Abstract
Deletion of chromosomal region 13q14 represents the most common genetic aberration in B-cell chronic lymphocytic leukemia (CLL). 13q14 deletions are commonly large and heterogeneous in size and affect multiple genes. We recently found that targeted deletion in mice of the 0.11 megabase (mb)–long minimal deleted region (MDR) encompassing the DLEU2/miR-15a/16-1 cluster recapitulates the spectrum of CLL-associated lymphoproliferations in humans, including CLL, CD5+ monoclonal B-cell lymphocytosis, and CD5− non-Hodgkin lymphomas. In the present study, we demonstrate that additional deletion of the 0.69-mb large genomic region telomeric to the MDR called the common deleted region (CDR) changed the spectrum of lymphoproliferations developing in CDR- versus MDR-deleted mice in that the number of CLL among B-cell lymphoproliferations was significantly elevated in the former. In addition, CDR-deleted mice seemed to succumb to their disease faster than MDR-deleted mice. Comparing HCDR3 regions of CD5+ lymphoproliferations derived from this and published CLL mouse models, 44% (29 of 66) of junctions could be assigned to 8 sets of highly similar HCDR3 regions, demonstrating that CLL developing in mice frequently expresses almost identical, stereotypic Ag receptors. These results suggest that the size of 13q14 deletions influences the phenotype of the developing lymphoproliferations and potentially the severity of disease, suggesting a tumor-suppressor function for genetic elements in addition to DLEU2/miR-15a/16-1.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is a neoplasm of mature B lymphocytes presenting with an indolent or aggressive disease course. The phenotype of the tumor cells and features of the Ab-binding regions suggest a derivation from Ag-experienced B cells.1-3 CLL is preceded by CD5+ monoclonal B-cell lymphocytosis (MBL)4,5 and occasionally progresses to diffuse large B-cell lymphoma. CLL is associated with recurrent genomic aberrations including chromosomal gains (trisomy 12) and deletions (17p, 11q, and 13q14)6 and, at a lower frequency, with recurrent somatic mutations in NOTCH1 and other genes.7,8 Deletions of 13q14 are the most common aberration in CLL (55%)6,9 and are found at a similar percentage in MBL5 and less frequently in diffuse large B-cell lymphoma, multiple myeloma, and several types of non-B cell tumors. The extensive characterization of 13q14 deletions has demonstrated that the break points are heterogenous and that the deleted region can comprise multiple genes.10-17 These studies led to the identification of a minimal deleted region (MDR)12,13 that comprises the deleted in leukemia 2 (DLEU2) gene encoding a sterile RNA transcript and the microRNA 15a/16-1 cluster18 that is located in an intron of DLEU2 (Figure 1); this DLEU2/miR-15a/16-1 locus is invariably affected by deletions in all CLL with 13q14 aberrations. Functional evidence has long suggested an involvement of miR-15a/16-1 deletion in CLL development,19-21 and the causative role of miR-15a/16-1 in CLL pathogenesis has been demonstrated recently in vivo in a knockout mouse model in which these microRNAs were specifically deleted in B cells.22 This same study also demonstrated that a larger deletion (MDR) affecting the host gene of these microRNAs, dleu2, as well as the dleu5 and kcnrg genes (that in the mouse overlap with the dleu2/miR-15a/16-1 locus), shows a significantly more aggressive disease course.22 These results therefore suggest that additional genetic elements within the 13q14 locus in addition to miR-15a/16-1 may act as tumor suppressors, and that their concomitant deletion with miR-15a/16-1 may exacerbate the disease course.
Whereas the analysis of the MDR has led to the definition of the minimal targets of the 13q14 deletion, it must be noted that the deletions are usually significantly larger in CLL. In particular, a 0.9-megabase (mb) region telomeric to the DLEU2/miR-15a/16-1 cluster is frequently deleted in combination with the MDR and is therefore referred to as the common deleted region (CDR; Figure 1). The deleted region telomeric to DLEU2/miR-15a/16-1 comprises 3 genes, namely DLEU1, DLEU7, and RNASEH2B (Figure 1), of which only the latter 2 genes are conserved across species. DLEU1 encodes a sterile transcript of unknown function and, because of its lack of evolutionary conservation, has not been considered a candidate tumor-suppressor gene. Functional studies of DLEU7, a protein-encoding gene,23 in non-B cells suggest that it may act as an inhibitor of several signaling pathways that promote cell-cycle entry.24 In the majority of CLL cases analyzed, DLEU7 expression was low and associated with promoter methylation.23 It is presently unclear whether DLEU7 is methylated specifically in CLL and to what extent promoter methylation plays a role in CLL pathogenesis. Mutations in RNASEH2B (AGS2/DLEU8/FLJ11712), which encodes the ribonuclease H2 subunit B that is involved in eliminating RNA-DNA intermediates during replication, are associated with Aicardi-Goutières syndrome,25 a rare recessive neurologic disorder. The expression pattern and function of RNASEH2B in B cells is unknown.
The present study is a continuation of our efforts to elucidate the tumor-suppressor function of the 13q14 locus that shows extensive heterogeneity in the sizes of the deleted regions. We constructed a CDR conditional knockout mouse that, in addition to the h13q14/m14qC3 MDR,22 has the genomic region telomeric to the MDR deleted. We sought to determine whether the loss of genes in the h13q14/m14qC3 locus in addition to those encoded in the MDR change the spectrum of lymphoproliferations and/or the disease course. We also compared the rearranged Ig heavy chain–variable region genes of the CD5+ B-cell lymphoproliferations developing in the CDR-deleted mice with those observed in miR-15a/16-1, MDR-deleted,22 and TCL1-Tg mice26 to determine whether similar to human CLL, independent CLL mouse models are characterized by the expression of highly identical, stereotypic Ag receptors.
Methods
Generation of CDR-conditional and CDR-null mice
The 2 targeting vectors used to flank the CDR with loxP and frt sites were derivatives of pEmod227 containing either a phosphoglycerate kinase (PK)– neomycin-resistance (5′tag) or a PK-hygromycin-resistance (3′tag) poly(A) cassette, the herpes simplex virus thymidine kinase gene (both tags), a loxP and a frt site (both tags), a promoterless gene encoding enhanced green fluorescent protein (eGFP) and immediately preceding a triple simian virus 40 poly(A) site in 5′tag, a PK promoter (3′tag), and multiple unique restriction sites for cloning 14qC3 segments corresponding to the homology arms (supplemental Figure 1-A1 and A2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The vectors were constructed so that in the appropriate cell type, Flp-mediated recombination produced deletion of the whole targeted region (supplemental Figure 1-A3) and Cre-mediated recombination produced deletion of the CDR with simultaneous activation of eGFP expression (supplemental Figure 1-A4). Successively inserted into the cloning sites of the corresponding 5′tag and 3′tag vectors were 2 DNA fragments of the 129/Sv-14qC3 locus comprising the following: (5′tag) a 2.4-kb (primers: forward, 5′-AGCTTTAGGCATGCACTCAC-3′; reverse, 5′-GAGGCTGGTTGCACAAAGAC-3′) and a 2.8-kb (primers: forward, 5′-GGAAGGATGAGCTAGGACAG-3′; reverse, 5′-TCTGGAATCACTTTGTGGTCTC-3′) PCR-generated fragment of a transcriptionally inactive region 20 kb upstream of exon 1 of dleu5; (3′tag) a 2.5 kb (primers: forward, 5′-CAGTCATCCATCGACAGCTG-3′; reverse, 5′-TATCAACACCATGCTCTTAAAC-3′) and a 2.3 kb (primers: forward, 5′-TCCACTGGCGTCCACTGAAAG-3′; reverse, 5′-GAGCTGTGGTTTTAGATTATGC-3′) PCR-generated fragment downstream of the last exon of Gucy1b2. The linearized 5′tag vector was electroporated into W9.5 embryonic stem (ES) cells derived from 129/SvEvTac, and correctly targeted ES cell colonies were identified by Southern blot analysis after selection with ganciclovir and G418 (supplemental Figure 1-A1). The identified ES cells were electroporated with the linearized 3′tag vector, and correctly targeted ES cell colonies were identified by Southern blot analysis after selection with ganciclovir and hygromycin (supplemental Figure 1-A2). ES clones with both targetings on the same chromosome were identified by Adeno-Cre (a gift from J. Kitajewsky, Columbia University, New York, NY) transduction and identification of eGFP-expressing ES cells by flow cytometry (supplemental Figure 2); correct deletion of the loxP-flanked MDR was verified in the corresponding cultures by Southern blot analysis (Figure 1C and supplemental Figure 1-A4). Chimeras were obtained after injection of the targeted ES clones into blastocysts derived from C57BL/6 mice. From the chimeras, we obtained mice with the loxP-flanked CDR allele in germline CDRfl/+, as determined by Southern blot analysis. The chimeras were also crossed with 129/SvCAGGS-Flpe mice (The Jackson Laboratory) to generate a null allele without eGFP expression (supplemental Figure 1-A3). The obtained CDR+/− mice were crossed to C57BL/6 mice, and the resulting F1 generation was intercrossed to obtain CDR−/− mice. Because CDR deficiency led to embryonic lethality, we interbred CDR+/− with littermate wild-type mice to establish the experimental mice of the “constitutional” CDR+/− knockout cohort, which were therefore of a 129/Sv-C57BL/6 mixed background. CDRfl/+ chimeras were bred with C57BL/6 mice, and these mice were intercrossed with CD19-Cre mice for establishing the experimental mice of the conditional knockout cohort and were therefore of the 129/Sv-C57BL/6 mixed background. In vivo deletion of the loxP-flanked CDR allele was verified by Southern blot analysis of purified B cells derived from CDRfl/+CD19-Cre mice (Figure 1C). Mice were housed and treated according to the guidelines of the Institute of Comparative Medicine at Columbia University (New York, NY).
Cell isolation, flow cytometry, and histology
Blood was withdrawn from the hearts of killed mice with a heparin-coated syringe and erythrocytes were subjected to 2 consecutive hypotonic lyses. Peritoneal cavity cells were isolated by flushing with 0.5% BSA in PBS. BM cells were flushed out of the tibia or femur using a syringe filled with PBS/BSA. Spleen cell suspensions were subjected to hypotonic lysis. The Abs and labels used for single-cell suspensions from spleen, peripheral blood (PB), peritoneal cavity, and BM have been described previously.22 Data were acquired on a FACSCalibur (BD Biosciences) using CellQuest software; data were analyzed using CellQuest Version 3.3 or FlowJo Version 9.3.2 software (TreeStar). Four-micrometer-thick, formalin-fixed, paraffin-embedded tissue sections were stained with H&E for morphologic analysis. Micrographs were taken with an Olympus BX41 bright-field microscope with the lens type UPlanFL. For a total magnification of 100×, the objective magnification was 10× and the numeric aperture was 0.30; for a total magnification of 200×, the objective magnification was 20× and the numeric aperture was 0.50. Photomicrographs were taken with a digital camera (Qcolor 3; Olympus) and Qcapture Version 2.90.1 software (Quantitative Imaging Corporation). The TIFF image format was converted to JPEG format using Adobe Photoshop Elements Version 2.0. Both flow cytometric and histological analyses were performed for all mice of the 15- to 18-month CDR cohort.
Event-free survival and statistical analysis
Tumor-watch studies were conducted on animals in a 129/Sv-C57BL/6 mixed background (ie, the first and second backcross generations). Mice were monitored for tumor incidence biweekly and were killed when visibly ill. Within the transgenic line, comparable numbers of age-matched wild-type littermates were controlled for possible differences in tumor incidence. Animals were monitored for up to 20 months. Statistical analysis was performed on the Prism Version 5 software program (GraphPad) using Kaplan-Meier cumulative survival and the log-rank (Mantel-Cox) test to determine whether differences were significant (Figure 5C). The χ2 and/or the Fisher exact probability tests were used to compare B-cell lymphoproliferation incidence between the different genotypes of the CDR cohorts (Figures 4 and 5B) and to determine differences in the spectrum of lymphoproliferations among the CDR, MDR, and miR-15a/16-1 cohorts (Figure 6). The Wilcoxon rank-sum test was used to determine differences in the CD5+ B-cell lymphoproliferations among the corresponding fractions (Figures 2 and 5A).
IgV gene sequence analysis
RNA was purified from PBMCs, splenic tissue, or lymph nodes from mice that were diagnosed with CD5+ or CD5− B-cell lymphoproliferations after cell lysis with TRIzol reagent, and reverse transcribed into cDNA using (poly)dT oligonucleotides and Superscript II (Invitrogen). Rearranged VH sequences were amplified by PCR using forward primers that anneal to the framework region I of the mouse VH families, and reverse primers positioned in the JH1, JH2, JH3, and JH4 segments, as described previously.22 PCR products were gel-purified and sequenced directly (Genewiz). Sequences were compared with the ImMunoGeneTics database for sequence analysis (http://www.imgt.org). The MultiAlin sequence alignment algorithm28 was used for hierarchical clustering of HCDR3 regions. IgV gene sequences are available upon request.
Results
Construction of mice deleting the CDR specifically in B cells and in germline
We have previously generated mice that have the 14qC3-MDR deleted in the germline and also specifically in B cells using CD19-Cre mice22 (Figure 1A). The deleted region encompasses the dleu2/miR-15a/16-1 cluster and the Kcnrg and dleu5 genes that in mice, in contrast to humans, are located intronic of (Kcnrg) or overlap with (dleu5) dleu2. To investigate the consequences of deleting the entire CDR, which in addition comprises the protein-coding genes dleu7 and Rnaseh2b, we generated a conditional mouse allele that on Cre- or Flpe-mediated deletion mimics the human CDR (Figure 1B). To generate a conditional CDR allele, we inserted 2 loxP and 2 frt sites each by consecutive ES cell targetings into transcriptionally silent regions located 0.8 mb apart (Figure 1B; for details of the targeting strategy, see supplemental Figure 1-A1 and A2). The ES cell clone with the 5′ site targeted is the same that was used in the generation of the conditional MDR allele22 ; the 5′ site is located approximately 20 kb centromeric to dleu5. The 3′ site is located between the Rnaseh2b and Gucy1b2 genes, approximately 2 kb centromeric to the last exon of Gucy1b2. The placement of an eGFP mini gene in the centromeric (5′ site), and a PGK promoter in the telomeric (3′ site) targeting vectors that enable eGFP expression on Cre-mediated deletion of the loxP-flanked CDR (Figure 1B-C) allows screening for ES cell clones in which both targetings occurred on the same chromosome. Supplemental Figure 2A through D shows the general strategy (panel A), identification (panels B-C), and verification (panel D) of homologous recombination of the consecutive targetings. Correct homologous recombination and Cre-mediated deletion of the loxP-flanked region was confirmed by Southern blot analysis (Figure 1C and supplemental Figure 1-A1, A2, and A4). Mice harboring the CDRloxP−frt/+ allele were bred with Flp-transgenic mice, and the correct deletion of the frt-flanked region was confirmed by Southern blot analysis (Figure 1C and supplemental Figure 1-A3); these mice are further on designated as CDR+/− mice. All animal experiments were approved by the institutional animal care and use committee review board of Columbia University.
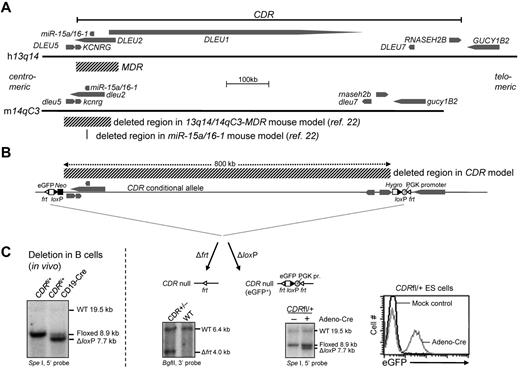
B-cell specific deletion of the 13q14-CDR in mice. (A) Schematic representation of the human 13q14 and mouse 14qC3 locus. Genes and their 5′-3′ orientation are indicated by thick arrows. The deleted regions in the MDR and miR-15a/16-1 mouse models described previously22 are indicated. Because in humans, KCNRG is encoded within an intron of DLEU2, the former gene is considered part of the human MDR. In mice, and in contrast to what is found in humans, dleu2, in addition to Kcnrg, also overlaps with dleu5 (exon 3 of dleu5 is located in the last intron of dleu2), thus making dleu5 part of the mouse MDR. (B) Schematic representation of the CDR targeting strategy (for details, see supplemental Figure 1-A1 to A4). Indicated are the expected fragments detected by Southern blot analysis after Flp-mediated recombination in mice, which generates an CDR-null allele, and after Adeno-Cre–mediated recombination in CDRfl/+ ES cells, which shows a ΔloxP fragment in addition to the targeted fragment, demonstrating the feasibility of deleting the 0.8-mb conditional CDR allele. Flow cytometric analysis of Adeno-Cre–treated CDRfl/+ ES cells shows that deletion of the CDR is accompanied by eGFP expression (for details, see supplemental Figures 1-2). (C) Deletion of the 0.8-mb loxP-flanked CDR allele in vivo. Southern blot analysis of SpeI-digested DNA from purified CD19+ B cells of CDRfl/+CD19-Cre and CDRfl/+ mice. CDRfl/+CD19-Cre mice show the WT allele and the allele after loxP-mediated deletion.
B-cell specific deletion of the 13q14-CDR in mice. (A) Schematic representation of the human 13q14 and mouse 14qC3 locus. Genes and their 5′-3′ orientation are indicated by thick arrows. The deleted regions in the MDR and miR-15a/16-1 mouse models described previously22 are indicated. Because in humans, KCNRG is encoded within an intron of DLEU2, the former gene is considered part of the human MDR. In mice, and in contrast to what is found in humans, dleu2, in addition to Kcnrg, also overlaps with dleu5 (exon 3 of dleu5 is located in the last intron of dleu2), thus making dleu5 part of the mouse MDR. (B) Schematic representation of the CDR targeting strategy (for details, see supplemental Figure 1-A1 to A4). Indicated are the expected fragments detected by Southern blot analysis after Flp-mediated recombination in mice, which generates an CDR-null allele, and after Adeno-Cre–mediated recombination in CDRfl/+ ES cells, which shows a ΔloxP fragment in addition to the targeted fragment, demonstrating the feasibility of deleting the 0.8-mb conditional CDR allele. Flow cytometric analysis of Adeno-Cre–treated CDRfl/+ ES cells shows that deletion of the CDR is accompanied by eGFP expression (for details, see supplemental Figures 1-2). (C) Deletion of the 0.8-mb loxP-flanked CDR allele in vivo. Southern blot analysis of SpeI-digested DNA from purified CD19+ B cells of CDRfl/+CD19-Cre and CDRfl/+ mice. CDRfl/+CD19-Cre mice show the WT allele and the allele after loxP-mediated deletion.
Homozygous germline deletion of the CDR leads to embryonic lethality
Interbreeding CDR+/− mice did not yield any CDR−/− offspring. Of 214 mice born alive, 138 (64.5%) were CDR+/− and 76 (35.5%) wild-type. Because MDR−/− mice are born at Mendelian frequencies, this suggests that the additional 0.69 mb of DNA in the CDR encodes a genetic element or elements required for embryonic development, which is abrogated at an unknown time point by an unknown mechanism. Therefore, study of the role of the CDR in leukemogenesis requires a cell type–specific knockout approach.
Mice with CDR deletion specifically in B cells develop CLL
To determine the extent to which deletion of the CDR affects B-cell development and physiology, we generated mice that have the CDR in B cells specifically deleted by crossing CDRloxP−frt/+ mice with mice in which the Cre recombinase is under transcriptional control of the B cell–specific CD19 gene (CD19-Cre mice); these mice are further on referred to as CDRfl/+CD19-Cre mice. Southern blot analysis of purified CD19+ B cells demonstrated the ability of the conditional CDR allele to recombine in vivo at an approximately 90% deletion efficiency (Figure 1C).
We generated cohorts of CDRfl/−CD19-Cre and CDRfl/flCD19-Cre mice to achieve homozygous deletion of the CDR specifically in B cells, and the corresponding heterozygous and wild-type controls, CDRfl/+CD19-Cre and CD19-Cre mice. Two- to 3-month-old mice with homozygous deletion of the CDR in B cells showed normal percentages of B-cell subpopulations (supplemental Figure 3), and histologic analysis showed normal development of lymphoid organs (data not shown). These results suggest that homozygous deletion of the CDR does not impair B-cell development in young mice. In contrast, at approximately 12 months of age, CDRfl/−CD19-Cre and CDRfl/flCD19-Cre mice began to develop CD5+B220lo clonal lymphoproliferations in the PB (Figure 2), virtually all of which had the characteristics of CLL, BM and peripheral tissue involvement by CD5+B220lo tumor cells (Figure 3A-B). The clonality of these lymphoproliferations was confirmed by PCR for rearranged IgV genes (supplemental Table 1). Clonal CD5+ lymphoproliferations occurring in the PB were IgMhiIgD+ (Figure 3A), and infiltration of lymphoid and other organs was observed frequently (Figure 3B). In the spleen, the white pulp was enlarged as the result of an accumulation of small lymphocytes that showed histopathologic features of human CLL/small cell lymphocytic leukemia (Figure 3B top). Aggregates of small lymphocytes were also noted in the BM (Figure 3B middle), a finding consistent with the appearance of a B220+eGFP+ population in the BM by flow cytometric analysis (Figure 3A bottom) and in the liver (Figure 3B bottom). In summary, the phenotype of the tumor cells and the histopathological features were consistent with CLL/small cell lymphocytic leukemia.
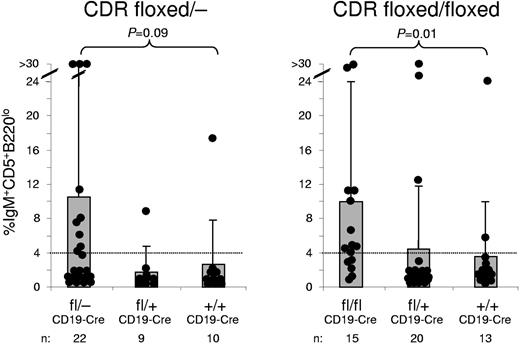
CDR-deleted mice develop CD5+ B-cell expansions in the PB. Percentages of CD5+B220lo cells among mononuclear cells of mice from CDRfl/−CD19-Cre and CDRfl/flCD19-Cre cohorts. P values (Wilcoxon rank-sum test) are indicated; the dotted line demarks the upper threshold for the normal range of CD5+B220lo cells in a panel of 3- to 6-month-old wild-type mice determined by the average percentage ± 3σ; percentages above 4% are considered CD5+ lymphocytosis. Data are shown both as actual values (●) and as mean ± SD.
CDR-deleted mice develop CD5+ B-cell expansions in the PB. Percentages of CD5+B220lo cells among mononuclear cells of mice from CDRfl/−CD19-Cre and CDRfl/flCD19-Cre cohorts. P values (Wilcoxon rank-sum test) are indicated; the dotted line demarks the upper threshold for the normal range of CD5+B220lo cells in a panel of 3- to 6-month-old wild-type mice determined by the average percentage ± 3σ; percentages above 4% are considered CD5+ lymphocytosis. Data are shown both as actual values (●) and as mean ± SD.
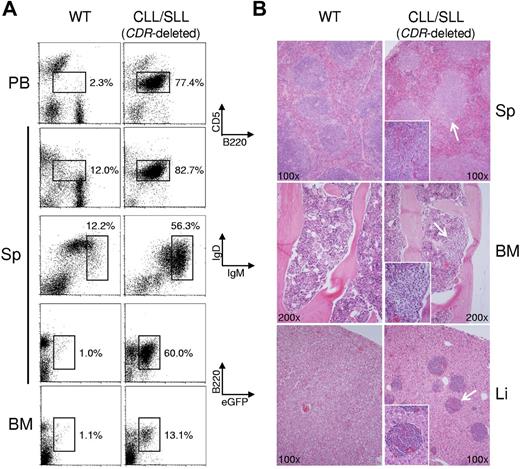
CDR-deleted mice develop CLL. (A) Flow cytometry of PBMC (PB), splenic (Sp), or BM cells from a mouse presenting with CLL/small cell lymphocytic leukemia (SLL) and a wild-type (WT) mouse as control for CD5 and the B-cell marker B220 (top), IgM and IgD (middle), and eGFP and B220 (bottom). The CLL/SLL case shows a predominant CD5+B220lo population in the spleen and PB that was IgMhiIgD+/low and eGFP+, demonstrating that the tumor clone is indeed derived from a CDR-deleted B cell. (B) Representative H&E–stained spleen, BM, and liver sections from CDR-deleted mice presenting with CLL/SLL (right) and an age-matched WT mouse (left). CLL/SLL section shows enlargement of the splenic white pulp by the expansion or accumulation of small B cells with architectural and morphologic features of CLL/SLL (top), aggregates of small lymphocytes in the BM (middle) and liver (bottom). Both flow cytometric and histological analysis were performed for all mice of the 15- to 18-month-old CDR cohort (for numbers of mice analyzed, see Figures 4 and 5; for details regarding micrographs, see “Cell isolation, flow cytometry, and histology”).
CDR-deleted mice develop CLL. (A) Flow cytometry of PBMC (PB), splenic (Sp), or BM cells from a mouse presenting with CLL/small cell lymphocytic leukemia (SLL) and a wild-type (WT) mouse as control for CD5 and the B-cell marker B220 (top), IgM and IgD (middle), and eGFP and B220 (bottom). The CLL/SLL case shows a predominant CD5+B220lo population in the spleen and PB that was IgMhiIgD+/low and eGFP+, demonstrating that the tumor clone is indeed derived from a CDR-deleted B cell. (B) Representative H&E–stained spleen, BM, and liver sections from CDR-deleted mice presenting with CLL/SLL (right) and an age-matched WT mouse (left). CLL/SLL section shows enlargement of the splenic white pulp by the expansion or accumulation of small B cells with architectural and morphologic features of CLL/SLL (top), aggregates of small lymphocytes in the BM (middle) and liver (bottom). Both flow cytometric and histological analysis were performed for all mice of the 15- to 18-month-old CDR cohort (for numbers of mice analyzed, see Figures 4 and 5; for details regarding micrographs, see “Cell isolation, flow cytometry, and histology”).
Mice with deletion of the CDR show a penetrance of lymphoproliferations similar to that of MDR-deleted mice
Forty-two percent of 15- to 18-month-old CDRfl/−CD19-Cre and 67% of CDRfl/flCD19-Cre mice developed clonal lymphoproliferations (Figure 4). The higher penetrance of the phenotype in CDRfl/flCD19-Cre compared with CDRfl/−CD19-Cre mice was not significant, likely because of the smaller number of mice analyzed in the former cohort. Overall, the frequency of lymphoproliferations in mice with CDR deletion in B cells was higher than that reported previously for miR-15a/16-1 deletion (26% and 34% in miR-15a/16-1−/− and miR-15a/16-1fl/−CD19-Cre mice, respectively)22 and in the same range as that of the MDR deletion (each 42% in the MDR−/− and MDRfl/−CD19-Cre mice, respectively).22
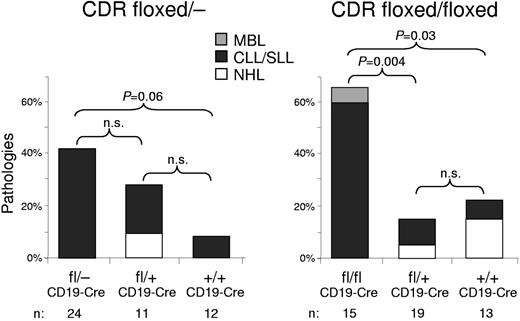
Mice with deletion of CDR develop lymphoproliferations. Percentages of B-lymphoid pathologies observed in 15- to 18-month-old mice of the CDRfl/−CD19-Cre and CDRfl/flCD19-Cre cohorts. The percentages of MBL, CLL/small cell lymphocytic leukemia (SLL), and non-Hodgkin lymphoma (NHL) in each genotype are color coded. P values (χ2 test of association) among the genotypes are indicated; n.s. indicates not significant.
Mice with deletion of CDR develop lymphoproliferations. Percentages of B-lymphoid pathologies observed in 15- to 18-month-old mice of the CDRfl/−CD19-Cre and CDRfl/flCD19-Cre cohorts. The percentages of MBL, CLL/small cell lymphocytic leukemia (SLL), and non-Hodgkin lymphoma (NHL) in each genotype are color coded. P values (χ2 test of association) among the genotypes are indicated; n.s. indicates not significant.
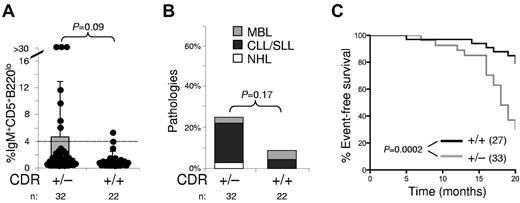
Mice with deletion of the CDR exhibit an indolent disease course but succumb to their disease faster than MDR-deleted mice
In agreement with the notion that heterozygous deletion of the chromosome 13q14 tumor-suppressor region is sufficient to promote lymphoproliferations, we observed previously that MDR+/− mice, in contrast to wild-type mice, showed a trend toward developing clonal lymphoproliferations.22 We therefore established a cohort of CDR+/− mice and wild-type littermates of sufficient size to permit statistical evaluation and analyzed 15- to 18-month-old mice for the occurrence of CD5+ lymphoproliferations in the PB, B-cell pathologies, and survival (Figure 5A-C). CDR+/− mice developed PB CD5+ lymphoproliferations more frequently than wild-type mice (Figure 5A) and also showed a trend toward harboring B-cell malignancies at an elevated frequency (Figure 5B; 25% vs 8% observed in wild-type mice). The event-free survival curves showed that CDR+/− mice died sooner than their wild-type littermates (P = .0002; Figure 5C). Comparing survival of the CDR and MDR cohorts, 70% of CDR+/− versus 45% of MDR+/− mice had died at 20 months (supplemental Figure 4), suggesting that once CDR+/− mice develop lymphoproliferations, they succumbed to their disease faster than MDR+/− mice. In summary, CDR+/− mice developed lymphoproliferations at a similar frequency compared with MDR+/− mice22 (25% vs 24%, respectively), with a similar time of disease onset and a trend toward a more aggressive disease course compared with that observed in MDR+/− mice.
Mice with heterozygous deletion of the CDR develop lymphoproliferations and show an indolent disease course. (A) Percentages of CD5+B220lo cells among mononuclear cells of mice from the CDR+/− cohort. P values are indicated. For further details, see the legend to Figure 2. (B) Percentages of B-lymphoid pathologies observed in 15- to 18-month-old mice of the CDR+/− cohort. For further details, see the legend to Figure 4. (C) Percentage of event-free survival in the CDR+/− cohort. Mice were followed for 20 months. Events comprised illness or mice identified as moribund or sick (palpable tumor or visible ascites), which were killed. P values between the CDR+/− and wild-type mice are indicated. The number of mice of each genotype is indicated in brackets. Mice shown in panels B and C correspond to different cohorts.
Mice with heterozygous deletion of the CDR develop lymphoproliferations and show an indolent disease course. (A) Percentages of CD5+B220lo cells among mononuclear cells of mice from the CDR+/− cohort. P values are indicated. For further details, see the legend to Figure 2. (B) Percentages of B-lymphoid pathologies observed in 15- to 18-month-old mice of the CDR+/− cohort. For further details, see the legend to Figure 4. (C) Percentage of event-free survival in the CDR+/− cohort. Mice were followed for 20 months. Events comprised illness or mice identified as moribund or sick (palpable tumor or visible ascites), which were killed. P values between the CDR+/− and wild-type mice are indicated. The number of mice of each genotype is indicated in brackets. Mice shown in panels B and C correspond to different cohorts.
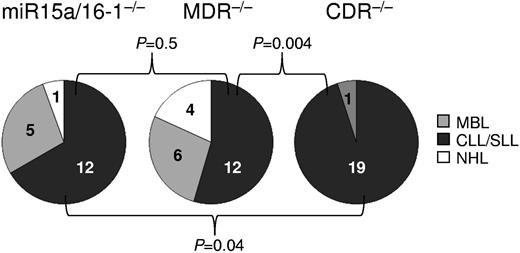
Mice with deletion of the CDR display a different spectrum of lymphoproliferations compared with MDR and miR-15a/16-1–deleted mice
Fifteen- to 18-month-old CDRfl/−CD19-Cre and CDRfl/flCD19-Cre mice developed clonal CD5+ lymphoproliferations, almost all of which represented CLL (Figure 4). This spectrum of lymphoproliferations differed significantly from those observed in the miR-15a/16-1– and MDR-deleted mice,22 which, in addition to CLL, also showed sizable frequencies of MBL and CD5− non-Hodgkin lymphoma (Figure 6). These results suggest that a concomitant deletion of the genetic elements encoded in the chromosomal region telomeric to the MDR, while not elevating disease penetrance, does change the spectrum of B-cell malignancies. Because CDR-deleted mice show a diminished fraction of MBL, the presumed precursor of CLL, compared with miR-15a/16-1 and MDR-deleted mice,22 this finding is suggestive of a faster transition of MBL to CLL or the bypass of a precursor stage in CDR-deleted mice. Together with the results from the event-free survival analysis (Figure 5C and supplemental Figure 4), this observation suggests that CDR-deleted mice develop more aggressive lymphoproliferations and succumb to their disease earlier than their MDR counterparts.
Spectrum of lymphoproliferations in mice with homozygous deletion of the CDR, MDR, and miR-15a/16-1. The fraction of CLL among the various types of B-lymphoproliferations in mice with homozygous deletion of the CDR, MDR, and miR-15a/16-1 was statistically evaluated among the respective cohorts. CDR−/− group: CDRfl/−CD19-Cre and CDRfl/flCD19-Cre mice; MDR−/− group: MDR−/− and MDRfl/−CD19-Cre mice22 ; miR-15a/16-1−/− group: miR-15a/16-1−/− and miR-15a/16-1fl/−CD19- Cre mice.22
Spectrum of lymphoproliferations in mice with homozygous deletion of the CDR, MDR, and miR-15a/16-1. The fraction of CLL among the various types of B-lymphoproliferations in mice with homozygous deletion of the CDR, MDR, and miR-15a/16-1 was statistically evaluated among the respective cohorts. CDR−/− group: CDRfl/−CD19-Cre and CDRfl/flCD19-Cre mice; MDR−/− group: MDR−/− and MDRfl/−CD19-Cre mice22 ; miR-15a/16-1−/− group: miR-15a/16-1−/− and miR-15a/16-1fl/−CD19- Cre mice.22
Stereotypic Ag receptors in CLL derived from different transgenic mice
Sequence analysis of PCR-amplified IgV genes from lymphopro-liferations of homo- or heterozygous CDR-deleted mice from CDRfl/flCD19-Cre, CDRfl/−CD19-Cre, and CDR+/− cohorts demonstrated that CD5+ tumors expressed unmutated IgV genes and CD5− tumors somatically mutated IgV genes (supplemental Table 1). The same distribution was observed previously in the MDR and miR-15a/16-1 cohorts.22
CLL in humans is characterized by the expression of stereotypic Ag receptors; that is, the expression of structurally highly similar or identical BCR heavy chain complementary determining regions (HCDR3s) that arise from the rearrangement of the Ab gene segments (VDJ recombination) between unrelated individuals.29-32 Comparing the altogether 27 amino acid sequences of the HCDR3 regions among the different lymphoproliferations derived from the CDR cohorts, we identified several clonal CD5+ B-cell proliferations that expressed stereotypic HCDR3 regions that are defined by displaying 80% or more homology at the protein level (Table 1). Comparison with 30 previously published HCDR3 regions from MDR and miR-15a/16-1 cohorts22 revealed several distinct clusters of stereotypic HCDR3 regions (supplemental Table 2). TCL1-transgenic mice develop CLL similar to the miR-15a/16-1–, MDR-, and CDR-deleted mice.26 Comparing all HCDR3 regions derived from MBL or CLL cases in our studies (46 from the present study and from Klein et al22 ) and of the published TCL1-transgenic study26 (20 sequences), we observed that 44% (29 of 66) of the junctions can be assigned to 8 sets of highly similar HCDR3 regions among the sequence collection (Table 2). Several sequences of the CDR, MDR, or miR-15a/16-1 cohorts could be assigned to 4 of 5 sets of stereotypic HCDR3 regions defined previously by Chiorazzi and colleagues.26 Among the different transgenic mouse cohorts, 33% (2 of 6) of the sequences of the miR-15a/16-1, 53% (8 of 15) of the MDR, 36% (9 of 25) of the CDR, and 50% (10 of 20) of the TCL1 cohorts could be assigned to the 8 sets of stereotypic HCDR3 regions. The higher fraction of stereotypic HCDR3 regions in the TCL1 and MDR compared with any of the other cohorts did not reach statistical significance in a Fisher exact probability test. These observations suggest that the same Ags drive the expansion of the tumor cells in the 13q14 and TCL1 transgenic mouse cohorts. Interestingly, in the clustering analysis of the HCDR3 regions of all 4 cohorts (Table 2), the TCL1_005 and TCL1_006 HCDR3 regions, that together comprised one stereotypic cluster in the analysis of Yan et al,26 could be assigned to 2 different clusters (stereotype set IV and V in Table 2) characterized by the usage of different D genes (DFL16.1 and DSP2.x/DSP2.1, respectively). This implies that expanding the set of HCDR3 sequences derived from the various CLL mouse models can lead to a more precise definition of stereotypic HCDR3 regions in mice. In summary, the occurrence of highly similar and identical (Table 2: CDR+/+#114, CDR+/−#181, and MDR−/−#138; CDR+/−CD19-Cre#225, MDR+/−#27 and TCL1_006; CDRfl/flCD19-Cre#202 and TCL1_010) HCDR3 regions between different transgenic mice that develop CLL provide additional evidence for a critical role for common Ags or auto-Ags in the clonal expansion of the tumor cells. In accordance, the similarity of Abs of the stereotypic sets to murine Abs of known structure and Ag-binding specificities has been noted by Yan et al.26
HCDR3 rearrangements of lymphoproliferations from CDR-deleted and wild-type mice
| Animal . | VH . | VH . | N . | DH . | N . | JH . | JH . | CDR3 length . | Diagnosis . |
|---|---|---|---|---|---|---|---|---|---|
| CDR+/− #40 | V286 | SR | GVE | IL | N | W | 3 | 9 | DLBCL |
| CDR+/+ #51 | V265 | TR | R | GSSY | YYAMDY | 4 | 13 | CLL/SLL | |
| CDR+/− #179-J4 | >V251 | ARD | P | NWD | Y | YYAMDY | 4 | 14 | CLL/SLL |
| CDRfl/−CD19-Cre #39 | V054 | AR | S | DYR | SQY | YYAMDY | 4 | 15 | CLL/SLL |
| CDRfl/+CD19-Cre #47 | V186 | AR | IYDGYY | YFDY | 2 | 12 | CLL/SLL | ||
| CDRfl/−CD19-Cre #224 | V122 | ARD | DGYY | YFDY | 2 | 11 | CLL/SLL | ||
| CDR+/−CD19-Cre #328 | V332 | AR | E | EA | G | YYAMDY | 4 | 12 | CLL/SLL |
| CDRfl/−CD19-Cre #304-J1 | V304 | AS | HDYD | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #308-J2 | V126 | AR | G | YDYD | DYFDY | 2 | 12 | CLL/SLL | |
| CDR+/− #117 | V188 | LR | EDK | YDG | FDF | 2 | 11 | DLBCL | |
| CDRfl/−CD19-Cre #184-J4 | V027 | VR | D | DYD | AMDY | 4 | 10 | CLL/SLL | |
| CDR+/− #147 | V054 | AR | C | TTVVAT | KE | NAMDY | 4 | 16 | CLL/SLL |
| CDR+/− #67 | V332 | AR | R | DYGSSY | WYFDV | 1 | 14 | CLL/SLL | |
| CDRfl/flCD19-Cre #202 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | CLL/SLL | |
| CDRfl/flCD19-Cre #27 | V120 | ARD | HYGSSY | AWFAY | 3 | 14 | CLL/SLL | ||
| CDRfl/flCD19-Cre #178 | V126 | AR | DP | YYYGSS | L | YYFDY | 2 | 16 | CLL/SLL |
| CDR+/+ #114 | V235 | MRY | GNY | WYFDV | 1 | 11 | MBL | ||
| CDR+/− #181 | V235 | MRY | GNY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDR+/−CD19-Cre #225 | V235 | MRY | SNY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #184-J1 | V163 | AS | YYGY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #308-J1 | V328 | AR | PS | LPY | WYFDV | 1 | 12 | CLL/SLL | |
| CDR+/− #174 | V313 | AR | GVY | YDGYY | PP | FDV | 1 | 15 | CLL/SLL |
| CDRfl/−CD19-Cre #330 | V286 | AR | ERL | DYGY | E | AWFAY | 3 | 15 | CLL/SLL |
| CDR+/− #179-J1 | V190 | AK | E | PYYSNY | D | YWYFDV | 1 | 16 | CLL/SLL |
| CDRfl/−CD19-Cre #304-J2 | V346 | AR | DYSN | YDY | 2 | 9 | CLL/SLL | ||
| CDRfl/flCD19-Cre #14 | V563 | A | G | NW | DFDY | 2 | 8 | CLL/SLL | |
| CDR+/+CD19-Cre #30 | V364 | AR | WDFDV | 1 | 7 | CLL/SLL |
| Animal . | VH . | VH . | N . | DH . | N . | JH . | JH . | CDR3 length . | Diagnosis . |
|---|---|---|---|---|---|---|---|---|---|
| CDR+/− #40 | V286 | SR | GVE | IL | N | W | 3 | 9 | DLBCL |
| CDR+/+ #51 | V265 | TR | R | GSSY | YYAMDY | 4 | 13 | CLL/SLL | |
| CDR+/− #179-J4 | >V251 | ARD | P | NWD | Y | YYAMDY | 4 | 14 | CLL/SLL |
| CDRfl/−CD19-Cre #39 | V054 | AR | S | DYR | SQY | YYAMDY | 4 | 15 | CLL/SLL |
| CDRfl/+CD19-Cre #47 | V186 | AR | IYDGYY | YFDY | 2 | 12 | CLL/SLL | ||
| CDRfl/−CD19-Cre #224 | V122 | ARD | DGYY | YFDY | 2 | 11 | CLL/SLL | ||
| CDR+/−CD19-Cre #328 | V332 | AR | E | EA | G | YYAMDY | 4 | 12 | CLL/SLL |
| CDRfl/−CD19-Cre #304-J1 | V304 | AS | HDYD | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #308-J2 | V126 | AR | G | YDYD | DYFDY | 2 | 12 | CLL/SLL | |
| CDR+/− #117 | V188 | LR | EDK | YDG | FDF | 2 | 11 | DLBCL | |
| CDRfl/−CD19-Cre #184-J4 | V027 | VR | D | DYD | AMDY | 4 | 10 | CLL/SLL | |
| CDR+/− #147 | V054 | AR | C | TTVVAT | KE | NAMDY | 4 | 16 | CLL/SLL |
| CDR+/− #67 | V332 | AR | R | DYGSSY | WYFDV | 1 | 14 | CLL/SLL | |
| CDRfl/flCD19-Cre #202 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | CLL/SLL | |
| CDRfl/flCD19-Cre #27 | V120 | ARD | HYGSSY | AWFAY | 3 | 14 | CLL/SLL | ||
| CDRfl/flCD19-Cre #178 | V126 | AR | DP | YYYGSS | L | YYFDY | 2 | 16 | CLL/SLL |
| CDR+/+ #114 | V235 | MRY | GNY | WYFDV | 1 | 11 | MBL | ||
| CDR+/− #181 | V235 | MRY | GNY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDR+/−CD19-Cre #225 | V235 | MRY | SNY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #184-J1 | V163 | AS | YYGY | WYFDV | 1 | 11 | CLL/SLL | ||
| CDRfl/−CD19-Cre #308-J1 | V328 | AR | PS | LPY | WYFDV | 1 | 12 | CLL/SLL | |
| CDR+/− #174 | V313 | AR | GVY | YDGYY | PP | FDV | 1 | 15 | CLL/SLL |
| CDRfl/−CD19-Cre #330 | V286 | AR | ERL | DYGY | E | AWFAY | 3 | 15 | CLL/SLL |
| CDR+/− #179-J1 | V190 | AK | E | PYYSNY | D | YWYFDV | 1 | 16 | CLL/SLL |
| CDRfl/−CD19-Cre #304-J2 | V346 | AR | DYSN | YDY | 2 | 9 | CLL/SLL | ||
| CDRfl/flCD19-Cre #14 | V563 | A | G | NW | DFDY | 2 | 8 | CLL/SLL | |
| CDR+/+CD19-Cre #30 | V364 | AR | WDFDV | 1 | 7 | CLL/SLL |
The order of cases is according to the alignment obtained after hierarchical clustering of all HCDR3 sequences. Cases that showed ≥ 80% amino acid sequence homology in the CDR3 are grouped.
Stereotypic HCDR3 rearrangements of lymphoproliferations from CDR, MDR, and miR-15a/16-1–deleted and TCL1-transgenic26 mice
| Animal . | VH . | VH . | N . | DH . | N . | JH . | JH . | CDR3 length . | Stereotype set . |
|---|---|---|---|---|---|---|---|---|---|
| MDR+/− #175-PB | V261 | AG | DR | YGY | WYFDV | 1 | 12 | I | |
| TCL1_001 | V261 | AG | DRRGY | WYFDV | 1 | 12 | I | ||
| TCL1_002 | V261 | AG | DRTGY | WYFDV | 1 | 12 | I | ||
| CDRfl/flCD19-Cre #178 | V126 | AR | DP | YYYGSS | L | YYFDY | 2 | 16 | II |
| MDRfl/+CD19-Cre #211 | V126 | AR | YYYGSSY | YFDY | 2 | 13 | II | ||
| TCL1_003 | V4S1 | AR | HYYGSSY | FDY | 2 | 12 | II | ||
| TCL1_004 | V4S1 | AR | HYYGSSY | FDV | 1 | 12 | II | ||
| TCL1_007 | V332 | AR | IYYYGSSY | AMDY | 4 | 14 | III | ||
| TCL1_008 | V1S61 | AR | SYYDGSYY | AMDY | 4 | 14 | III | ||
| CDR+/+ #114 | V235 | MRY | GNY | WYFDV | 1 | 11 | IV | ||
| CDR+/− #181 | V235 | MRY | GNY | WYFDV | 1 | 11 | IV | ||
| MDR−/− #138 | V153 | MR | YGNY | WYFDV | 1 | 11 | IV | ||
| CDR+/−CD19-Cre #225 | V235 | MRY | SNY | WYFDV | 1 | 11 | IV | ||
| MDR+/− #27 | V235 | MR | YSNY | WYFDV | 1 | 11 | IV | ||
| TCL1_006 | V153 | MR | YSNY | WYFDV | 1 | 11 | IV | ||
| CDR+/− #67 | V332 | AR | R | DYGSSY | WYFDV | 1 | 14 | V | |
| CDRfl/flCD19-Cre #202 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | V | |
| TCL1_010 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | V | |
| MDRfl/−CD19-Cre #219 | V128 | YYGSSY | WYFDV | 1 | 11 | V | |||
| TCL1_005 | V153 | MR | YGSSY | WYFDV | 1 | 12 | V | ||
| TCL1_009 | V332 | AR | R | YYGSS | WYFDV | 1 | 13 | V | |
| miRfl/−CD19-Cre #10 | V328 | A | IYYGNY | WYFDV | 1 | 12 | prov. VI | ||
| CDRfl/−CD19-Cre #184-J1 | V163 | AS | YYGY | WYFDV | 1 | 11 | prov. VI | ||
| MDRfl/−CD19Cre #99 | V328 | AR | YYSNY | WYFDV | 1 | 12 | prov. VI | ||
| miRfl/−CD19Cre #234 | V328 | AR | GEK | YSNY | WYFDV | 1 | 14 | prov. VI | |
| CDRfl/+CD19-Cre #47 | V186 | AR | IYDGYY | YFDY | 2 | 12 | prov. VII | ||
| CDRfl/−CD19-Cre #224 | V122 | ARD | DGYY | YFDY | 2 | 11 | prov. VII | ||
| MDRfl/−CD19-Cre #197 | V227 | A | SP | NWD | WYFDV | 1 | 11 | prov. VIII | |
| MDR+/+CD19-Cre #212 | V186 | AR | NWD | WYFDV | 1 | 10 | prov. VIII |
| Animal . | VH . | VH . | N . | DH . | N . | JH . | JH . | CDR3 length . | Stereotype set . |
|---|---|---|---|---|---|---|---|---|---|
| MDR+/− #175-PB | V261 | AG | DR | YGY | WYFDV | 1 | 12 | I | |
| TCL1_001 | V261 | AG | DRRGY | WYFDV | 1 | 12 | I | ||
| TCL1_002 | V261 | AG | DRTGY | WYFDV | 1 | 12 | I | ||
| CDRfl/flCD19-Cre #178 | V126 | AR | DP | YYYGSS | L | YYFDY | 2 | 16 | II |
| MDRfl/+CD19-Cre #211 | V126 | AR | YYYGSSY | YFDY | 2 | 13 | II | ||
| TCL1_003 | V4S1 | AR | HYYGSSY | FDY | 2 | 12 | II | ||
| TCL1_004 | V4S1 | AR | HYYGSSY | FDV | 1 | 12 | II | ||
| TCL1_007 | V332 | AR | IYYYGSSY | AMDY | 4 | 14 | III | ||
| TCL1_008 | V1S61 | AR | SYYDGSYY | AMDY | 4 | 14 | III | ||
| CDR+/+ #114 | V235 | MRY | GNY | WYFDV | 1 | 11 | IV | ||
| CDR+/− #181 | V235 | MRY | GNY | WYFDV | 1 | 11 | IV | ||
| MDR−/− #138 | V153 | MR | YGNY | WYFDV | 1 | 11 | IV | ||
| CDR+/−CD19-Cre #225 | V235 | MRY | SNY | WYFDV | 1 | 11 | IV | ||
| MDR+/− #27 | V235 | MR | YSNY | WYFDV | 1 | 11 | IV | ||
| TCL1_006 | V153 | MR | YSNY | WYFDV | 1 | 11 | IV | ||
| CDR+/− #67 | V332 | AR | R | DYGSSY | WYFDV | 1 | 14 | V | |
| CDRfl/flCD19-Cre #202 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | V | |
| TCL1_010 | V332 | AR | I | YYGSSY | WYFDV | 1 | 14 | V | |
| MDRfl/−CD19-Cre #219 | V128 | YYGSSY | WYFDV | 1 | 11 | V | |||
| TCL1_005 | V153 | MR | YGSSY | WYFDV | 1 | 12 | V | ||
| TCL1_009 | V332 | AR | R | YYGSS | WYFDV | 1 | 13 | V | |
| miRfl/−CD19-Cre #10 | V328 | A | IYYGNY | WYFDV | 1 | 12 | prov. VI | ||
| CDRfl/−CD19-Cre #184-J1 | V163 | AS | YYGY | WYFDV | 1 | 11 | prov. VI | ||
| MDRfl/−CD19Cre #99 | V328 | AR | YYSNY | WYFDV | 1 | 12 | prov. VI | ||
| miRfl/−CD19Cre #234 | V328 | AR | GEK | YSNY | WYFDV | 1 | 14 | prov. VI | |
| CDRfl/+CD19-Cre #47 | V186 | AR | IYDGYY | YFDY | 2 | 12 | prov. VII | ||
| CDRfl/−CD19-Cre #224 | V122 | ARD | DGYY | YFDY | 2 | 11 | prov. VII | ||
| MDRfl/−CD19-Cre #197 | V227 | A | SP | NWD | WYFDV | 1 | 11 | prov. VIII | |
| MDR+/+CD19-Cre #212 | V186 | AR | NWD | WYFDV | 1 | 10 | prov. VIII |
Order of cases is according to the alignment obtained after hierarchical clustering of all HCDR3 sequences. Cases that showed ≥ 80% amino acid sequence homology in the CDR3 to the next neighbor/s are grouped. MDR and miR cases are derived from Klein et al,22 TCL1 cases from Yan et al.26 Stereotype sets defined by Yan et al (sets I-V26 and provisional [prov.] sets VI-VIII identified in the present work) are indicated in the right column.
Discussion
Cre/loxP-mediated conditional deletion of a large chromosomal region in somatic cells
Large chromosomal deletions have been generated previously in mouse ES cells,33-35 and chromosomal translocations have been mimicked in somatic cells using Cre/loxP site-specific recombination.36,37 To our knowledge, the in vivo B cell–specific deletion of a 0.8-mb chromosomal region described here represents the largest conditional deletion that has been accomplished in somatic cells thus far. The deletion efficiency in B cells was high (∼ 90%), possibly because of a good accessibility of the 14qC3 locus for the Cre recombinase. We have established a transgenic system in which the recombination-dependent juxtaposition of a promoter to a gene encoding a fluorescent marker signals Cre/loxP-mediated deletion and simultaneously permits the direct identification of ES cell clones when the 2 targetings occurred on the same chromosome (supplemental Figure 2). This system has the added advantage that such deleted cells can be detected and traced in vivo using fluorescence-based analysis (Figures 1 and 3), a feature that makes the cells amenable to functional and molecular analyses after FACS. The targeting approach described in the present work should be highly suitable for the analysis of chromosomal deletions that occur with lower deletion efficiencies than that observed for the CDR, which would therefore be difficult to study without a fluorescent marker that unequivocally identifies the Cre/loxP-deleted cells.
Extent of h13q14/m14qC3 deletions affects phenotype of lymphoproliferations and disease course
We demonstrated previously that mice with deletion of the h13q14/m14qC3 MDR, while developing the same spectrum of lymphoproliferations, show a higher penetrance of the phenotype and a more severe disease course compared with miR-15a/16-1–deleted mice.22 The present study found that mice with deletion of the CDR develop a different spectrum of lymphoproliferations compared with both MDR- and miR-15a/16-1–deleted mice,22 and suggests that CDR-deleted mice succumb to their disease earlier than MDR-deleted mice. However, both the time of disease onset and disease penetrance were similar to those of MDR-deleted mice.22 Because the miR-15a/16-1, MDR, and CDR cohorts were set up over a similar time period and housed in the same animal room, and because the different cohorts were analyzed over largely the same time span, we consider it unlikely that environmental or seasonal influences could account for the observed differences. We note that the similarity of the late disease onset (∼ 1 year) and low penetrance (∼ 20%-40%) of the miR-15a/16-1-, MDR-, and CDR-deleted mice suggests a role for additional genetic mutations7,8 and/or predisposing factors38,39 in the development of CLL in these mouse models mimicking the heterogeneous 13q14 deletions. It will be interesting to determine the nature of these factors.
The present study focused on 13q14 deletions affecting the MDR and the telomeric region of this tumor-suppressor locus. Subtypes of 13q14 deletions have been identified with loss of DNA centromeric to the DLEU2/mir-15a/16-1 cluster, a fraction of which includes the retinoblastoma (Rb1) tumor-suppressor gene,16 and that have a less favorable prognosis.40-43 Inactivation of miR-15a/16-1, the expression of which is invariably ablated in all CLL with 13q14 aberrations, has functionally overlapping effects with Rb1 deletion, because miR-15a/16-1 down-regulates the expression levels of G0-G1/S-phase–promoting proteins that normally lead to Rb phosphorylation and result in cell-cycle entry. A concomitant deletion of both miR-15a/16-1 and Rb1 may accelerate the disease course.
These observations suggest that, whereas deletion of the miR-15a/16-1 cluster is the critical mechanism in the pathogenesis of CLL with 13q14 aberrations, the additional loss of genetic elements encoded in the 13q14 tumor-suppressor locus can significantly influence the penetrance of the phenotype, the spectrum of the lymphoproliferations, and the severity of the disease course.
Narrowing down the critical genetic elements in the 13q14 tumor-suppressor locus
The results obtained from the investigation of the tumor-suppressor function of the 13q14 locus through miR-15a/16-1, MDR, and CDR conditional knockout mouse models collectively demonstrate that inactivation of the microRNA cluster miR-15a/16-1 alone can cause lymphoproliferations, and that the concomitant deletion of other genetic elements located in the MDR and CDR leads to more aggressive phenotypes. The high conservation between the human 13q14 and murine 14qC3 loci with regard to both the identity of genes and the gene order (Figure 1A; DLEU1 represents the only exception), along with a similar spectrum of diseases developing in both human and mouse, strongly imply that the 14qC3 mouse models faithfully recapitulate the heterogeneous 13q14 deletions observed in humans.
h13q14/m14qC3 MDR
The genetic elements located in the MDR include DLEU2, which produces a long noncoding RNA and the protein-encoding genes DLEU5 and KCNRG. DLEU2 is the host gene of miR-15a/16-1, and its essential role in providing the primary transcript of these microRNAs is demonstrated by the complete ablation of miR-15a/16-1 production on specific deletion of the DLEU2/dleu2 promoter region44 (and our own unpublished observations); this particular deletion that leaves the miR-15a/16-1 gene cluster intact has been described to occur in vivo in several CLL cases.17,40,44 An unresolved issue is whether DLEU2 has any additional roles, because experiments aimed at identifying an independent function for the DLEU2 transcript, the sequence of which does not display homology to any other long noncoding RNA, have not yielded any insights.22,44 A further question regarding a potential function of DLEU2 transcripts is whether the processed pseudogene DLEU2L, encoded on chromosomal region 1p22, may compensate for the loss of DLEU2 in CLL with 13q14 deletions. Likewise, it remains to be determined to what extent expression of the miR-15b/16-2 cluster complements the biologic effects of h13q14/m14qC3 deletions and, in turn, whether genetic mutations in this region exacerbate the pathogenesis of CLL with h13q14/m14qC3 deletions. Whereas both DLEU5 and KCNRG are considered to be located outside of the MDR in humans, these genes are deleted in a sizable fraction of CLL cases and therefore their functional inactivation may potentially contribute to the aggressiveness of the phenotype. The biologic function of KCNRG is only beginning to be understood45 ; however, its low or absent expression in mature B-cell subpopulations (unpublished observations) would argue against its being a candidate tumor suppressor in CLL. DLEU5 encodes a protein of unknown function. In the mouse, the dleu5 gene overlaps with dleu2, and the genes are located in opposite transcriptional orientations (Figure 1A). In the human, the exons encoding the major form of the polyadenylated DLEU2 transcript do not overlap with the DLEU5 exons, although rare transcripts have been reported that use putative DLEU2 downstream exons overlapping with the DLEU5 gene.46 Such alternative DLEU2 transcripts were suggested to function as an antisense RNA for DLEU5,46 invoking a regulatory function of the DLEU2 gene for DLEU5 mRNA expression. Future investigations will need to clarify the functional interaction of the DLEU2 and DLEU5 genes and their respective roles in B-cell physiology and neoplastic transformation.
h13q14/m14qC3 CDR
The observation that CDR-deleted mice show a diminished fraction of MBL compared with MDR-deleted mice suggests that the corresponding genomic region contains a genetic element the deletion of which either causes a faster transition of MBL to CLL or the bypassing of the CLL precursor stage in CDR-deleted mice. The known genes encoded in the region telomeric to the MDR are RNASEH2B and DLEU7. There are no confirmed microRNAs or other small noncoding RNAs in this region in humans (Basso et al47 and K. Basso and R.D.-F., unpublished observations; miRBase Release 17) or mouse (miRBase Release 17). Whereas the function of RNASEH2B in B cells is unknown, DLEU7 was recently suggested to play a role in the negative regulation of cell-cycle entry based on functional assays performed in nonlymphoid cells,24 implying that DLEU7 is a 13q14 candidate tumor-suppressor gene that may contribute to the severity of disease. It cannot be excluded that the CDR contains an as-yet-unidentified regulatory DNA element that may control or modify the expression of genes encoded in the h13q14/m14qC3 locus. In addition, it remains to be determined how deletion of DLEU1, which is not conserved in the mouse genome, influences CLL pathogenesis.
In summary, the present study contributes to characterizing and redefining the critical genetic elements in the h13q14/m14qC3 tumor-suppressor locus. Because 13q14 deletions in humans are usually heterogeneous, these findings provide a clear rationale for determining the leukemogenic role of the individual genetic elements affected by deletions of the 13q14 locus in addition to that of miR-15a/16-1.
Clusters of stereotypic Ag receptors among independent CLL mouse models
The striking similarities of Ab HCDR3 regions derived from tumor cases of different CLL mouse models underscores the validity of previously identified clusters of stereotypic Ag receptors in TCL1-transgenic mice26 and identifies additional clusters (Table 2). These findings provide a solid rationale for defining clusters of stereotypic Ag receptors in CLL-prone mice, which may potentially lead to the identification of their cognate Ags in a way similar to what has been accomplished for human CLL Abs that have a common stereotypic rearrangement.48-50 Knowing the identity of these Ags would provide the basis for establishing suitable animal models aimed at studying the dynamics of Ag dependency in CLL pathogenesis in vivo.
Correlative data strongly suggest a role for extrinsic Ags or auto-Ags in human CLL development. The present observations that transgenic mice mimicking CLL-associated genetic alterations develop lymphoproliferations with stereotypic Ab receptors support this concept by providing functional evidence that genetic aberrations disrupting the control of cell growth and survival and chronic Ag stimulation cooperate in the clonal expansion of CLL tumor cells.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
Acknowledgments
The authors thank Thomas Ludwig for advice on the generation of the transgenic mice, Lauren Bertin for help with the IgV gene analysis, and Kristie Gordon and Chenhong Liu of the Herbert Irving Comprehensive Cancer Center Flow Cytometry Shared Resource for expert cell isolation.
Authorship
Contribution: M.L. cloned the targeting constructs, screened for homologous recombinants, organized the establishment of cohorts, and genotyped the mice; A.C. performed the IgV gene analysis, flow cytometric analysis of blood samples, and in vitro stimulation assays; H.T. performed flow cytometric analysis of lymphoid and tumor tissue and prepared tissue samples for histology; Q.S. oversaw the ES cell work and performed blastocyst injections; T.M. established and maintained the cohorts; G.B. performed the pathology analysis and revised the manuscript; R.D.-F. designed the research and cowrote the manuscript; and U.K. devised the targeting strategy, designed the research, analyzed the data, and cowrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
IgV gene sequences are available upon request.
Correspondence: Ulf Klein, Herbert Irving Comprehensive Cancer Center, 1130 St Nicholas Ave R312, New York, NY 10032; e-mail: uk30@columbia.edu.