an anti-V5 antibody.
Luciferase assay
Human HEY1 promoter (full-length, 3.9 kb)/pGL3 and Myc-N1ICD/pEFBOS were transfected with pRL-TK plasmid (Promega) into HUVECs. Twenty-four hours after transfection, the luciferase activities were measured using a dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Chromatin immunoprecipitation assay
A chromatin immunoprecipitation (ChIP) assay was performed according to the manufacturer's recommendation (Upstate) with the exception that the protein-DNA complex was precipitated by anti-CBF1 antibodies (Millipore) bound with Dynabeads-ProteinG (Invitrogen). The used primer sets are as follows: human HEY1 promoter, 5′-aattcagcggcgcgaga-3′ (forward) and 5′-ctcacgctttgcctctggtta-3′ (reverse); human HEY2 promoter, 5′-cttgcggccgagcagagttg-3′ (forward) and 5′-gcttcatcccggcgaagacc-3′ (reverse). The precipitated genome DNA fragments were detected by ABI Prism 7300 Fast Real-Time PCR system with Power SYBR Green PCR Master Mix (Applied Biosystems).
Immunostaining
Immunostaining of HUVEC.
HUVEC were fixed by 4% paraformaldehyde (PFA) and further treated with ice-cold methanol. Triton X-100–permiabilized cells were stained with anti-CBF1 (Abcam), anti-CUL3 (clone: 2236C1a, Bio-Matrix Research), anti-PML (clone: PG-M3, Santa Cruz Biotechnology), anti-SC35 (clone: SC-35, Sigma-Aldrich), and anti-Nucleolin (clone: 3G4B2, Upstate) antibodies. The nuclei were stained by Hoechst33342. Images were acquired by ImageXpress Micro (Molecular Devises), a confocal laser microscope A1 (Nikon) or a structured illumination microscope N-SIM (Nikon). Data were analyzed using MetaXpress (Molecular Devises) or NIS-Elements (Nikon).
Immunostaining of aortic frozen section.
The cultured aortic pieces in type I collagen gel were sectioned by a cryostat and stained with an anti–mouse CD31 antibody (clone: MEC13.3, BioLegend) and Hoechst33342. Images were acquired by a confocal laser microscope Nikon A1.
Immunostaining of retina.
Retinas for whole-mount immunostaining were fixed by 4% PFA overnight. Autoclaving process at 105°C for 15 minutes was performed as an antigen retrieval treatment of BAZF. After fixation, retinas were incubated in methanol at −30°C for 15 minutes. Retinas were stained with isolectin B4–Alexa Flour 647 in the presence or absence of anti-BAZF, anti-CBF1, or anti-HEY1 (Chemicon) antibodies. Images were acquired by a confocal laser microscope A1. Data were analyzed using ImagePro Plus 4.5.1 (Roper).
Mass spectrometry
Mass Spectrometry analysis was performed according to the procedure described in supplemental Methods.
In vivo analysis for retinal angiogenesis
Mice were treated according to the guidelines of Ehime University. This procedure was modified from a published procedure.4 Generation of BAZF knockout mice was reported previously.22 For the assessment of retinal angiogenesis in BAZF+/+ and BAZF−/− littermate mice, the retinas were analyzed at P5. Eyes were fixed in 4% PFA overnight at 4°C. For single staining, retinas were incubated in PBlec buffer (1% Triton X-100, 1mM CaCl2, 1mM MgCl2, and 1mM MnCl2 in PBS (pH 6.8) containing 10% FCS for 1 hour. The retinas were incubated overnight in PBlec buffer containing isolectin B4–Alexa Fluor 647 (Invitrogen). Image was obtained by a confocal laser microscopy A1. Quantification of diameter of vascular area, isolectin B4+ area, branch points, and numbers of filopodia was performed with ImagePro Plus.
Angiogenesis analysis in wound healing
Wound healing experiments were performed in BAZF+/+ (n = 13) and BAZF–/– (n = 13) littermate mice according to a published procedure.23 The wounded skin tissues were fixed with 10% formalin in phosphate buffer (pH 7.2) and embedded in paraffin. The sections were stained with hematoxylin and eosin. For anti-CD31 staining, the sections were autoclaved in 20mM citrate buffer (pH 6.0) for 20 minutes at 121°C, and the sections were stained with an anti-CD31 antibody (clone: SZ31, Dianova). Sections were further reacted with an anti–rat IgG-HRP antibody, and visualized by a DAB substrate kit (Vector). CD31+ area, avascular area and leading edge ratio were measured by ImagePro Plus 4.5.1.
Statistical analysis
Data are based on a minimum of 3 independent experiments, or 3 mutant or control animals for each stage. The results are represented as mean ± SE. The 2 groups were compared using the student t test. ANOVA with Bonferroni posthoc test was used for multiple comparisons. P < .05 was considered statistically significant.
Further information is also described in supplemental Methods.
Results
VEGF-A up-regulates BAZF in HUVEC
Down-regulation of Notch signaling by knockdown of the related molecules or a treatment of γ-secretase inhibitor increased sensitivity to VEGF-A stimulation in endothelial network formation (supplemental Figure 1A-C). As shown in supplemental Figure 1D, Hairy/enhancer-of-split related with YRPW motif protein 1 (HEY1) and HEY2, target genes of Notch signaling, were also significantly down-regulated as an early response of VEGF-A stimulation. Therefore, we hypothesized that angiogenesis sprouting might require transient disconnection of Notch signaling. To characterize genes up-regulated by VEGF-A stimulation, we compared the gene expression profiles of control and VEGF-A–treated HUVECs using a DNA microarray representing 458 of the C2H2 ZnF transcriptional repressor genes, resulting in the identification of BAZF as being up-regulated in response to VEGF-A exposure (supplemental Figure 1E). Quantitative RT-PCR and Western blotting confirmed that BAZF was transiently induced at the level of both mRNA (maximum induction approximately 2- to 2.5-fold) and protein (maximum induction approximately 3-fold) in HUVECs (Figure 1A-B). BAZF mRNA was up-regulated after 2 hours of VEGF-A stimulation and rapidly returned to the basal level (Figure 1A), and BAZF protein increased up to 4 hours before gradually decreasing to the basal level at 12 hours (Figure 1B). Interestingly, BCL6, known as a hetero-binding partner of BAZF,24 was undetectable at the mRNA level and had no response to VEGF-A stimulation in HUVECs (supplemental Figure 1E). These results demonstrate that BAZF is a target of VEGFR signal transduction in HUVECs.
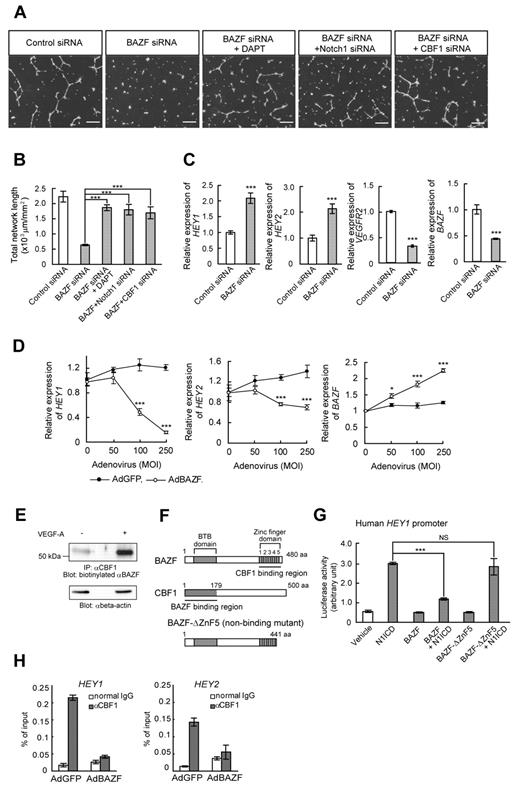
BAZF regulates EC network formation in culture. (A) Expression pattern of BAZF mRNA after VEGF-A (50 ng/mL) stimulation in HUVECs. Quantitative RT-PCR of BAZF mRNA was performed using a TaqMan-gene expression system. The results represent mean values from 3 independent experiments. Error bars represent ± SD. (B) Analysis of BAZF protein level in HUVECs after VEGF-A stimulation by Western blotting using an anti-BAZF antibody. Relative intensity of each band was measured by Lane Analyzer 3.0 software (ATTO) and normalized relative to β-actin (loading control). (C) Angiogenic sprouting from AdBAZF-infected or AdGFP-infected HUVECs. Arrows show sprouting cells. Left panels represent microscopic images, and right panel shows total number of sprouting cells in 50 beads. Scale bar: 100 μm (***P < .001). (D) Protein level of BAZF in HUVEC transfected with BAZF or control siRNA with or without VEGF-A stimulation. BAZF protein was detected by Western blotting using an anti-BAZF antibody. (E) BAZF knockdown disrupts VEGF-A–induced network formation of HUVECs on the Matrigel. The cells were stained with Calcein-AM and the images were acquired. Scale bar: 500 μm. (F) Quantification of the network formation of HUVEC transfected with BAZF or control siRNA. The total length of the network per field (mm2) was measured by ImagePro Plus software 4.5. (Roper). The results represent mean values from 3 experiments. Error bars represent ± SD (***P < .001). (G) Time-lapse imaging of the network formation in HUVEC treated with BAZF or control siRNA. Scale bar: 10 μm.
BAZF regulates EC network formation in culture. (A) Expression pattern of BAZF mRNA after VEGF-A (50 ng/mL) stimulation in HUVECs. Quantitative RT-PCR of BAZF mRNA was performed using a TaqMan-gene expression system. The results represent mean values from 3 independent experiments. Error bars represent ± SD. (B) Analysis of BAZF protein level in HUVECs after VEGF-A stimulation by Western blotting using an anti-BAZF antibody. Relative intensity of each band was measured by Lane Analyzer 3.0 software (ATTO) and normalized relative to β-actin (loading control). (C) Angiogenic sprouting from AdBAZF-infected or AdGFP-infected HUVECs. Arrows show sprouting cells. Left panels represent microscopic images, and right panel shows total number of sprouting cells in 50 beads. Scale bar: 100 μm (***P < .001). (D) Protein level of BAZF in HUVEC transfected with BAZF or control siRNA with or without VEGF-A stimulation. BAZF protein was detected by Western blotting using an anti-BAZF antibody. (E) BAZF knockdown disrupts VEGF-A–induced network formation of HUVECs on the Matrigel. The cells were stained with Calcein-AM and the images were acquired. Scale bar: 500 μm. (F) Quantification of the network formation of HUVEC transfected with BAZF or control siRNA. The total length of the network per field (mm2) was measured by ImagePro Plus software 4.5. (Roper). The results represent mean values from 3 experiments. Error bars represent ± SD (***P < .001). (G) Time-lapse imaging of the network formation in HUVEC treated with BAZF or control siRNA. Scale bar: 10 μm.
BAZF regulates network formation of HUVECs in vitro
To determine the effect of BAZF overexpression, HUVECs were infected with a BAZF-encoding adenovirus (AdBAZF). AdBAZF-infected HUVECs exhibited filopodia protrusion without VEGF-A stimulation on the Matrigel in a dose-dependent manner up to a multiplicity of infection (MOI) of 250 (supplemental Figure 1F-G). Further, we performed a HUVEC fibrin gel bead assay to examine the effect of BAZF on EC sprouting. As shown in Figure 1C, much abundant filopodia protrusion was induced from AdBAZF-infected HUVECs than GFP-encoding adenovirus (AdGFP)–infected HUVECs (27 vs 9 cells with filopodia formation on 50 beads).
To confirm endogenous BAZF function in HUVEC, we knocked down the level of the BAZF mRNA by siRNA transfection of cells. BAZF siRNA reduced not only the BAZF mRNA by more than 80% (supplemental Figure 2A), but the BAZF protein as well (Figure 1D). VEGF-A–induced EC network formation was completely abrogated by BAZF siRNA (Figure 1E-G). However, BAZF knockdown had no effect on cell adhesion and spreading on the Matrigel (supplemental Figure 2B-C). Time-lapse image analysis also showed that there was no difference in the mobility and the incidence of cell-cell contact for the control and BAZF siRNA-transfected HUVECs (data not shown). In addition, we used BAZF siRNA transfection to examine the role of BAZF on network formation by different type of ECs, including human coronary artery, dermal microvascular, and retinal ECs in culture. BAZF knockdown completely abrogated the network formation in these EC cultures despite the presence of VEGF-A (supplemental Figure 2D-E). These results suggest that under certain conditions, BAZF might play a crucial role for EC filopodia protrusion, elongation, and network formation.
Inhibition of Notch signaling rescues the disruption of EC network formation by BAZF knockdown
If BAZF indeed plays a role in VEGFR cross-talk with Notch signaling, we hypothesized that blockade of Notch signaling would rescue the disruption of EC network formation by BAZF siRNA. Treatment of BAZF siRNA-transfected HUVEC cultures with the γ-secretase inhibitor DAPT resulted in the recovery of VEGF-A–induced network formation (Figure 2A-B). Similar to the effects of DAPT, Notch1, or CBF1 knockdown also rescued the disruption of EC network formation by BAZF siRNA (Figure 2A-B). In addition, the levels of the HEY1 and HEY2 mRNAs increased more than 2-fold in BAZF knocked down HUVECs. On the other hand, the level of VEGFR2 mRNA, one of down-regulating target genes of Notch signaling,25,26 decreased in BAZF knocked down HUVEC (Figure 2C). In contrast, BAZF-overexpressing HUVECs by AdBAZF infection significantly reduced the levels of HEY1 and HEY2 transcripts (Figure 2D). These data suggest that BAZF modulates the Notch signaling pathway as a negative regulator to promote angiogenesis by VEGF-A.
BAZF down-regulates Notch signaling by targeting CBF1. (A-B) Disruption of network formation by BAZF knockdown was rescued by DAPT, Notch1 siRNA, or CBF1 siRNA. (A) HUVECs were transfected with indicated combinations with siRNAs and DAPT. After the transfection and serum-starvation, the cells were seeded on the Matrigel with or without DAPT (1μM) in the presence of VEGF-A. Scale bar: 100 μm. (B) Total network length per mm2 was quantified by ImagePro Plus. The results represent mean values from 3 experiments. Error bars represent ± SD. (C) BAZF knockdown increases the expression of Notch target genes, HEY1 and HEY2. The expression levels of the HEY1, HEY2, VEGFR2, and BAZF mRNAs were analyzed by quantitative RT-PCR. (D) BAZF overexpression down-regulates HEY1 and HEY2 expression. The expression levels of HEY1 and HEY2 mRNAs were analyzed by quantitative RT-PCR (*P < .05, ***P < .001). (E) Interaction of BAZF with CBF1. The cell lysates were prepared from VEGF-A–treated (6 hours) or non-treated HUVECs and immunoprecipitated with an anti-CBF1 antibody. The immunoprecipitates were analyzed by Western blotting with a biotinylated anti-BAZF antibody. Beta-actin was detected as a loading control. (F) A molecular schema of BAZF-CBF1 interaction designed by the data in supplemental Figure 3. BAZF-ΔZnF5 is a mutant lacking CBF1-binding capacity. (G) A reporter assay of HEY1 promoter activity. HUVECs were cotransfected with a luciferase reporter plasmid containing human HEY1 promoter, along with the plasmids encoding N1ICD, BAZF, and/or BAZF-ΔZnF5. Luciferase activities are represented as arbitrary units. Error bars show ± SD of 5 samples from 3 independent experiments (***P < .001). NS indicates not significant. (H) A ChIP assay using an anti-CBF1 antibody in HUVECs infected with AdBAZF or AdGFP. The CBF1 binding element on human HEY1 and HEY2 promoters was detected in the immunoprecipitates by quantitative RT-PCR.
BAZF down-regulates Notch signaling by targeting CBF1. (A-B) Disruption of network formation by BAZF knockdown was rescued by DAPT, Notch1 siRNA, or CBF1 siRNA. (A) HUVECs were transfected with indicated combinations with siRNAs and DAPT. After the transfection and serum-starvation, the cells were seeded on the Matrigel with or without DAPT (1μM) in the presence of VEGF-A. Scale bar: 100 μm. (B) Total network length per mm2 was quantified by ImagePro Plus. The results represent mean values from 3 experiments. Error bars represent ± SD. (C) BAZF knockdown increases the expression of Notch target genes, HEY1 and HEY2. The expression levels of the HEY1, HEY2, VEGFR2, and BAZF mRNAs were analyzed by quantitative RT-PCR. (D) BAZF overexpression down-regulates HEY1 and HEY2 expression. The expression levels of HEY1 and HEY2 mRNAs were analyzed by quantitative RT-PCR (*P < .05, ***P < .001). (E) Interaction of BAZF with CBF1. The cell lysates were prepared from VEGF-A–treated (6 hours) or non-treated HUVECs and immunoprecipitated with an anti-CBF1 antibody. The immunoprecipitates were analyzed by Western blotting with a biotinylated anti-BAZF antibody. Beta-actin was detected as a loading control. (F) A molecular schema of BAZF-CBF1 interaction designed by the data in supplemental Figure 3. BAZF-ΔZnF5 is a mutant lacking CBF1-binding capacity. (G) A reporter assay of HEY1 promoter activity. HUVECs were cotransfected with a luciferase reporter plasmid containing human HEY1 promoter, along with the plasmids encoding N1ICD, BAZF, and/or BAZF-ΔZnF5. Luciferase activities are represented as arbitrary units. Error bars show ± SD of 5 samples from 3 independent experiments (***P < .001). NS indicates not significant. (H) A ChIP assay using an anti-CBF1 antibody in HUVECs infected with AdBAZF or AdGFP. The CBF1 binding element on human HEY1 and HEY2 promoters was detected in the immunoprecipitates by quantitative RT-PCR.
The ZnF domain of BAZF interacts with the amino terminal region of CBF1
To determine whether there is a physical interaction between BAZF and CBF1, we performed an assay of immunoprecipitation Western blotting between BAZF and CBF1 in HUVEC, resulting in the successful detection of endogenous BAZF coimmunoprecipitated with CBF1 in HUVEC in a VEGF-A–dependent fashion (Figure 2E). To analyze more precisely the interaction between BAZF and CBF1, we performed glutathione S-transferase (GST) pull-down assays. We constructed various V5-tagged deletion mutants for both BAZF and CBF1 (supplemental Figure 3D-F), which were then expressed in HT1080 cells. A GST-CBF1 or a GST-BAZF fusion protein was used to capture the various V5-tagged mutants from HT1080 cell extracts, and the captured proteins were analyzed by Western blotting using an anti-V5 antibody (supplemental Figure 3A-F). The results showed that the N-terminal region (residues 1-178 aa) corresponding to the LAG1-DNA binding region, but not the center or the C-terminal region of CBF1, bound the ZnF domain of BAZF (Figure 2F). These results suggested that BAZF might interfere with Notch signaling by competing with the intracellular domain of Notch (NICD) for binding to CBF1. To test this hypothesis, we analyzed the effect of overexpressing V5-tagged BAZF (V5-BAZF) on the ability of Myc-tagged Notch1 ICD (Myc-N1ICD) to be coimmunoprecipitated with FLAG-tagged CBF1 (FLAG-CBF1). As shown in supplemental Figure 3G, the overexpression of V5-BAZF did not interfere with the FLAG-CBF1-Myc-N1ICD interaction, consistent with the fact that BAZF and NICD bind to different regions of CBF1.27 BAZF knockdown had no major effects on N1ICD production (supplemental Figure 3H).
We next examined the functional consequences of the interaction between BAZF and CBF1 using a luciferase reporter assay (Figure 2G). For this assay, HUVEC were transfected with a reporter plasmid in which expression of luciferase was controlled by the human HEY1 promoter. Luciferase activity increased under enforced coexpression of N1ICD. The luciferase activity induction by N1ICD was blocked if BAZF was also overexpressed; however, overexpression of the nonCBF1-binding mutant, BAZF-ΔZnF5 (Figure 2F) had no effect on N1ICD-induced reporter activation, suggesting that BAZF suppressed Notch signaling by regulation of CBF1 transcriptional activity (Figure 2G). To investigate the mechanism of the transcriptional repression by BAZF, we performed a ChIP assay in HUVECs using an anti-CBF1 antibody to determine the relative extent to which the HEY1 and HEY2 promoter regions were bound to CBF1. CBF1 binding on the HEY1 or HEY2 promoters was clearly detected in control HUVECs that had been infected with AdGFP. However, BAZF overexpression dramatically abrogated the CBF1 binding on these promoter regions (Figure 2H). These data indicate that BAZF suppresses the transcriptional activation in response to Notch signaling by releasing CBF1 from the targeted gene promoter regions.
Up-regulation of BAZF leads to degradation of CBF1 via recruitment of cullin3
We noticed that the immunostaining intensity of endogenous CBF1 got weaker in a time-dependent manner in HUVECs after VEGF stimulation or BAZF, but not BAZF-ΔZnF5 overexpression (Figure 3A-B), whereas the level of CBF1 mRNA was constant on a BAZF-overexpressing condition (supplemental Figure 3I).
BAZF regulates polyubiquitination and degradation of CBF1. (A-B) Immunocytostaining of endogenous CBF1 in HUVECs with and without VEGF-A. Scale bar: 5 μm. (A) HUVECs were treated with VEGF-A for indicated hours and stained with anti-CBF1. (B) HUVECs were transfected with Mock, BAZF wild-type (WT), or BAZF-ΔZnF5. After transfection, the cells were stained with anti-CBF1. The images were acquired by ImageXpress Micro (Molecular Devices). The signal intensity of CBF1 staining in a hundred cells was measured by ImagePro Plus. Error bars show ± SD of independent triplicates (*P < .05, **P < .01). (C-D) Turnover of CBF1 in HUVECs. (C) After HUVEC were infected with AdBAZF or AdGFP, CHX was added to cell cultures at a concentration of 10μM. Cells were lysed at indicated hours post-CHX addition. Endogenous CBF1 in the chromatin-enriched nuclear matrix fraction was detected by Western blotting with an anti-CBF1 antibody. (D) HUVECs were treated with VEGF-A for indicated hours in the presence or absence of MG132. (E) Western blotting of CBF1 and BAZF in cytosol, nucleoplasm (soluble factions), and chromatin-enriched nuclear matrix (insoluble fraction) of AdBAZF or AdGFP infected HUVECs in the presence or absence of MG132. After infection and treatment of MG132, the cells were then harvested and fractionated to cytosol, nucleoplasm, and chromatin-enriched nuclear matrix fractions according to the protocol described in supplemental Methods. CBF1 and BAZF were detected by Western blotting with antibodies against each protein. Immunoprecipitation was performed using an anti-CBF1 antibody. Ubiquitinated CBF1 was detected by probing Western blots of the precipitates with an anti-ubiquitin antibody. Beta-tubulin, lamin A/C, and histone H3 were detected as fraction markers.
BAZF regulates polyubiquitination and degradation of CBF1. (A-B) Immunocytostaining of endogenous CBF1 in HUVECs with and without VEGF-A. Scale bar: 5 μm. (A) HUVECs were treated with VEGF-A for indicated hours and stained with anti-CBF1. (B) HUVECs were transfected with Mock, BAZF wild-type (WT), or BAZF-ΔZnF5. After transfection, the cells were stained with anti-CBF1. The images were acquired by ImageXpress Micro (Molecular Devices). The signal intensity of CBF1 staining in a hundred cells was measured by ImagePro Plus. Error bars show ± SD of independent triplicates (*P < .05, **P < .01). (C-D) Turnover of CBF1 in HUVECs. (C) After HUVEC were infected with AdBAZF or AdGFP, CHX was added to cell cultures at a concentration of 10μM. Cells were lysed at indicated hours post-CHX addition. Endogenous CBF1 in the chromatin-enriched nuclear matrix fraction was detected by Western blotting with an anti-CBF1 antibody. (D) HUVECs were treated with VEGF-A for indicated hours in the presence or absence of MG132. (E) Western blotting of CBF1 and BAZF in cytosol, nucleoplasm (soluble factions), and chromatin-enriched nuclear matrix (insoluble fraction) of AdBAZF or AdGFP infected HUVECs in the presence or absence of MG132. After infection and treatment of MG132, the cells were then harvested and fractionated to cytosol, nucleoplasm, and chromatin-enriched nuclear matrix fractions according to the protocol described in supplemental Methods. CBF1 and BAZF were detected by Western blotting with antibodies against each protein. Immunoprecipitation was performed using an anti-CBF1 antibody. Ubiquitinated CBF1 was detected by probing Western blots of the precipitates with an anti-ubiquitin antibody. Beta-tubulin, lamin A/C, and histone H3 were detected as fraction markers.
Next, we examined CBF1 protein stability in the cell lysates collected from cultures in which new translation was blocked by the presence of cycloheximide (CHX). Western blotting examinations showed that CBF1 protein rapidly disappeared with a half-life of approximately 12 hours in cells infected with AdBAZF, although it was constant (up to 24 hours) after AdGFP infection (Figure 3C). Therefore, we further analyzed whether the disappearance of CBF1 depends on protein degradation or not by Western blotting of cell lysates with a treatment of a proteasome inhibitor MG132. The level of CBF1 protein apparently increased (Figure 3D). Next, we fractionated cell lysates into cytosol, nucleoplasm, and chromatin-enriched nuclear matrix (nuclear matrix) fractions, and performed Western blotting to detect CBF1 (Figure 3E). In the absence of MG132, whereas CBF1 was dominantly detected in the nuclear matrix (Figure 3E top panel, lane 5), the level of CBF1 dramatically decreased in the nuclear matrix after AdBAZF infection (Figure 3E top panel, compare lanes 5-6). However, addition of MG132 restored CBF1 protein stability in the nucleoplasm in AdBAZF-infected cells (Figure 3E third panel, lane 4). Given the results, we next analyzed polyubiquitination of endogenous CBF1 by immunoprecipitation Western blotting (Figure 3E 5th panel). Polyubiquitinated CBF1 was only detected in the nucleoplasm, when AdBAZF, but not the AdGFP, was infected in HUVECs (Figure 3E 5th panel, compare lanes 3-4). On the other hand, BAZF was primarily detected in the nuclear matrix (Figure 3E 2nd and 4th panels, lanes 5-6), but was essentially undetectable in the cytosol or the nucleoplasm (Figure 3E 2nd and 4th panels, lanes 1-4) in the presence or absence of MG132 regardless of whether the cells were infected with AdBAZF. These data strongly suggest that BAZF, when overexpressed, triggered CBF1 polyubiquitination as well as subcellular translocalization from the nuclear matrix fraction to the nucleoplasmic fraction and proteasome-dependent degradation. It is important to note that the level of overexpression achieved with AdBAZF infection (2- to 3-fold over the basal level) was similar to the level of induction accomplished by VEGF-A stimulation.
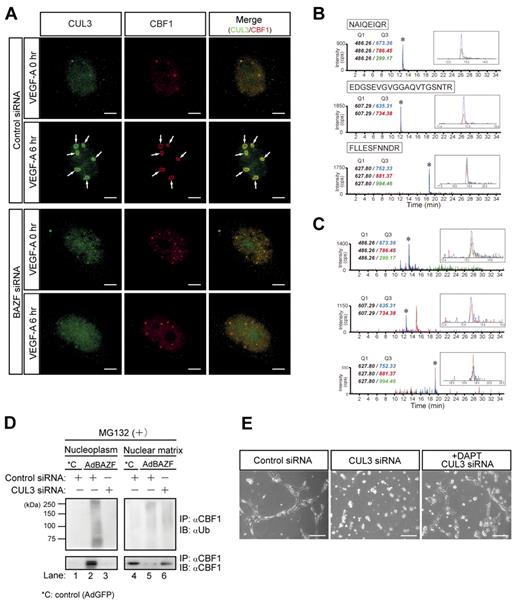
BTB domains can interact with cullin3 (CUL3), a main component of the CUL3-based E3 ubiquitin ligase complex.28-32 To examine the possible interaction between CUL3 and BTB domain of BAZF, we performed immunoprecipitation and Western blotting using BAZF deletion mutants, and revealed that CUL3 interacted primarily with the BTB-center domain and faintly with the ZnF domain of BAZF (supplemental Figure 4A-B). Then, we performed a simultaneous immunostaining analysis of endogenous CUL3 and CBF1 in BAZF siRNA or control siRNA-treated HUVEC under conditions of VEGF-A stimulation. Under control siRNA-treatment, CUL3 and CBF1 distributed throughout the nucleus in the absence of VEGF-A stimulation. However, VEGF-A addition dramatically triggered their colocalization with ring shape at the PML nuclear body after 6 hours of incubation (supplemental Figure 4A-C). In contrast, under BAZF siRNA-treatment, the both localization was neither altered nor merged after 6 hours of incubation with VEGF-A (Figure 4A). We also analyzed CUL3-CBF1 complex formation by a combination assay of immunoprecipitation and multiple reaction monitoring (MRM) analysis by mass spectrometry (MS). Based on tandem MS information obtained from tryptic digests of recombinant CUL3 protein, we selected Q1 (precursor)/Q3(fragment) ion pairs for 3 peptides (NAIQEIQR, EDGSEVGVGGAQVTGSNTR and FLLESFNNDR; supplemental Figure 5A-C), and measured recombinant CUL3 (Figure 4B) and the immunoprecipitates by an anti-CBF1 antibody (Figure 4C) in MRM mode. Three Q1/Q3 ion pairs were clearly detected in the immunoprecipitates as well as recombinant CUL3, which indicates that an anti-CBF1 antibody coprecipitated CUL3. Taken together, these data indicate that BAZF can mediate a trimer complex formation with CUL3 and CBF1.
CUL3 mediates BAZF-induced degradation of CBF1. (A) Super-resolution imaging of colocalization of endogenous CUL3 and CBF1 on VEGF-A–induced ring structures. HUVEC were transfected with BAZF or control siRNA. After transfection and serum starvation, the cells were stimulated with VEGF-A for 6 hours. The cells were immunostained with anti-CUL3 (green) and anti-CBF1 (red) antibodies. The images were acquired by a structured illumination microscopy N-SIM (Nikon) to analyze colocalization of endogenous CUL3 and CBF1. Arrows: VEGF-A–induced ring structures including CUL3 and CBF1. Scale bar: 5 μm. (B-C) MRM analysis of CUL3 using hybrid triple quadrupole/linear ion trap mass spectrometer. (B) Representative MRM profiles of in-gel tryptic peptides from recombinant CUL3. Gel-separated CUL3 tryptic peptides in supplemental Figure 5 were subjected to MRM analysis. * indicates detected CUL3 ion peaks (enlarged view shown in the inset). MRM transitions for the targeted CUL3 peptides were as follows: NAIQEIQR (m/z 486.26→673.36, 786.45, and 299.17), EDGSEVGVGGAQVTGSNTR (m/z 607.29→635.31, and 734.38), and FLLESFNNDR (m/z 627.80→752.33, 881.37, and 994.46). (C) MRM analysis of endogenous CUL3 coimmunoprecipitated with CBF1. Tryptic digests of the immunoprecipitates were subjected to MRM analysis. * indicates Ion peaks assigned to the CUL3 peptide (an enlarged view is shown in the inset). CBF1 was also detected from immunoprecipitates by MRM analysis (data not shown). (D) Effect of CUL3 knockdown on polyubiquitination and degradation of CBF1 induced by AdBAZF infection. HUVECs were transfected with CUL3 siRNA or control siRNA. After transfection, HUVECs were infected with AdBAZF or AdGFP. And then, MG132 was added to the cell culture (50μM). The cells were harvested and fractionated into nucleoplasm and chromatin-enriched nuclear matrix fractions. Immunoprecipitation was performed using an anti-CBF1 antibody. Polyubiquitinated or nonpolyubiquitinated CBF1 was detected using anti-ubiquitin and anti-CBF1 antibodies. (E) Network formation of CUL3 knocked down HUVECs in the presence or absence of DAPT. HUVECs were transfected with CUL3 siRNA or control siRNA. The cells were seeded on the Matrigel with VEGF-A with or without DAPT. Twelve hours later, the images were acquired by a microscope. Scale bar: 20 μm.
CUL3 mediates BAZF-induced degradation of CBF1. (A) Super-resolution imaging of colocalization of endogenous CUL3 and CBF1 on VEGF-A–induced ring structures. HUVEC were transfected with BAZF or control siRNA. After transfection and serum starvation, the cells were stimulated with VEGF-A for 6 hours. The cells were immunostained with anti-CUL3 (green) and anti-CBF1 (red) antibodies. The images were acquired by a structured illumination microscopy N-SIM (Nikon) to analyze colocalization of endogenous CUL3 and CBF1. Arrows: VEGF-A–induced ring structures including CUL3 and CBF1. Scale bar: 5 μm. (B-C) MRM analysis of CUL3 using hybrid triple quadrupole/linear ion trap mass spectrometer. (B) Representative MRM profiles of in-gel tryptic peptides from recombinant CUL3. Gel-separated CUL3 tryptic peptides in supplemental Figure 5 were subjected to MRM analysis. * indicates detected CUL3 ion peaks (enlarged view shown in the inset). MRM transitions for the targeted CUL3 peptides were as follows: NAIQEIQR (m/z 486.26→673.36, 786.45, and 299.17), EDGSEVGVGGAQVTGSNTR (m/z 607.29→635.31, and 734.38), and FLLESFNNDR (m/z 627.80→752.33, 881.37, and 994.46). (C) MRM analysis of endogenous CUL3 coimmunoprecipitated with CBF1. Tryptic digests of the immunoprecipitates were subjected to MRM analysis. * indicates Ion peaks assigned to the CUL3 peptide (an enlarged view is shown in the inset). CBF1 was also detected from immunoprecipitates by MRM analysis (data not shown). (D) Effect of CUL3 knockdown on polyubiquitination and degradation of CBF1 induced by AdBAZF infection. HUVECs were transfected with CUL3 siRNA or control siRNA. After transfection, HUVECs were infected with AdBAZF or AdGFP. And then, MG132 was added to the cell culture (50μM). The cells were harvested and fractionated into nucleoplasm and chromatin-enriched nuclear matrix fractions. Immunoprecipitation was performed using an anti-CBF1 antibody. Polyubiquitinated or nonpolyubiquitinated CBF1 was detected using anti-ubiquitin and anti-CBF1 antibodies. (E) Network formation of CUL3 knocked down HUVECs in the presence or absence of DAPT. HUVECs were transfected with CUL3 siRNA or control siRNA. The cells were seeded on the Matrigel with VEGF-A with or without DAPT. Twelve hours later, the images were acquired by a microscope. Scale bar: 20 μm.
To explore the functional consequences of the interaction between BAZF and CUL3, we performed CUL3 knockdown using a CUL3-specific siRNA (supplemental Figure 4D). Polyubiquitination of endogenous CBF1 in HUVEC infected with AdBAZF or AdGFP was analyzed in the presence or absence of CUL3 siRNA with a treatment of MG132 (Figure 4D). Polyubiquitinated CBF1 was mainly detected in the nucleoplasm (Figure 4D lane 2) in the AdBAZF-infected HUVECs. However, the polyubiquitination of CBF1 in the nucleoplasm was completely abrogated by CUL3 knockdown (Figure 4D lane 3). CBF1 protein primarily existed in the nuclear matrix (Figure 4D compare lanes 1-4) in the AdGFP-infected HUVEC. AdBAZF-infection altered the subcellular localization of CBF1 from the nuclear matrix to the nucleoplasm (Figure 4D lanes 2 vs 5), which was also completely abrogated by CUL3 knockdown (Figure 4D lanes 3 vs 6).
We next investigated the role of CUL3 in VEGF-A–induced HUVEC network formation on the Matrigel. When the cells were transfected with CUL3 siRNA before plating on the Matrigel, VEGF-A failed to induce network formation. Network formation was partially restored by the addition of DAPT to the cultures (Figure 4E).
Taken together, these results strongly suggest that in angiogenic response to VEGF-A stimulation, BAZF makes a complex with CUL3 and CBF1 at the PML nuclear body, which triggers CUL3-dependent CBF1 polyubiquitination, subcellular translocalization, and degradation.
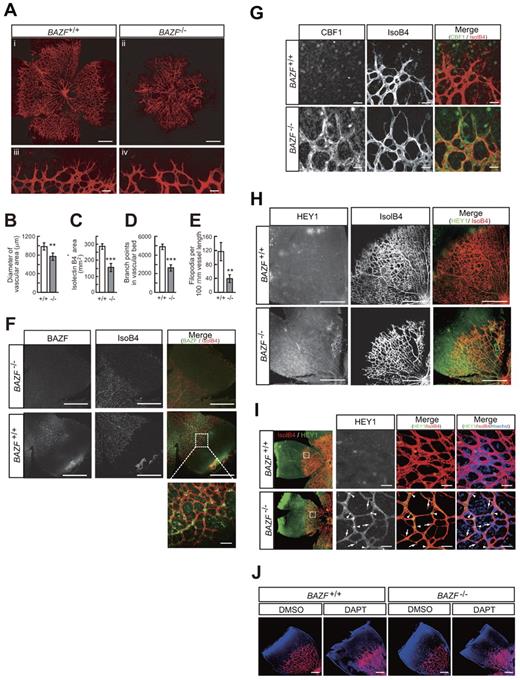
BAZF−/− mice show angiogenic retardation and up-regulation of Notch signaling in the developing retina
It has been reported that BAZF−/− mice are viable, healthy, and normal in size.22,33 Vasculogenesis and angiogenesis abnormalities in BAZF−/− mice have not been reported. However, close examination of the retinas of mice with gene disruption of BAZF showed reduction in vascular density and sprouts in the peripheral part of the developing retinal vascular plexus at P5 (Figure 5A-E, supplemental Figure 6A). A tissue immunostaining assay with an anti-BAZF antibody revealed that BAZF was expressed in the developing retinal vascular plexus in BAZF+/+ mice (Figure 5F). Immunostaining of CBF1 and HEY1 revealed that their protein levels markedly up-regulated in the retinas of BAZF−/− mice (Figure 5G-I). It was similar to a phenotype identical to that reported for the ECs of the retinas in mice with an excessively active Notch signaling.2 On the contrary, inhibition of Notch signal by injection of DAPT into BAZF−/− and BAZF+/+ retinas resulted in the formation of hyperfused plexus similarly (Figure 5J). Taken together, these findings suggest that endogenous BAZF regulates angiogenesis by inactivating Notch signaling during development of the mouse retina.
Up-regulation of Notch signaling and angiogenic suppression in BAZF knockout mouse retina. Littermate mice of representing 2 genotypes (BAZF+/+ and BAZF−/−) were assessed for retinal angiogenesis and Notch signaling at P5. The retinas were stained with isolectin B4, and then flat-mounted for confocal laser microscopic analysis. (A) Whole retinal vascular bed (i and ii, scale bar: 200 μm) and front (iii and iv, scale bar: 20 μm) of BAZF+/+ and BAZF−/− mice. Quantitative analyses of the diameter of the vascular area (B), isolectin B4+ area (C), branching points (D), and the number of filopodia (E) in BAZF+/+ (n = 9) and BAZF−/− (n = 3) retinas. Error bars represent ± SD (**P < .01, ***P < .001). (F) BAZF immunostaining in whole-mount retina. Both BAZF+/+ and BAZF−/− retinas were stained with isolectin B4 and anti-BAZF. Scale Bar: 500 μm (50 μm in the enlarged image). (G) CBF1 immunostaining in whole-mount retina. Retinas were stained with isolectin B4 and anti-CBF1 or normal rabbit IgG as a negative control. Scale bar: 15 μm. (H) HEY1 immunostaining in whole-mount retinal vasculature. Retinas were stained with isolectin B4 and anti-HEY1 antibody. Scale bar: 200 μm. (I) HEY1 localization in retinal vascular plexus. Nuclear localization (arrowheads) of HEY1 in stalk ECs, whereas faint signals in sprouting tip cells (arrows). Endothelia were also stained with isolectin B4. Scale bar: 20 μm. (J) Formation of hyperfused plexus by Notch signal inhibition in retinas. After administration of DMSO or DAPT (100 mg/kg) at P3 and P4, retinas were collected at P5. Retinas were stained with isolectin B4 and Hoechst33342. Images were obtained by a confocal laser microscopy. Scale bar: 200 μm.
Up-regulation of Notch signaling and angiogenic suppression in BAZF knockout mouse retina. Littermate mice of representing 2 genotypes (BAZF+/+ and BAZF−/−) were assessed for retinal angiogenesis and Notch signaling at P5. The retinas were stained with isolectin B4, and then flat-mounted for confocal laser microscopic analysis. (A) Whole retinal vascular bed (i and ii, scale bar: 200 μm) and front (iii and iv, scale bar: 20 μm) of BAZF+/+ and BAZF−/− mice. Quantitative analyses of the diameter of the vascular area (B), isolectin B4+ area (C), branching points (D), and the number of filopodia (E) in BAZF+/+ (n = 9) and BAZF−/− (n = 3) retinas. Error bars represent ± SD (**P < .01, ***P < .001). (F) BAZF immunostaining in whole-mount retina. Both BAZF+/+ and BAZF−/− retinas were stained with isolectin B4 and anti-BAZF. Scale Bar: 500 μm (50 μm in the enlarged image). (G) CBF1 immunostaining in whole-mount retina. Retinas were stained with isolectin B4 and anti-CBF1 or normal rabbit IgG as a negative control. Scale bar: 15 μm. (H) HEY1 immunostaining in whole-mount retinal vasculature. Retinas were stained with isolectin B4 and anti-HEY1 antibody. Scale bar: 200 μm. (I) HEY1 localization in retinal vascular plexus. Nuclear localization (arrowheads) of HEY1 in stalk ECs, whereas faint signals in sprouting tip cells (arrows). Endothelia were also stained with isolectin B4. Scale bar: 20 μm. (J) Formation of hyperfused plexus by Notch signal inhibition in retinas. After administration of DMSO or DAPT (100 mg/kg) at P3 and P4, retinas were collected at P5. Retinas were stained with isolectin B4 and Hoechst33342. Images were obtained by a confocal laser microscopy. Scale bar: 200 μm.
BAZF−/− mice show the impaired angiogenesis in skin wound healing
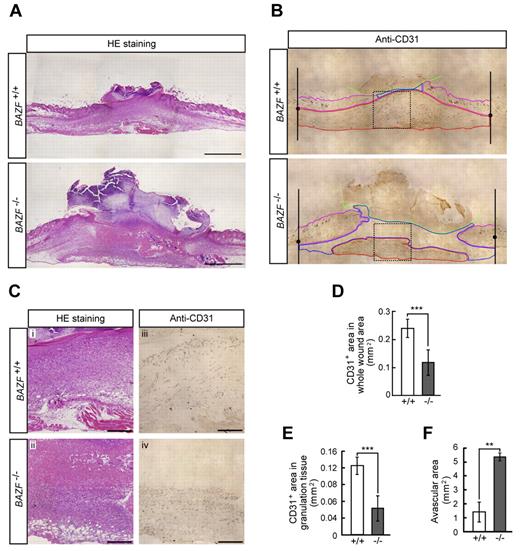
Because it is well known that wounded tissues require angiogenesis for repair, we analyzed skin wound healing in BAZF−/− mice. H&E staining of wounded skin section of BAZF−/− mice at day 7 revealed the impairment of healing (Figure 6A and C). Immunostaining of CD31 in the section showed that CD31 positive vascular area in wound sites, especially in granulation sites of BAZF−/− mice was much smaller than that of BAZF+/+ mice (Figure 6B-E), and that the larger avascular area was necrotic tissue in BAZF−/− mice (Figure 6C and F). In consequence, delayed wound healing in BAZF−/− mice was observed with statistically significant at 3 and 7 days after wounding (supplemental Figure 6B-D). These data suggest that wound-induced angiogenic response was impaired in BAZF−/− mice, resulting in the delay of wound healing.
Impaired angiogenesis in skin wounded BAZF−/− mice. Eight-mm punch biopsies were made in the skin of the backs of 8 to 9-week-old BAZF+/+or BAZF−/− mice. (A) H&E staining of wound section in BAZF+/+ and BAZF−/− mice at day 7. Scale bar: 2 mm. (B) Anti-CD31 staining of serial section in panel A. Green line: border of scab, magenta line: border of skin tissue, blue line: border of avascular area, red line: vascularized area in granulation tissue. (C) Magnified images of dot square regions in Figure 6B. Subpanels i and ii: H&E staining, iii and iv: anti-CD31 staining. Scale bar: 400 μm. Quantitative analysis of CD31+ area in whole wound area (D), CD31+ area in granulation tissue (E), and avascular area (F) in wound section of BAZF+/+ (n = 3) and BAZF−/− (n = 3) mice was carried out using ImagePro Plus 4.5.1.
Impaired angiogenesis in skin wounded BAZF−/− mice. Eight-mm punch biopsies were made in the skin of the backs of 8 to 9-week-old BAZF+/+or BAZF−/− mice. (A) H&E staining of wound section in BAZF+/+ and BAZF−/− mice at day 7. Scale bar: 2 mm. (B) Anti-CD31 staining of serial section in panel A. Green line: border of scab, magenta line: border of skin tissue, blue line: border of avascular area, red line: vascularized area in granulation tissue. (C) Magnified images of dot square regions in Figure 6B. Subpanels i and ii: H&E staining, iii and iv: anti-CD31 staining. Scale bar: 400 μm. Quantitative analysis of CD31+ area in whole wound area (D), CD31+ area in granulation tissue (E), and avascular area (F) in wound section of BAZF+/+ (n = 3) and BAZF−/− (n = 3) mice was carried out using ImagePro Plus 4.5.1.
Discussion
BAZF is a protein most closely related to BCL6, and has BTB and ZnF domains in the N and the C-terminal regions, respectively.24 It has been suggested that BAZF and BCL6 bind an identical DNA sequence similar to a Stat binding motif,24,34,35 and that BAZF functions as a transcriptional repressor by recruiting a corepressor complex such as mSin3A/histone deacetylase 1 through association with BCL6.36 Tissue expression of BAZF mRNA is restricted to heart and lung, whereas BCL6 mRNA is expressed ubiquitously.24,37 However, BCL6 mRNA was barely detectable in HUVECs, whereas BAZF mRNA was easily detected. It has been reported that several BTB-domain–containing proteins bind specifically to CUL3, a key scaffold protein of the recently identified CUL3-based E3 ubiquitin ligase complexes, and serve as linker-adaptor proteins between CUL3 and substrates.28-32 BAZF indeed bound CUL3 at its BTB domain, unlike CBF1 which bound BAZF at its ZnF domain. Super-resolution imaging also revealed the complete colocalization of CBF1 and CUL3 at the PML nuclear body in ECs under VEGF-A stimulation, which was completely disrupted by BAZF knockdown. The PML nuclear body has multiple functions including proteasomal degradation,38 suggesting that CBF1 degradation might be processing at the PML nuclear body. Furthermore, we successfully detected CBF1-CUL3 complex formation by a combination assay of immunoprecipitation and MRM analysis. These findings indicate that BAZF has a unique, BCL6-independent function in ECs. In this study, we have clearly demonstrated that BAZF functions in ECs as a linker-adaptor protein of CUL3-based E3 ubiquitin ligase complex formation targeting CBF1 as summarized in Figure 7.
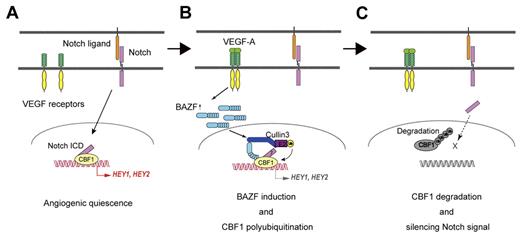
Model for BAZF-containing, CUL3-based E3 ligase complex as a silencer of Notch signaling for angiogenic quiescence-to-sprouting transition. (A) Degradation of CBF1 leads to silencing Notch signal and supports angiogenic sprouting. Notch signaling maintains angiogenic quiescence. Notch1 signaling is transmitted via N1ICD to CBF1. CBF1-N1ICD recruits coactivators for expression of anti-angiogenic genes. (B) BAZF-containing, CUL3-based E3 ligase complex induces CBF1 polyubiquitination. VEGF-A induces BAZF expression, and make a complex with CUL3 and CBF1 for CBF1 polyubiquitination. BAZF binding to CBF1 does not compete with N1ICD binding. However, in this step it is still unclear whether BAZF coexists with N1ICD on CBF1. (C) Degradation of polyubiquitinated CBF1. Polyubiquitinated CBF1 is released from DNA, and is degraded by proteasomes. Degradation of CBF1 leads to silencing Notch signal and supports angiogenic sprouting.
Model for BAZF-containing, CUL3-based E3 ligase complex as a silencer of Notch signaling for angiogenic quiescence-to-sprouting transition. (A) Degradation of CBF1 leads to silencing Notch signal and supports angiogenic sprouting. Notch signaling maintains angiogenic quiescence. Notch1 signaling is transmitted via N1ICD to CBF1. CBF1-N1ICD recruits coactivators for expression of anti-angiogenic genes. (B) BAZF-containing, CUL3-based E3 ligase complex induces CBF1 polyubiquitination. VEGF-A induces BAZF expression, and make a complex with CUL3 and CBF1 for CBF1 polyubiquitination. BAZF binding to CBF1 does not compete with N1ICD binding. However, in this step it is still unclear whether BAZF coexists with N1ICD on CBF1. (C) Degradation of polyubiquitinated CBF1. Polyubiquitinated CBF1 is released from DNA, and is degraded by proteasomes. Degradation of CBF1 leads to silencing Notch signal and supports angiogenic sprouting.
In terms of phenotypic analyses in vivo, BAZF−/− mice exhibited less vessel sprouting and filopodia protrusion of tip cells as well as fewer vessel branches in the developmental retina. These phenotypic features are highly reflective of those seen in retinas of CBF1−/− mice39 and comparable with those of mouse retinas treated with a soluble Jagged1 peptide to activate Notch,2 implicating BAZF as a specific regulator of Notch signaling in angiogenesis. However, Notch active transgenic mice showed abnormal vasculature and the impaired dorsal aorta in embryo, which seemed to be normal in BAZF−/− mice (supplemental Figure 6E-F), suggesting that BAZF might be less effective for angiogenesis in embryonic development. On the other hand, the skin-wounded BAZF−/− mice showed impaired angiogenesis, resulting in the enhanced tissue damage. Taken together, these findings suggest that BAZF functionally mediates VEGFR and Notch cross-signaling in postnatal angiogenesis involved in the tissue remodeling and repair rather than the developmental process, and that other factors to compensate BAZF might be exist in developmental stage.
Angiogenic sprouting is led by tip cells with filopodia formation and nonproliferative feature. Recent study demonstrated elegantly that dynamic position shuffling of tip and stalk cells is tightly regulated by the VEGFR-Dll4-Notch signaling circuit,3 which consists in part of (1) Dll4 expression in ECs by VEGFR signaling,7,14 (2) Dll4 activates Notch signaling in adjacent ECs,2,6,10-12 (3) Nrarp-mediated negative regulation of Notch signaling,40-43 and (4) Jagged1 expression in stalk cells, resulting in antagonizm of Dll4-mediated Notch activation in tip cells.4 However, it is still obscure how such fine-tuned “tug-of-war” balance of VEGFR-Dll4-Notch signaling waves in tip and stalk cells would be constantly achieved.
In addition to the regulation of gene expression, the posttranslational modification of crucial factors plays a key role in regulation of cell siganlling. NICD, a key player in Notch signaling, has been shown to be acetylated, which prolonged its turnover and amplified the duration of Notch responses. NAD-dependent deacetylase SIRT1 negatively modulates Notch signaling.44 Moreover, CBF1 is also an essential transcription cofactor of NICD, and its disruption results in spontaneous angiogenesis in retina, cornea, and internal organs such as liver and lung,39 indicating that Notch-CBF1 signaling is an important pathway for regulation of angiogenesis, and may play an essential role in the maintenance of vascular homeostasis in adults. However, no regulation of CBF1 protein modification has been reported yet. Here we described a novel molecular mechanism underling CBF1 protein turnover regulated by VEGFR inducing BAZF-CUL3-based polyubiquitination, which might be involved in making the fine-tuned angiogenic “tug-of-war” balance between VEGFR and Notch signaling in ECs. However, it is still unclear when and how BAZF-mediated CBF1 regulation is integrated into VEGFR-Notch signaling circuit in the emergence of tip cells and filopodia formation. To understand this mechanism more precisely, the further study of the regulatory network would be essential in temporal axis.
Currently, in anti-angiogenic therapies for diabetic retinopathy, age-related macular degeneration, retinopathy of prematurity, and solid-tumor growth, Dll4-Notch signaling is becoming a prominent molecular target. Based on our studies presented here, we propose BAZF as an alternative target of Dll4-Notch signaling for future anti-angiogenesis strategies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Dr Matsushita for insightful discussions, Mr Kawahito and Ms Ouchi (Nikon Corporation) for super-resolution imaging microscopic analysis, Mr Yokoyama (AB SCIEX) for MRM analysis, and Drs Shirakata, Iwabuki, and Yang, and Ms Tsuda for technical assistance. They are grateful to Immuno-Biologic Laboratories for providing anti-BAZF antibodies. They are also grateful to Dr Judith A. Abraham (Novartis Institutes for BioMedical Research) for helpful comments and editing the paper.
This work was supported by Grant-in-Aid for Scientific Research (20390082, 1701468, and 23112513 to S.H. and 21780312 to H.O.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan, (7610010302J to S.H.) from Japan Science and Technology Agency, (6003842 to S.H.) from Strategic Young Researcher Overseas Visiting Program for Accelerating Brain Circulation, JSPS, Japan, from Incubation Project for Seeds, Ehime Industrial Promotion Foundation, and (021000 to H.O.) from Support Project for Research and Development in Ehime University.
Authorship
Contribution: H.O. performed most of the experiments assisted by H.I., H.N., S.F., and D.M.; N.T. performed mass spectrometry analysis; E.N. helped analyze gene expression by a cDNA microarray system; M.H. and T.T. provided BAZF−/− mice; Y.E. and M.N. helped design the experiments, and advised on the results; and S.H. and H.I. conceived the project, designed the experiments with H.O. and wrote the paper assisted by H.O.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ehime University Graduate School of Medicine, Shitsukawa, Toon, Ehime, Japan 791-0295; e-mail: shigeki@m.ehime-u.ac.jp.
References
Author notes
H.O. and H.I. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal