Abstract
Since the introduction of highly active antiretroviral therapies (ART), the prognosis for HIV-1 patients has improved immensely. However, approximately 25% of patients can experience a variety of inflammatory symptoms that are collectively known as immune reconstitution inflammatory syndrome (IRIS). Studying the etiology and immunopathology of IRIS has been hampered by the fact that the symptoms and associated opportunistic infections are highly varied. We hypothesized that there is a common mechanism underlying IRIS pathogenesis and investigated a patient group with IRIS related to different pathogens. Functional and phenotypic characterization of PBMC samples was performed by polychromatic flow cytometry after in vitro stimulation with relevant antigenic preparations. In most patients, IRIS events were characterized by the robust increase of preexisting polyfunctional, highly differentiated effector CD4+ T-cell responses that specifically targeted the antigens of the underlying co-infection. T-cell responses to HIV-1 or other underlying infections were not affected and did not differ between IRIS and non-IRIS patients. These data suggest that patients with IRIS do not have a generalized T-cell dysfunction; instead, IRIS represents a dysregulated CD4+ T-cell response against residual opportunistic infection antigen. These studies were registered at www.clinical-trials.gov as NCT00557570 and NCT00286767.
Introduction
Immune reconstitution inflammatory syndrome (IRIS) is a severe clinical complication that manifests in approximately 25% of HIV-1 patients initiating highly active antiretroviral therapy (ART).1 Previous studies showed that the incidence of IRIS is higher in patients with advanced immunodeficiency who also have underlying opportunistic infections (OIs).2 With ART becoming more widely available in populations with a high incidence of OIs, the occurrence of IRIS is likely to further increase in the near future. Thus, a better understanding of the immunopathology and identification of predictive biomarkers are of great clinical importance to develop targeted interventions.
Different types of IRIS have been identified, including “unmasking” IRIS, defined as a previously subclinical, undiagnosed OIs occurring shortly after ART initiation, which cannot be explained by a de novo infection; “paradoxical” IRIS, defined as a worsening of a known condition for which the patient has been treated successfully (microbial cultures are often negative)3,4 ; and “autoimmune” IRIS, encompassing immune responses to self-antigens. This last category has received less attention in the literature, although there have been several reports of Graves' disease associated with IRIS.5-7 In all 3 types, there is inflammation occurring during immune reconstitution that cannot be explained by drug toxicity or de novo infection. IRIS is largely a diagnosis of exclusion, but some criteria have been outlined by the AIDS Clinical Trials Group,8 as well as the International Network for the study of HIV-associated IRIS.9,10
The immunopathogenesis of IRIS remains unclear. IFN-γ–producing T cells appear to be the main players driving the dysregulated inflammatory response,11 but these findings are not consistent with those of other groups.12,13 In a study of Cryptococcus-associated IRIS, a paucity of immune responses before ART appeared to predispose to IRIS, possibly because of defective antigen (Ag) clearance and accumulation.14 Severely immunocompromised patients who are ART-naive at OI diagnosis1 and those with low CD4 counts appear to be most susceptible to IRIS.2,15,16 Other risk factors include high HIV-1 RNA levels before ART,2 a more rapid initial HIV-1 RNA suppression,1,17 a high OI-derived antigen load at the time of ART initiation,3,18 a short time interval between diagnosis and/or treatment start of an OI and ART initiation,1,3,16 and a low body mass index.15 Nonetheless, reports are not consistent, and it has been suggested that risk factors might differ by mode of IRIS presentation and etiology.13,15
Most studies to date trying to elucidate potentially diagnostic risk factors and/or biomarkers of IRIS have focused on patient groups with a particular opportunistic infection, such as Mycobacterium tuberculosis (TB),16,19-21 Mycobacterium avium, and intracellulare complex (MAC),22 Cryptococcus neoformans,3,14,18 or CMV.12 However, we hypothesized that there might be some common mechanisms underlying the development of IRIS in HIV-1 patients on ART, despite the highly variable clinical manifestations and variety of agents that can be associated with the syndrome.
We recently reported elevated frequencies of PD-1+ cells before ART, both within the CD4+ and CD8+ T-cell compartments,23 showing evidence of antigenic stimulation and a propensity for Th1 cytokine production. It was unclear whether the increased T-cell activation was the result of HIV-1, the IRIS-associated pathogen, or other co-pathogens. We addressed this question using a cohort of HIV-1–infected IRIS patients with variable IRIS-causing OIs and HIV-1–infection contemporary controls. To exclude the baseline CD4+ T-cell count and persistent HIV-1 viremia as factors influencing outcome, only patients with less than 100 CD4+ cells/μL at baseline and less than 500 HIV-1 RNA copies/mL after one year of treatment were included in the present study. We found that most IRIS patients manifest an upsurge of preexisting Ag-specific CD4+ T-cell responses that are polyfunctional and are targeting exclusively the IRIS-associated pathogen and not HIV-1 itself or other co-infections.
Methods
Human subjects and sample collection
A total of 67 HIV-1–infected subjects were enrolled and provided written informed consent at the Clinical Center of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under an institutional review board-approved protocol. Patient inclusion criteria have been described previously.24 Briefly, all patients (1) were ART-naive or had interrupted treatment for at least 1 year (n = 4, plus n = 2 who had previously received brief mono- or dual-therapy; 1 of these was an IRIS patient) with a viral rebound of more than 10 000 copies/mL, (2) had less than or equal to 100 CD4+ T cells/μL at baseline; (3) suppressed their HIV-1 viral load to less than 500 copies/mL within 1 year of ART; and (4) had available PBMC samples taken before ART as well as after 1, 3, 6, and 12 months of ART. Nineteen patients developed IRIS episodes after commencement of ART, whereas 48 underwent uneventful immune reconstitution (Table 1). IRIS was defined according to the AIDS Clinical Trials Group criteria.8 The median time to IRIS was 1.4 months (interquartile range, 0.9-2.6 months). Baseline characteristics, use of non-nucleoside reverse-transcriptase inhibitors or protease inhibitors, and ethnicity distribution were comparable between the IRIS and non-IRIS groups; the frequency of co-infections was more elevated in IRIS patients (Table 1). A subset of this cohort has been previously described in detail.24
Patient cohort characteristics
| . | IRIS . | Non-IRIS . |
|---|---|---|
| N | 19 | 48 |
| Age at ART initiation, y* | 35.5 (30.4-43.3) | 36.5 (32.6-40) |
| Male, % | 63.2 | 83.3 |
| Pre-ART | ||
| PVL, log10* | 5.11 (4.76-5.39) | 5.11 (4.7-5.38) |
| CD4+ cells/μL* | 14 (6-47) | 22 (10-39) |
| CD8+ cells/μL* | 333 (230-692) | 391 (238-639) |
| Coinfections, % (no. of IRIS events)† | 94.7 | 64.6 |
| CMV | 15.8 (1u) | 12.5 |
| C neoformans | 10.5 (1p, 1u) | 6.3 |
| EBV | 5.3 (1u) | 2.1 |
| HBV | 5.3 (1p) | 12.5 |
| HHV-8 | 10.5 (1p, 1u) | 2.1 |
| H capsulatum | 10.5 (1p) | 4.2 |
| HPV | 26.3 (1p) | 16.7 |
| JCV | 5.3 (1u) | 0 |
| MAC | 31.6 (2p, 4u) | 6.3 |
| S stercoralis | 10.5 (1u) | 6.3 |
| TB | 21.1 (2p) | 16.7 |
| VZV | 15.8 (1p) | 20.8 |
| ART regimen component, % | ||
| NNRTIs | 68.4 | 52.1 |
| PIs | 31.6 | 47.9 |
| Ethnicity, % | ||
| Black | 52.6 | 47.9 |
| American Indian | 0 | 2.1 |
| Asian | 0 | 2.1 |
| White | 15.8 | 20.8 |
| Mixed | 10.5 | 12.5 |
| . | IRIS . | Non-IRIS . |
|---|---|---|
| N | 19 | 48 |
| Age at ART initiation, y* | 35.5 (30.4-43.3) | 36.5 (32.6-40) |
| Male, % | 63.2 | 83.3 |
| Pre-ART | ||
| PVL, log10* | 5.11 (4.76-5.39) | 5.11 (4.7-5.38) |
| CD4+ cells/μL* | 14 (6-47) | 22 (10-39) |
| CD8+ cells/μL* | 333 (230-692) | 391 (238-639) |
| Coinfections, % (no. of IRIS events)† | 94.7 | 64.6 |
| CMV | 15.8 (1u) | 12.5 |
| C neoformans | 10.5 (1p, 1u) | 6.3 |
| EBV | 5.3 (1u) | 2.1 |
| HBV | 5.3 (1p) | 12.5 |
| HHV-8 | 10.5 (1p, 1u) | 2.1 |
| H capsulatum | 10.5 (1p) | 4.2 |
| HPV | 26.3 (1p) | 16.7 |
| JCV | 5.3 (1u) | 0 |
| MAC | 31.6 (2p, 4u) | 6.3 |
| S stercoralis | 10.5 (1u) | 6.3 |
| TB | 21.1 (2p) | 16.7 |
| VZV | 15.8 (1p) | 20.8 |
| ART regimen component, % | ||
| NNRTIs | 68.4 | 52.1 |
| PIs | 31.6 | 47.9 |
| Ethnicity, % | ||
| Black | 52.6 | 47.9 |
| American Indian | 0 | 2.1 |
| Asian | 0 | 2.1 |
| White | 15.8 | 20.8 |
| Mixed | 10.5 | 12.5 |
PVL indicates plasma viral load; p, paradoxical IRIS; u, unmasking IRIS; NNRTIs, non-nucleoside reverse-transcriptase inhibitors; and PIs, protease inhibitors.
Median (interquartile range).
Only those OIs associated with IRIS events in this study were considered. P = .0138. None of the other parameters was statistically different between patient groups; individual infections were not tested for statistical significance.
Ethical approval
All patients provided written informed consent before inclusion in accordance with the Declaration of Helsinki, and the study was performed according to a National Institute of Allergy and Infectious Diseases institutional review board–approved protocol.
Determination of plasma viral load, CD4+ and CD8+ T-cell counts
Plasma HIV-1 viral loads, as well as CD4+ and CD8+ T-cell counts were determined in a Clinical Laboratory Improvement Amendment-approved laboratory. The plasma viral load was measured using the ultrasensitive Quantiplex HIV-1 bDNA Version 3.0 (Bayer).
CD4+ and CD8+ T-cell counts were determined by 4-color flow cytometry. The BD Multitest (BD Biosciences) that was used includes the following Abs: CD3FITC (clone SK7), CD4APC (clone SK3), CD8PE (clone SK1), and CD45PerCP (clone 2D1). Samples were acquired on a FACSCanto (BD Biosciences). CD4+ T-cell counts were calculated as percentage CD4+ CD3+ cells within CD45+ lymphocytes divided by 1% of the white blood cell count. The corresponding calculation was performed for CD8+ T-cell counts.
Sample preparation and Ag stimulation
Cryopreserved PBMCs were thawed in prewarmed RPMI 1640, 10% FCS, 2mM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Invitrogen; this medium will hereafter be referred to as complete RPMI), in the presence of 20 μg/mL benzonase nuclease (Novagen). Cells were rested in complete RPMI for 4 to 6 hours at 37°C, 5% CO2 and either left unstimulated (mock control) or stimulated overnight in 200 μL RPMI complete with different Ag preparations (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article): 2 μg/mL CMV pp65 peptide pool,25 2.5 μg/mL HBV surface Ag peptide pool,26 0.8 μg/mL HHV-8 K12 and K15 peptide pool,27 2.5 μg/mL HIV-1 Gag peptide pool (NIH AIDS Research and Reference Reagent Program), protein pool of HPV6 (1.6 μg/mL), HPV11 (3.2 μg/mL), HPV16 (3.2 μg/mL), and HPV18 (1.6 μg/mL; Gardasil, Merck), 5.7 μg/mL JC Virus VP1 peptide pool, 2.5 μg/mL VZV IE62 peptide pool (New England Peptide), 5 μg/mL MAC bacterial lysate,28 2.5 μg/mL TB PPD (Statens Serum Institute), 25 μg/mL C neoformans–secreted mannoprotein,29 5 μg/mL Histoplasma capsulatum cell wall extract,30 or 100 μg/mL Strongyloides stercoralis sodium deoxycholate-soluble proteins from parasite lysate.31,32 Stimulation cultures contained anti-CD49d and anti-CD28Cy5-PE mAb (BD Biosciences). Monensin and brefeldin A (BD Biosciences) were added after 2 hours of stimulation. Healthy donor PBMCs were stimulated with staphylococcal enterotoxin B (Sigma-Aldrich) to serve as a positive control.
Flow cytometry
The reagent panel used in the present study was developed based on that described by Mahnke and Roederer33 and is given in supplemental Table 2. The following reagents were used: anti-CD3APC-Cy7 (clone SK7), anti-CD14PacBlu (clone M5E2), anti-CD28PE-Cy5 (clone CD28.2), anti–IL-2APC (clone MQ1-17H12; BD Biosciences PharMingen), anti–IFN-γAx488 (clone B27; Reametrix), anti-CD31PE-Cy7 (clone WM59), anti–PD-1biotin (clone EH12.2H7; BioLegend), anti-CD127PE-Cy5.5 (clone R34.34; Immunotech Coulter), anti–TIM-3PE (clone 344823; R&D Systems), and streptavidinQD605 (Invitrogen). Anti-CD4 (clone M-T477), anti-CD7 (clone M-T701), anti-CD8 (clone RPA-T8), anti-CD19 (clone HIB19), anti-CD27 (clone M-T271), anti-CD45RO (clone UCHL1), anti-CD57 (clone NK-1), anti-TNF (clone MAb11; BD Biosciences PharMingen), and anti-CCR7 (clone 150503; R&D Systems) were conjugated in-house to QD800, QD705, QD585, Pacific blue, QD655, QD545, QD565, Ax594, and Ax680 (Invitrogen), respectively. Dead cells were detected with the live/dead fixable violet dead cell stain (Invitrogen). For intracellular staining, cells were treated with BD Cytofix/Cytoperm Permeabilization Solution (BD Biosciences). Data were acquired on an LSR II (BD Biosciences).
Data analysis
Data were analyzed using FlowJo Version 9.2 (TreeStar), Pestle (by M.R.), and SPICE Version 5.1.34 The gating scheme is illustrated in supplemental Figure 3. All cytokine measurements were background subtracted, taking into account the frequency of cells producing cytokines in the absence of antigenic stimulation (mock control). For the phenotypic analysis of Ag-specific cells, only those samples with more than 10 cytokine-positive events and response magnitudes more than 3 times that of the corresponding mock control were considered.34
Statistical analysis
A 2-tailed t test was used to compare the proportion of individuals using steroids between the 2 patient groups. Nonparametric tests were used for all other analyses (SAS Version 9.2). Changes from baseline within each group (paired differences) were evaluated using the Sign test. Data comparisons of single measurements between groups (IRIS vs non-IRIS vs other IRIS) were performed using the Wilcoxon rank-sum test. Statistical comparisons of pie charts were performed in SPICE Version 5.1 software using 10 000 permutations.34 Given the exploratory nature of this study, there was no adjustment for multiple comparisons.
Results
Longitudinal analysis of total CD4+ and CD8+ T cells during ART
Because IRIS episodes were experienced at different time points by individual patients, we defined time ranges for IRIS patients synchronized to the initiation of ART and their respective IRIS events (supplemental Table 3).
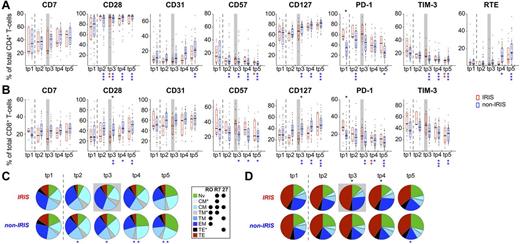
We first investigated the evolution of T-cell activation phenotypes and T-cell differentiation stages over the course of ART in IRIS and non-IRIS patients to determine whether these groups experienced differences in the overall reconstitution of T cells during ART. Some of the phenotypes were previously investigated using different aliquots of a subset of these samples.23 Here we significantly extended those studies, synchronizing samples to the IRIS event (supplemental Table 3) and characterizing the pathogen-specific responses.
Even though statistically significant changes from baseline were observed over time in non-IRIS patients for many of the differentiation and activation markers investigated, comparable changes in IRIS patients were only rarely significant (Figure 1). Nevertheless, no statistically significant differences were detected between the overall activation status or T-cell subset distribution of CD4+ T cells between IRIS and non-IRIS patients over the course of the study, except for the previously described elevated frequencies of PD-1+ cells before ART in eventual IRIS patients (Figure 1A,C).23 PD-1+ cells were also elevated before ART in CD8+ T cells, although the only statistically significant differences at the time of IRIS (tp3) were observed in the frequencies of CD28+ and CD127+ cells, both of which were decreased in IRIS patients (Figure 1B). Concordantly, a significantly more elevated proportion of highly differentiated (mainly TEM) CD8+ T cells was found in IRIS patients during and early after (tp3-tp4) clinical IRIS diagnosis (Figure 1D). Together, these data confirm our previous observations that IRIS events are associated with highly activated T cells.
Longitudinal analysis of total CD4+ and CD8+ T-cell phenotypes of IRIS and non-IRIS patients during ART. Characteristics of total CD4+ (A,C) and CD8+ T cells (B,D) were analyzed in PBMC samples from IRIS (red) and non-IRIS patients (blue) before as well as after 1, 3, 6, and 12 months of ART. (A-B) Activation phenotype and presence of recent thymic emigrants (RTE; in CD4+ T cells only). (C-D) Representation of T-cell differentiation states. T-cell differentiation subsets were defined by expression of CD45RO (“RO”), CCR7 (“R7”), and CD27 (“27”). TNV indicates naive; TCM, central memory; TTM, transitional memory; TEM, effector memory; and TTE, terminal effector. TCM*, TTM*, and TTE* represent phenotypically defined populations that are not described in the literature but that arise by this gating scheme; their activation phenotype and cytokine potential most closely resemble that of TCM, TTM, and TTE, respectively; hence their nomenclature. Graphs represent interquartile ranges, median bars, as well as individual data points. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. All time points were compared between patient groups (results indicated in black above bars/pies), as well as with corresponding pre-ART measurements within each patient group (results are color-coded and indicated below bars/pies). *P < .01; **P < .001; ***P < .0001.
Longitudinal analysis of total CD4+ and CD8+ T-cell phenotypes of IRIS and non-IRIS patients during ART. Characteristics of total CD4+ (A,C) and CD8+ T cells (B,D) were analyzed in PBMC samples from IRIS (red) and non-IRIS patients (blue) before as well as after 1, 3, 6, and 12 months of ART. (A-B) Activation phenotype and presence of recent thymic emigrants (RTE; in CD4+ T cells only). (C-D) Representation of T-cell differentiation states. T-cell differentiation subsets were defined by expression of CD45RO (“RO”), CCR7 (“R7”), and CD27 (“27”). TNV indicates naive; TCM, central memory; TTM, transitional memory; TEM, effector memory; and TTE, terminal effector. TCM*, TTM*, and TTE* represent phenotypically defined populations that are not described in the literature but that arise by this gating scheme; their activation phenotype and cytokine potential most closely resemble that of TCM, TTM, and TTE, respectively; hence their nomenclature. Graphs represent interquartile ranges, median bars, as well as individual data points. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. All time points were compared between patient groups (results indicated in black above bars/pies), as well as with corresponding pre-ART measurements within each patient group (results are color-coded and indicated below bars/pies). *P < .01; **P < .001; ***P < .0001.
Longitudinal analysis of HIV-1–specific T-cell responses during ART
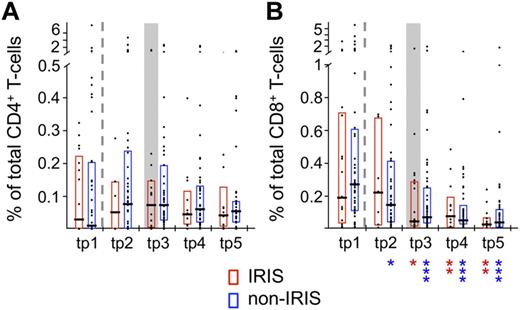
We then set out to determine whether the HIV-1–specific T-cell response varied between the 2 patient groups. No significant differences were observed in terms of response magnitude, cytokine pattern, or T-cell subset distribution within HIV-1 Gag-specific cells in either CD4+ (Figures 2A and 3) or CD8+ T cells (Figures 2B and 3C-D) at any of the time points analyzed. The same was true for the activation phenotype of HIV-1–reactive CD4+ or CD8+ T cells in IRIS and non-IRIS patients (P > .2 for CD28, CD31, CD57, CD127, PD-1, and TIM-3). Statistically significant decreases were observed over time in the magnitude of the HIV-1–specific CD8+ T-cell response after ART initiation in both patient groups (Figure 2B). Although the relative frequency of IFN-γ+ IL-2− TNF− cells decreased (Figure 3A) and that of TTM cells among HIV-1–specific CD4+ T cells increased (Figure 3B) over time in non-IRIS patients, the changes from baseline were not statistically different between patient groups.
The magnitude of HIV-1 Gag-specific T cells does not differ significantly between IRIS and non-IRIS patients. The total response magnitude, measured by production of IFN-γ and/or IL-2 and/or TNF, generated by HIV-1 Gag reactive CD4+ (A) and CD8+ T cells (B) of IRIS and non-IRIS patients were compared at the 5 analysis time points. Graphs represent interquartile ranges, median bars, as well as individual data points. Dashed lines separate pre-ART from on-ART samples. Gray boxes indicate samples from IRIS patients within 3 months of clinical IRIS onset. All time points were compared between patient groups (no statistically significant differences found) and to corresponding pre-ART measurements within each patient group: red asterisks for IRIS; and blue asterisks, non-IRIS (indicated below graphs). *P < .01; **P < .001.
The magnitude of HIV-1 Gag-specific T cells does not differ significantly between IRIS and non-IRIS patients. The total response magnitude, measured by production of IFN-γ and/or IL-2 and/or TNF, generated by HIV-1 Gag reactive CD4+ (A) and CD8+ T cells (B) of IRIS and non-IRIS patients were compared at the 5 analysis time points. Graphs represent interquartile ranges, median bars, as well as individual data points. Dashed lines separate pre-ART from on-ART samples. Gray boxes indicate samples from IRIS patients within 3 months of clinical IRIS onset. All time points were compared between patient groups (no statistically significant differences found) and to corresponding pre-ART measurements within each patient group: red asterisks for IRIS; and blue asterisks, non-IRIS (indicated below graphs). *P < .01; **P < .001.
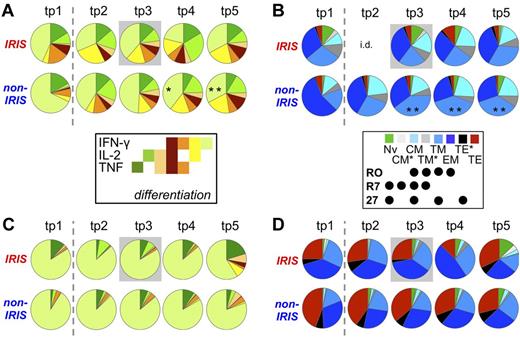
Cytokine pattern and phenotype of HIV-1 Gag-specific T cells do not differ significantly between patient groups. HIV-1 Gag-reactive CD4+ (A-B) and CD8+ T cells (C-D) of IRIS and non-IRIS patients were compared at the 5 analysis time points. (A,C) Cytokine pattern. Relative proportion of total HIV-1 Gag-reactive cells producing each possible combination of the cytokines measured. (B,D) Differentiation state. Dashed lines separate pre-ART from on-ART samples. Gray boxes indicate samples from IRIS patients within 3 months of clinical IRIS onset. All time points were compared between patient groups and with corresponding pre-ART measurements within each patient group (no statistically significant differences found). Individual pie segments were also compared between time points within each patient group (indicated within relevant pie segments): *P < .01; **P < .001. i.d. indicates insufficient data/(not enough samples met inclusion criteria).
Cytokine pattern and phenotype of HIV-1 Gag-specific T cells do not differ significantly between patient groups. HIV-1 Gag-reactive CD4+ (A-B) and CD8+ T cells (C-D) of IRIS and non-IRIS patients were compared at the 5 analysis time points. (A,C) Cytokine pattern. Relative proportion of total HIV-1 Gag-reactive cells producing each possible combination of the cytokines measured. (B,D) Differentiation state. Dashed lines separate pre-ART from on-ART samples. Gray boxes indicate samples from IRIS patients within 3 months of clinical IRIS onset. All time points were compared between patient groups and with corresponding pre-ART measurements within each patient group (no statistically significant differences found). Individual pie segments were also compared between time points within each patient group (indicated within relevant pie segments): *P < .01; **P < .001. i.d. indicates insufficient data/(not enough samples met inclusion criteria).
Elevated CD4+ T-cell responses to relevant Ags in patients with mycobacterial- or fungal-associated IRIS events
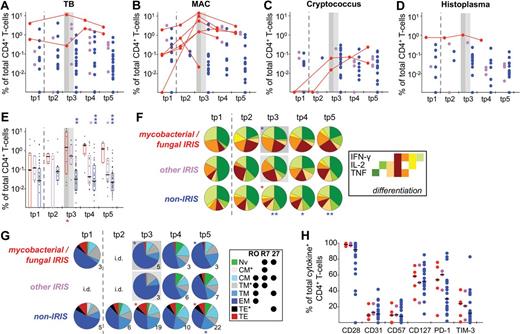
To obtain sufficient patient numbers to perform statistical analyses, the results of stimulations with mycobacterial (MAC, TB) or fungal (Cryptococcus, Histoplasma) Ags were grouped together, as similar trends were observed when analyzing the Ags separately (Figure 4A-D). The magnitude of the CD4+ T-cell response was significantly elevated in IRIS patients experiencing episodes associated with mycobacterial or fungal infections compared with samples from non-IRIS patients, starting at clinical IRIS diagnosis and lasting throughout follow-up (Figure 4E). Overall, the magnitude of these responses was significantly increased during IRIS (tp3) compared with pre-ART (tp1) in most IRIS patients. Patients with “other IRIS” events included patients with MAC-associated IRIS whose PBMCs were also tested for reactivity to TB, which explains the slight increase in CD4+ T-cell responses at tp3 as PBMC from MAC+ persons often show some measure of cross-reactivity to the TB Ag preparation (PPD) used. Nevertheless, CD4+ T-cell responses to the IRIS-associated Ag were significantly higher at later time points (tp4-tp5; Figure 4E) compared with nonassociated mycobacterial or fungal Ags (other IRIS). The change from baseline (tp1) to tp3 was significantly different between patients with mycobacterial or fungal IRIS and non-IRIS patients (P = .001), as well as other IRIS patients (P = .012). Differences in changes from baseline to tp4 (P = .002) and tp5 (P = .009) were also found to be statistically significant between IRIS and non-IRIS patients.
Elevated CD4+ T-cell responses to relevant Ags in patients with mycobacterial- or fungal-associated IRIS events. CD4+ T-cell responses to TB, MAC, C neoformans, and H capsulatum were analyzed. For this purpose, patients were divided into 3 groups: those with IRIS events to mycobacterial- or fungal-associated OI and stimulated with the relevant Ag (mycobacterial/fungal-associated IRIS; red), those with IRIS events to other Ags, which can include mycobacterial/fungal Ags, but stimulated here with IRIS-irrelevant Ags (other IRIS; pink), and non-IRIS patients (non-IRIS; blue). Each data point represents stimulation with one Ag only. Shown are the response magnitudes by Ag: TB (A), MAC (B), C neoformans (C), and H capsulatum (D), as well as grouped for all 4 Ags (E). (A-D) Longitudinal data points are only connected for IRIS patients with IRIS manifestations associated with the given Ag. Cytokine pattern (F), T-cell subset distribution (G), and activation phenotype (H; tp3 only) of CD4+ T cells reactive to TB, MAC, C neoformans, or H capsulatum were also determined. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. All groups were compared within time points with the mycobacterial/fungal-associated IRIS group (color-coded asterisks above bars or pies), as well as with corresponding pre-ART measurements within groups: red for IRIS; and blue asterisks, non-IRIS (indicated below graphs or pies). (G) The number of samples per pie are indicated (see “Data analysis”; these numbers also apply to data in panel H. i.d. indicates insufficient data. *P < .05; **P < .01; ***P < .001).
Elevated CD4+ T-cell responses to relevant Ags in patients with mycobacterial- or fungal-associated IRIS events. CD4+ T-cell responses to TB, MAC, C neoformans, and H capsulatum were analyzed. For this purpose, patients were divided into 3 groups: those with IRIS events to mycobacterial- or fungal-associated OI and stimulated with the relevant Ag (mycobacterial/fungal-associated IRIS; red), those with IRIS events to other Ags, which can include mycobacterial/fungal Ags, but stimulated here with IRIS-irrelevant Ags (other IRIS; pink), and non-IRIS patients (non-IRIS; blue). Each data point represents stimulation with one Ag only. Shown are the response magnitudes by Ag: TB (A), MAC (B), C neoformans (C), and H capsulatum (D), as well as grouped for all 4 Ags (E). (A-D) Longitudinal data points are only connected for IRIS patients with IRIS manifestations associated with the given Ag. Cytokine pattern (F), T-cell subset distribution (G), and activation phenotype (H; tp3 only) of CD4+ T cells reactive to TB, MAC, C neoformans, or H capsulatum were also determined. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. All groups were compared within time points with the mycobacterial/fungal-associated IRIS group (color-coded asterisks above bars or pies), as well as with corresponding pre-ART measurements within groups: red for IRIS; and blue asterisks, non-IRIS (indicated below graphs or pies). (G) The number of samples per pie are indicated (see “Data analysis”; these numbers also apply to data in panel H. i.d. indicates insufficient data. *P < .05; **P < .01; ***P < .001).
Within patients with TB-associated IRIS, one patient exhibited elevated IRIS-Ag–specific CD4+ T-cell responses at the sampling time point closest after clinical IRIS diagnosis (tp3), whereas in the other one the response was similar compared with pre-ART, rising again at a later time point (Figure 4A). This could be the result of the timing of PBMC sampling (23 vs 6 days after the onset of IRIS event), which could be indicative of variation of biologic phenomena or tissue redistribution of TB-specific CD4+ T cells.
The patient with Histoplasma-associated IRIS experienced concomitantly a MAC-IRIS event (cervical lymphadenopathy), and we cannot exclude that the IRIS occurrence was mainly driven by MAC. However, because fungal elements were detected in the biopsy material, this IRIS event had been formally classified as being driven by both OIs. In this patient (patient 56), the IRIS-associated Histoplasma-specific response demonstrated but a 1.3-fold increase from pre-ART (tp1), whereas the MAC-associated response increased 14.8-fold in the same time span (Figures 4D and 5A).
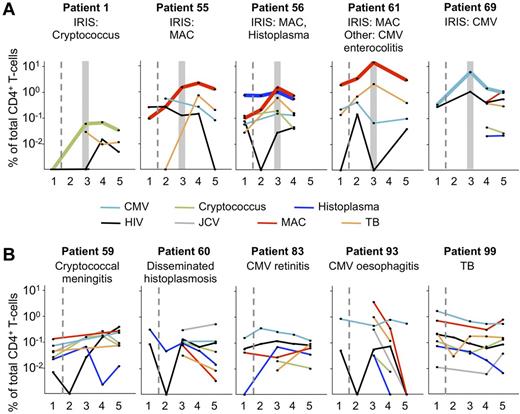
During ART, only frequencies of CD4+ cytokine-producing T cells specific to IRIS-associated Ags increase dramatically. PBMC samples from 5 IRIS patients (A) and 5 non-IRIS patients (B) were stimulated with MAC, H capsulatum, C neoformans, TB, CMV, JCV, or HIV-1. The IRIS-associated opportunistic infection and other known infecting pathogens tested for in the present assays are indicated for each patient. The number of stimulations performed with each PBMC sample was determined by the number of cells available, and priorities were given to those Ags to which a given patient was known to have been exposed. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. Bold lines indicate T-cell responses to IRIS-associated Ags. IRIS patients were selected for illustration if stimulation data were available for at least 2 Ags and at least 4 time points. If 2 Ags fulfilled these criteria, all other stimulations with at least 2 data points were shown for that patient. Non-IRIS patients were selected according to the aforementioned criteria, as well as having known to be exposed to at least one of the Ags being tested: 1 indicates tp1; 2, tp2; 3, tp3; 4, tp4; and 5, tp5.
During ART, only frequencies of CD4+ cytokine-producing T cells specific to IRIS-associated Ags increase dramatically. PBMC samples from 5 IRIS patients (A) and 5 non-IRIS patients (B) were stimulated with MAC, H capsulatum, C neoformans, TB, CMV, JCV, or HIV-1. The IRIS-associated opportunistic infection and other known infecting pathogens tested for in the present assays are indicated for each patient. The number of stimulations performed with each PBMC sample was determined by the number of cells available, and priorities were given to those Ags to which a given patient was known to have been exposed. Gray boxes indicate the first time point within 3 months of clinical manifestation of IRIS. Dashed lines separate pre-ART from on-ART samples. Bold lines indicate T-cell responses to IRIS-associated Ags. IRIS patients were selected for illustration if stimulation data were available for at least 2 Ags and at least 4 time points. If 2 Ags fulfilled these criteria, all other stimulations with at least 2 data points were shown for that patient. Non-IRIS patients were selected according to the aforementioned criteria, as well as having known to be exposed to at least one of the Ags being tested: 1 indicates tp1; 2, tp2; 3, tp3; 4, tp4; and 5, tp5.
IRIS patients also demonstrated a significantly higher fraction of polyfunctional IFN-γ+ cells during the IRIS episode than non-IRIS patients at a comparable sampling time point (Figure 4F), as well as a larger proportion of TEM cells (Figure 4G) among CD4+ T cells reactive to the IRIS-associated Ag. However, no marked differences were observed in their activation phenotypes (Figure 4H).
Unusually vigorous CD4+ T-cell responses to the IRIS-associated Ag during IRIS episodes
When we examined the evolution of CD4+ T-cell responses to different Ags in individual IRIS patients, we found dramatic increases only to the IRIS-associated Ag (with the exception of TB in MAC-IRIS patients because of the poor specificity of PPD; Figure 5A).35 A comparable analysis of non-IRIS patients did not reveal any significant changes in CD4+ T-cell responses over time, including pathogens with which the subjects were known to have been infected (Figure 5B).
CD8+ T-cell responses to the relevant Ag of IRIS patients with mycobacterial- or fungal-associated IRIS episodes did not show corresponding dramatic increases (supplemental Figure 2A-D) as observed in the CD4+ subset (supplemental Figure 1A-D), but that could partially be the result of the predominant use of proteins as stimulants. Interestingly, though, even in a case where IRIS was linked to CMV, a virus inducing both robust CD4+ and CD8+ T-cell responses,36 CMV-specific CD4+ T cells were substantially boosted in frequency (supplemental Figure 2A), whereas CMV-specific CD8+ T cells showed only a moderate increase during the IRIS episode (supplemental Figure 2B), even though a peptide pool was used for stimulation. Notably, both CD4+37 and CD8+38 T-cell responses are thought to be pivotal in controlling JCV. The patient with JCV-associated unmasking IRIS did not produce any detectable JCV-specific CD4+ T-cell responses before the IRIS episode; proximally to the IRIS event, more than 2% of total CD4+ T cells were JCV-specific (supplemental Figure 2C open symbol at tp4: 2.3 months after clinical IRIS diagnosis). JCV-specific CD8+ T-cell responses demonstrated a vigorous increase directly after commencing ART that dampened early during the IRIS event and increased again thereafter (supplemental Figure 2D). This indicates an involvement of CD8+ T cells in the pathogenesis of progressive multifocal encephalopathy-IRIS, although the increase in the CD8+ T-cell response during the IRIS event (tp3 to tp4) was lower in magnitude (2.1-fold) than the corresponding delayed CD4+ T-cell response (−0.3% to 2.1%). These data underscore a possible significant involvement of CD4+ T cells in IRIS pathogenesis in some cases linked to viral pathogens.
Other IRIS-associated OI-specific T-cell responses were investigated (HBV, HHV-8, HPV, Strongyloides, VZV) but did not yield conclusive data.
Finally, proportionately more IRIS patients (26%) received glucocorticosteroid treatment directly before ART or within the first 6 months of ART compared with patients with uneventful immunoreconstitution (6.3%; P = .0223). Such treatment could reduce the overall magnitude of cytokine responses measured. Notably, no significant changes were observed in HIV-1–specific CD8+ T-cell responses, whereas CD4+ T-cell responses were only moderately reduced in glucocorticosteroid-treated patients after 3 months (P = .013) and 12 months (P = .035) of ART. This indicates that the differences observed between IRIS and non-IRIS patients were not the result of glucocorticosteroid treatment.
Discussion
In this report, we show that, in the majority of cases, IRIS events are characterized by the expansion of highly differentiated, polyfunctional CD4+ T-cell responses directed against the underlying IRIS-associated infection. This exuberant CD4+ T-cell response is restricted to the OI-specific compartment, as T-cell responses directed against HIV-1 or other co-infections did not differ between IRIS and non-IRIS patients. Thus, IRIS does not appear to either be the result of, or result in, a global T-cell defect.
Early reports focusing on viral infections correlated the development of IRIS to the frequency or cell count of CD8+ cells.39,40 However, in our study, the CD4+ T-cell response to the IRIS-associated Ag was significantly boosted during the IRIS event, whereas CD8+ T-cell responses were only moderately affected, if at all, suggesting that IRIS occurs primarily because of a hyperactivation of CD4+ T cells. This was true even for CMV, to which strong CD8+ T-cell responses are typically observed: during CMV-associated unmasking IRIS, a far more dramatic amplification occurred in the CD4+ T-cell compartment.
In previous studies, IRIS was linked to increased IFN-γ+,10,11,19,41 IL-2+,11 and TNF+19 T-cell responses, especially in the case of TB-associated IRIS. Because some of these studies have been based on total PBMC ELISpot data, the association might have been underestimated, as CD4+ T cells appear to be the main players in the immunopathology of IRIS. In agreement with previous studies,10,11,19 we observed an exuberant CD4+ T-cell response to the IRIS-associated Ag. In the case of mycobacterial and fungal Ags, the response was significantly more elevated than in non-IRIS patients or in persons experiencing IRIS events related to other causative agents. The most important observation in terms of cytokine-producing CD4+ T cells appears to be the change in IFN-γ+ cells from baseline to the IRIS time point: a significant increase was detected in mycobacterial/fungal IRIS patients, resulting in increased proportions of polyfunctional cells (IFN-γ+ IL-2+ TNF+ and IFN-γ+ IL-2− TNF+), whereas in non-IRIS patients the proportion of IFN-γ+ (mainly single positive) cells decreased during the same time frame. Furthermore, as previously reported for TB-IRIS patients,19 we found that the frequency of TEM cells among CD4+ T cells reactive to the IRIS-associated OI was dramatically increased at the time of IRIS. Thus, the functional and phenotypic analyses support the conclusion that highly activated cells (polyfunctional, late differentiation stage) are enriched in IRIS patients' CD4+ T cells specific to the IRIS-OI-relevant Ag during clinical episodes. This was particularly evident in cases of mycobacterial and cryptococcal IRIS. However, no predictive alteration in Ag-specific T-cell responses was identified at baseline, and there was no apparent (antigen-specific or general) T-cell defect before IRIS.
The dysregulated inflammatory immune responses occurring during IRIS have been hypothesized to stem from a combination of poor clearance of OI-related Ags because of HIV-1 co-infection14 and a reversal of the HIV-1–induced CD4+ T-cell suppression when commencing ART.42 In this context, TB-specific CD4+ T-cell responses rapidly decrease during HIV-1 infection,43 which could contribute to the lack of Ag clearance. Furthermore, progressive multifocal encephalopathy was shown to occur in HIV-1− persons after withdrawal of unrelated immunosuppressive treatment.44 This as well as other examples of paradoxical inflammatory reactions45 demonstrate that IRIS-like phenomena can occur independently of lymphopenia and may instead be solely driven by changes in the activation status of T cells and their abrupt recovery from immunosuppression in the presence of accumulated residual Ag of a partially treated (paradoxical) or untreated (unmasking) infection.
We did not identify any phenotypic alterations at the time of IRIS in either total or Ag-specific (to HIV-1 or the IRIS-associated OI) CD4+ T cells. In contrast, total CD8+ T cells of IRIS patients demonstrated delayed recovery of CD28 and CD127 expression, which is probably a result, rather than a cause, of the inflammatory response occurring during IRIS. However, PD-1 expression was elevated before ART in IRIS patients both on CD4+ and CD8+ T cells, and many of those cells expressed the costimulatory ICOS and inhibitory CTLA-4 and LAG-3 molecules.23 Together, these data are consistent with a high level of activation and a potentially reduced functionality of CD4+ T cells in vivo before ART in IRIS patients.
Although altered levels of regulatory T cells have not been reported in IRIS patients,12,22,23 it has been proposed that the impairment of regulatory T cells to suppress proliferation of responder T cells reported during IRIS events could at least in part explain the exuberant inflammatory T-cell responses observed in IRIS patients.22 However, our finding that T-cell responses against other pathogens than the IRIS-associated one were not significantly altered is inconsistent with this hypothesis.
In conclusion, our data suggest a common mechanism for the development of many IRIS occurrences that is independent of the associated opportunistic infection, whereby rapidly expanded Ag-specific CD4+ T cells mount a polyfunctional inflammatory response to residual Ag of previously existing and inadequately controlled opportunistic infections once the HIV-1–induced functional inhibition is reversed by ART.
Presented in part in abstract format at the Conference on Retroviruses and Opportunistic Infections, San Francisco, CA, February 2010.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Joanne Yu for Ab conjugations and titrations, Jessica Hodge for assistance in panel development, Catherine Rehm and JoAnn Mican for help with PBMC samples, Brian O. Porter for providing clinical information, and Drs Joe Casazza, Guislaine Carcelain, Gene Major, Molly Perkins, Daniel Douek, Stuart Levitz, George Deepe, and David Abraham for their generosity in sharing their reagents.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: Y.D.M., L.R.V.A., A.S., M.R., and I.S. developed the study concept and experimental design; G.R. and I.S. were responsible for clinical supervision of patients and PBMC sampling; Y.D.M. and J.H.G. performed experiments and data analyses; Y.D.M. and R.D. performed statistical analyses; Y.D.M. and I.S. wrote the manuscript; and all authors were involved in manuscript review and editing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.R.V.A. is Laboratory of Immunopathology, René Rachou Research Center, Fiocruz, Belo Horizonte, Brazil.
Correspondence: Irini Sereti, Bldg 10, Magnuson Clinical Center, Rm11B07A, 10 Center Dr, Bethesda, MD 20892; e-mail: isereti@niaid.nih.gov.