Data on minimal residual disease (MRD) monitoring in acute promyelocytic leukemia (APL) are available only in the context of conventional all-trans retinoic acid plus chemotherapy regimens. It is recognized that the kinetics of leukemia clearance is different with the use of arsenic trioxide (ATO) in the treatment of APL. We undertook a prospective peripheral blood RT-PCR–based MRD monitoring study on patients with APL treated with a single agent ATO regimen. A total of 151 patients were enrolled in this study. A positive RT-PCR reading at the end of induction therapy was significantly associated on a multivariate analysis with an increased risk of relapse (relative risk = 4.9; P = .034). None of the good risk patients who were RT-PCR negative at the end of induction relapsed. The majority of the relapses (91%) happened within 3 years of completion of treatment. After achievement of molecular remission, the current MRD monitoring strategy was able to predict relapse in 60% of cases with an overall sensitivity and specificity of 60% and 93.2%, respectively. High-risk group patients and those that remain RT-PCR positive at the end of induction are likely to benefit from serial MRD monitoring by RT-PCR for a period of 3 years from completion of therapy.
Introduction
Treatment of acute promyelocytic leukemia (APL) with a conventional all-trans retinoic acid (ATRA) combined with chemotherapy has resulted in significant improvements in the clinical outcome.1 Major causes of failure with a conventional ATRA plus chemotherapy schedule includes early mortality (10%-30%) related to a hemorrhagic diathesis2 and subsequent relapse (10%-30% each).3,4 Monitoring minimal residual disease (MRD) by detection of the PML-RARA fusion transcript, in the peripheral blood or bone marrow, using a conventional RT-PCR has been shown to be an effective strategy to predict relapse.5,6 Further, it has been demonstrated that early intervention at the time of molecular relapse was associated with a significantly superior survival advantage compared with initiation of treatment at the time of frank hematologic relapse.7,8
Arsenic trioxide (ATO) has been successfully introduced in the treatment of newly diagnosed APL, both as a single agent and in combination with conventional agents.4,9,,–12 In addition, a number of cooperative groups have ongoing clinical trials that have incorporated ATO in upfront therapy.13 It is well recognized that the kinetics of leukemia clearance with the use of ATO, in induction, is significantly different from that of ATRA alone or ATRA plus chemotherapy combinations.1 Extrapolation of data generated from ATRA plus chemotherapy regimens may potentially not be valid when applied to regimens that use ATO in upfront therapy. At our center, we undertook a prospective MRD detection study in patients treated with a single-agent ATO-based regimen as reported by us previously.9
Methods
Patients and samples
From March 2000 to April 2010, newly diagnosed APL patients treated with a single-agent ATO regimen (as reported previously9,14 ) were enrolled for prospective MRD detection at our center, after getting written and informed consent. For enrollment, it was mandatory that, in addition to conventional diagnostic criteria, the patient should have had a positive RT-PCR for the PML-RARA fusion transcripts at diagnosis.
Sample time points for MRD monitoring
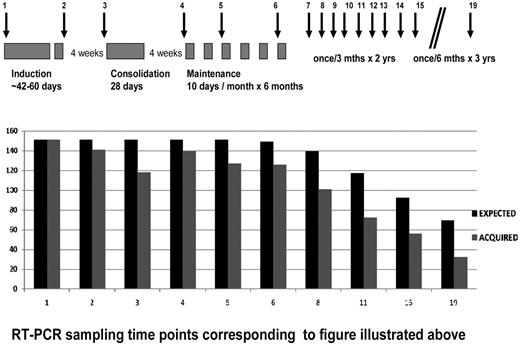
Peripheral blood samples were to be collected for RT-PCR at diagnosis, at the end of induction, beginning of consolidation, every 3 months during maintenance. After completion of maintenance therapy, peripheral blood samples were scheduled to be collected once in 3 months for the first 2 years and once in 6 months for the next 3 years (see Figure 1). RT-PCR analysis was done prospectively for all cases enrolled in this study.
We retrospectively checked the PML-RARA transcript levels by quantitative RT-PCR (RQ-PCR) on samples at molecular relapse and one sample point before the molecular relapse/frank hematologic relapse (within 3-4 months before documented relapse). The values generated in these cases were compared with twice the number of age and white blood cell (WBC) count at presentation matched controls that were greater than 2 years from completion of therapy and remained in continuous complete hematologic remission (CHR) and molecular remission (CRm) by RT-PCR.
Amplification of PML-RARA fusion transcripts
The PML-RARA fusion gene transcripts bcr1, bcr2, and bcr3 types were amplified by a 2-step (nested) qualitative RT-PCR. This was done using the recommendations of BIOMED-1 Concerted Action (supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).15 Quantification of the PML-RARA transcripts was done using Europe Against Cancer program protocols.16 The assays were run on an ABI PRISM 7000 DNA Sequence Detection System. Fusion transcripts were normalized with ABL17 and ΔCt method was used to generate normalized copy number using in-house plasmid standards for the bcr1 and bcr3 transcripts and commercial plasmids standards (Ipsogen) for the bcr2 transcript (supplemental Methods).15 All samples were tested in replicates, and the results were reported according to the Europe Against Cancer guidelines as the normalized copy number, derived by multiplying the PML-RARA copy number/ABL copy number ratio by 100. The RT-PCR and RQ-PCR sensitivity was assessed in-house using methodology as reported previously (supplemental Methods).18 Any value for PML-RARA copy number derived beyond a Ct of 40 was considered negative.16 A negative and a positive control were included in each run. Results were excluded if the Ct of ABL was more than 30. Samples with SD of less than 0.5 between duplicates were only included in the analysis (test was repeated if SD > 0.5) to exclude the variations because of technical errors.
Cytogenetic analysis and determination of FLT3 mutation status
The details of the methods and reporting criteria for cytogenetic analysis and the methods for molecular detection of FLT3 mutations are given in supplemental Methods.
Definition of outcomes
Definition of CHR, overall survival, event-free survival, CRm, and molecular relapse was as reported earlier by us for this regimen.14 Relapse-free survival (RFS) was calculated from time of achieving CR to the patient's last follow-up or an event defined as hematologic relapse only.
Statistical methods
The χ2 or Fisher exact test and Mann-Whitney U test were used to compare differences between groups for response to therapy. The probability of survival was estimated with the use of the product-limit method of Kaplan-Meier for overall survival, RFS, and cumulative incidence of relapse and was compared by the log-rank test. All survival estimates are reported ± SE. The relationships of clinical features to outcome were analyzed by Cox proportional hazard model. All P values were 2-sided, with values of .05 or less indicating statistical significance. Statistical analysis used the SPSS Version 16.0 software.
Results
Patient accrual and baseline characteristics
Between March 2000 and April 2010, 173 patients with PML-RARA+ APL were eligible to be enrolled for this study. Patients who died before starting treatment (n = 1), during induction (n = 18), those who were discharged against medical advice (n = 2), or those who were primary refractory (n = 1) were excluded from this study. The remaining 151 patients were enrolled after getting written and informed consent. The baseline characteristics and their impact on RFS of the patients enrolled are summarized in Table 1.
Baseline characteristics of patients and their impact on RFS
| Parameter . | N (%)/median (range) . | Impact on RFS (P) . |
|---|---|---|
| Age, y | ||
| < 10 | 18 (11.9) | |
| 11-15 | 15 (9.9) | NS |
| 16-55 | 111 (73.5) | |
| > 55 | 7 (4.6) | |
| Sex: male | 79 (52.3) | NS |
| Hemoglobin, × 10 g/Lt | 8.6 (2.1-16) | NS |
| WBC | ||
| < 5 × 109/Lt | 78 (51.7) | NS |
| ≥ 5 × 109/Lt | 73 (48.3) | |
| < 10 × 109/Lt | 98 (64.9) | NS |
| ≥ 10 × 109/Lt | 53 (35.1) | |
| Platelets | NS | |
| ≤ 20 × 109/Lt | 86 (57.0) | |
| > 20 × 109/Lt | 65 (43.0) | |
| Good-risk group* | 41 (27.2) | NS |
| High-risk group | 110 (72.8) | |
| RT-PCR | ||
| bcr1 | 92 (61.3) | |
| bcr2 | 9 (6.0) | NS |
| bcr3 | 49 (32.7) | |
| Lactate dehydrogenase, IU/Lt | 527 (75-2700) | NS |
| Serum creatinine, mg/100 mL | 0.8 (0.1-1.6) | NS |
| Additional CTG finding | ||
| Yes | 42 (31.1) | .001 |
| No | 93 (68.9) | |
| FLT3 mutation (ITD/TKD) | ||
| Yes | 36 (26.5) | NS |
| No | 100 (73.5) |
| Parameter . | N (%)/median (range) . | Impact on RFS (P) . |
|---|---|---|
| Age, y | ||
| < 10 | 18 (11.9) | |
| 11-15 | 15 (9.9) | NS |
| 16-55 | 111 (73.5) | |
| > 55 | 7 (4.6) | |
| Sex: male | 79 (52.3) | NS |
| Hemoglobin, × 10 g/Lt | 8.6 (2.1-16) | NS |
| WBC | ||
| < 5 × 109/Lt | 78 (51.7) | NS |
| ≥ 5 × 109/Lt | 73 (48.3) | |
| < 10 × 109/Lt | 98 (64.9) | NS |
| ≥ 10 × 109/Lt | 53 (35.1) | |
| Platelets | NS | |
| ≤ 20 × 109/Lt | 86 (57.0) | |
| > 20 × 109/Lt | 65 (43.0) | |
| Good-risk group* | 41 (27.2) | NS |
| High-risk group | 110 (72.8) | |
| RT-PCR | ||
| bcr1 | 92 (61.3) | |
| bcr2 | 9 (6.0) | NS |
| bcr3 | 49 (32.7) | |
| Lactate dehydrogenase, IU/Lt | 527 (75-2700) | NS |
| Serum creatinine, mg/100 mL | 0.8 (0.1-1.6) | NS |
| Additional CTG finding | ||
| Yes | 42 (31.1) | .001 |
| No | 93 (68.9) | |
| FLT3 mutation (ITD/TKD) | ||
| Yes | 36 (26.5) | NS |
| No | 100 (73.5) |
NS indicates not significant.
Defined as WBC < 5 × 109/Lt and platelets > 20 × 109/Lt.
Peripheral blood samples for MRD detection
RT-PCR was done prospectively on all patients. A total 1764 peripheral blood samples was collected from 151 patients (median, 12 patients; range, 5-19 patients) from a possible maximum of 2348, based on the planned schedule for MRD monitoring, for an overall compliance rate of 75.12%. Figure 1 summarizes the RT-PCR collection time points in relationship to the protocol used (as reported previously9,14 ) and the compliance rate at each of these time points.
Time points for evaluation of RT-PCR and compliance at each time point.
Response and relapses after therapy
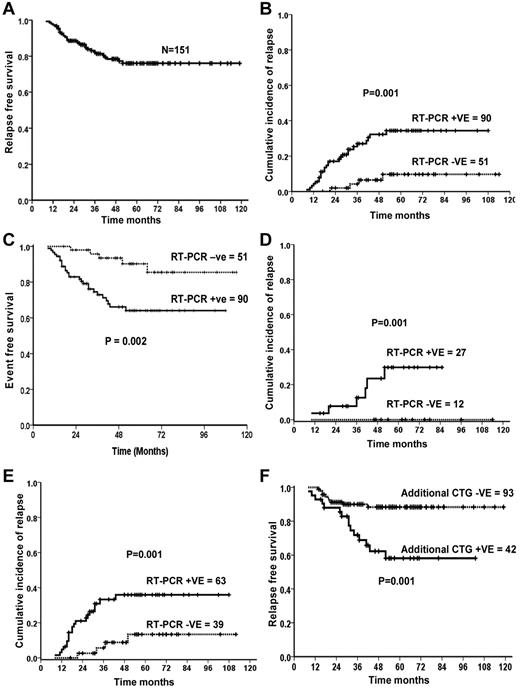
By definition, for this analysis, only patients who were documented to have achieved CHR were enrolled for the MRD monitoring part of the study. Among the 151 patients who were enrolled, CHR was attained at a median of 44 days (range, 24-99 days). All patients who achieved CHR went on to achieve a CRm before the start of maintenance therapy. Thirty-one patients (20.5%) had documented relapses (Figure 2A). Medullary relapse was noted in 30, with additional CNS disease in 3 at the time of relapse, and in one case there was an isolated CNS relapse. The median time to relapse was 15 months after completion of therapy (range, 0-41 months). Only 3 relapses happened beyond 3 years after completion of therapy. At a median follow-up of 50 months, the 5 year Kaplan-Meier estimate of overall survival and RFS of this cohort was 87.9% ± 3.1% and 76% ± 3.9%, respectively (Figure 2A).
Impact of additional karyotypic markers at diagnosis and after induction RT-PCR positively on relapse free survival. (A) RFS in entire cohort. (B) CIR of patients who were RT-PCR-positive or -negative at the end of induction. (C) Event-free survival of patients who were RT-PCR–positive or –negative at the end of induction. (D) CIR among good risk cases (WBC < 5 × 109/Lt and platelet count > 20 × 109/Lt at diagnosis) of those who were RT-PCR–positive or –negative at the end of induction. (E) CIR among high-risk group (not fulfilling good risk criteria) who were RT-PCR–positive or –negative at the end of induction. (F) RFS among those who had and those who did not have an additional cytogenetic finding at diagnosis.
Impact of additional karyotypic markers at diagnosis and after induction RT-PCR positively on relapse free survival. (A) RFS in entire cohort. (B) CIR of patients who were RT-PCR-positive or -negative at the end of induction. (C) Event-free survival of patients who were RT-PCR–positive or –negative at the end of induction. (D) CIR among good risk cases (WBC < 5 × 109/Lt and platelet count > 20 × 109/Lt at diagnosis) of those who were RT-PCR–positive or –negative at the end of induction. (E) CIR among high-risk group (not fulfilling good risk criteria) who were RT-PCR–positive or –negative at the end of induction. (F) RFS among those who had and those who did not have an additional cytogenetic finding at diagnosis.
Kinetics of PML-RARA clearance with an ATO-based induction regimen by RT-PCR
The RT-PCR done at the end of induction, beginning of consolidation, and at the beginning of the maintenance was assessed for evaluating the clearance of PML-RARA+ transcripts and its effect on the probability of relapse in an ATO-based regimen. The sensitivity of the qualitative RT-PCR assay, as assessed in-house, was 10−3 for the first round and 10−4 for the second round (supplemental Methods). All 151 cases (100%) were positive for PML-RARA at diagnosis. At the end of induction, the RT-PCR was positive in 90 of the 141 evaluated (63.8%) patients. By the beginning of consolidation, 23 of the 118 evaluated (19.5%) remained positive, whereas before first maintenance all 136 cases that were evaluated were negative.
Prediction of relapse based on RT-PCR clearance after initiation of therapy
At the end of induction, 90 patients (63.82%) remained RT-PCR-positive. These patients on a univariate analysis had an increased risk of relapse compared with those that were negative at this time point (relative risk = 4.8; 95% CI = 1.66-13.67; P = .004). On a step-wise forward multivariate analysis after adjusting for parameters that on a univariate analysis had a significant or trend to significance for increased risk of relapse or were well-established risk factors for relapse (Table 2), only the RT-PCR positivity at the end of induction retained its statistical significance (relative risk = 4.9; 95% CI = 1.13-21.20; P = .034). The sensitivity and specificity of a positive RT-PCR to predict relapse after induction were 86.7% and 42.3%, respectively. The cumulative incidence of relapse (CIR) and the 5-year Kaplan-Meier event-free survival was significantly superior for those who were RT-PCR–negative at this time point as illustrated in Figure 2B and C, respectively.
Univariate and multivariate analysis of parameters impacting relapse
| Variable . | Univariate analysis . | Multivariate analysis* . | ||||
|---|---|---|---|---|---|---|
| RR . | 95% Cl . | P . | RR . | 95% CI . | P . | |
| Age | 0.98 | 0.96-1.00 | NS | NA | NA | .388 |
| Hemoglobin | 0.97 | 0.85-1.10 | NS | NA | NA | .786 |
| WBC, × 109/Lt | ||||||
| > 5 | 1.24 | 0.61-2.52 | NS | NA | NA | .776 |
| < 5 | ||||||
| Platelet, × 109/Lt | ||||||
| < 20 | 0.99 | 0.49-2.03 | NS | NA | NA | .243 |
| > 20 | ||||||
| RT-PCR positive at end of induction | 4.76 | 1.66-13.67 | 0.004 | 4.89 | 1.13-21.19 | .034 |
| RT-PCR positive before consolidation | 2.29 | 0.99-5.32 | NS | NA | NA | .257 |
| Additional cytogenetics | 3.58 | 1.61-7.98 | .002 | NA | NA | .063 |
| Variable . | Univariate analysis . | Multivariate analysis* . | ||||
|---|---|---|---|---|---|---|
| RR . | 95% Cl . | P . | RR . | 95% CI . | P . | |
| Age | 0.98 | 0.96-1.00 | NS | NA | NA | .388 |
| Hemoglobin | 0.97 | 0.85-1.10 | NS | NA | NA | .786 |
| WBC, × 109/Lt | ||||||
| > 5 | 1.24 | 0.61-2.52 | NS | NA | NA | .776 |
| < 5 | ||||||
| Platelet, × 109/Lt | ||||||
| < 20 | 0.99 | 0.49-2.03 | NS | NA | NA | .243 |
| > 20 | ||||||
| RT-PCR positive at end of induction | 4.76 | 1.66-13.67 | 0.004 | 4.89 | 1.13-21.19 | .034 |
| RT-PCR positive before consolidation | 2.29 | 0.99-5.32 | NS | NA | NA | .257 |
| Additional cytogenetics | 3.58 | 1.61-7.98 | .002 | NA | NA | .063 |
RR indicates relative risk; NS, not significant; and NA, not applicable.
Forward step-wise multivariate analysis.
A similar analysis was done at the beginning of consolidation where 23 of 118 patients evaluated (19.5%) were still RT-PCR–positive, and this result did not have a significant impact on the risk of relapse.
The impact of additional cytogenetic findings, presence of FLT3 mutations, age, PML-RARA isoforms, WBC, and platelet counts at diagnosis and risk group stratification on the PML-RARA clearance after initiation of induction therapy and risk of relapse
The detailed results of the impact of an additional cytogenetic finding and presence of FLT3 mutations on molecular clearance and their impact on relapse are given in supplemental Methods. Figure 2F illustrates the impact of an additional cytogenetic finding on RFS. The detailed results of the impact of age, PML-RARA isoforms, and risk groups are given in supplemental Methods.
Among the good-risk group (good risk = WBC ≤ 5 × 109/Lt and ≥ 20 × 109/Lt; vs high-risk group = remaining patients), none of the patients who were RT-PCR–negative at the end of induction relapsed, whereas 22% of those who were RT-PCR–positive at the end of induction in this group relapsed (Figure 2D). Among the remaining patients (high-risk group), those who were RT-PCR–negative and –positive at the end of induction, 10% and 32%, respectively, relapsed (Figure 2E).
Prediction of relapse by MRD monitoring after achievement of CRm
Of the 31 hematologic relapses, an RT-PCR sample was positive 3 to 4 months before a frank hematologic relapse in 15 (48.4%) cases, negative in 10 (32.3%), and was not done at this time point in 6 (19.3%) cases. In an additional 8 of 151 (4.6%) cases, the RT-PCR was transiently positive, but these cases did not develop a hematologic relapse at a median follow-up of 49 months (range, 20-87.2 months). Based on these observations, the overall sensitivity, specificity, and positive predictive value of RT-PCR picking up a molecular relapse, by this MRD monitoring strategy, before a hematologic relapse was 60%, 93.2%, and 65.2%, respectively.
Kinetics of PML-RARA clearance by RQ-PCR in an ATO-based regimen
Unlike RT-PCR, the RQ-PCR was not done prospectively. Retrospective RQ-PCR analysis was only done on patients who had relapsed and a cohort of age and WBC count at diagnosis matched cases (controls) that were in continuous complete remission for greater than 2 years from completion of maintenance (n = 28 and 35, respectively; based on availability of banked cDNA). Table 3 compares the baseline characteristics of these 2 groups. Clearance of PML-RARA by RQ-PCR (normalized copy number = 0) at the end of induction, beginning of consolidation, and the beginning of maintenance was assessed for these cases. The sensitivity of the quantitative RQ-PCR assay, as assessed in-house, was 10−4, which was equivalent to a sensitivity of the nested RT-PCR (supplemental Appendix A3). Figure 3 summarizes the clearance of PML-RARA by RQ-PCR from start of therapy until the completion of consolidation treatment and compares it with clearance using RT-PCR.
Comparison of baseline characteristics of patients who had a documented relapse and that of age and WBC at diagnosis-matched controls that remain in CRm (beyond 2 years after completion of therapy)
| Parameter . | Relapsed cases (n = 31), N (%)/median (range) . | Controls (n = 35), N (%)/median (range) . | Impact on RFS (P) . |
|---|---|---|---|
| Age, y | |||
| < 10 | 7 (22.6) | 4 (11.4) | |
| 11-15 | 3 (9.7) | 6 (17.1) | NS |
| 16-55 | 21 (67.7) | 23 (65.7) | |
| > 55 | 0 | 2 (5.7) | |
| Sex: male | 20 (64.5) | 14 (40.0) | NS |
| Hemoglobin, × 10 g/Lt | 8.3 (2.1-16.0) | 8.6 (3.7-15.5) | NS |
| WBC | |||
| < 5 × 109/Lt | 15 (48.4) | 14 (40.0) | NS |
| ≥ 5 × 109/Lt | 16 (51.6) | 21 (60.0) | |
| < 10 × 109/Lt | 18 (58.1) | 21 (60.0) | NS |
| ≥ 10 × 109/Lt | 13 (41.9) | 14 (40.0) | |
| Platelets | |||
| ≤ 20 × 109/Lt | 18 (58.1) | 23 (65.7) | NS |
| > 20 × 109/Lt | 13 (41.9) | 12 (34.3) | |
| Good-risk group* | 6 (19.4) | 8 (22.9) | NS |
| High-risk group | 25 (80.6) | 27 (77.1) | |
| RT-PCR | |||
| bcr1 | 20 (64.5) | 21 (60) | |
| bcr2 | 2 (6.5) | 4 (11.4) | NS |
| bcr3 | 9 (29.0) | 10 (28.6) | |
| Lactate dehydrogenase, IU/Lt | 527 (280-2385) | 564 (226-2200) | NS |
| Serum creatinine, mg/100 mL | 0.9 (0.5-1.2) | 0.8 (0.5-1.6) | NS |
| Additional CTG finding | |||
| Yes | 15 (60.0) | 9 (29.0) | .030 |
| No | 10 (40.0) | 22 (71.0) | |
| FLT3 mutation (ITD/TKD) | |||
| Yes | 10 (35.7) | 11 (34.4) | NS |
| No | 18 (64.3) | 21 (65.6) |
| Parameter . | Relapsed cases (n = 31), N (%)/median (range) . | Controls (n = 35), N (%)/median (range) . | Impact on RFS (P) . |
|---|---|---|---|
| Age, y | |||
| < 10 | 7 (22.6) | 4 (11.4) | |
| 11-15 | 3 (9.7) | 6 (17.1) | NS |
| 16-55 | 21 (67.7) | 23 (65.7) | |
| > 55 | 0 | 2 (5.7) | |
| Sex: male | 20 (64.5) | 14 (40.0) | NS |
| Hemoglobin, × 10 g/Lt | 8.3 (2.1-16.0) | 8.6 (3.7-15.5) | NS |
| WBC | |||
| < 5 × 109/Lt | 15 (48.4) | 14 (40.0) | NS |
| ≥ 5 × 109/Lt | 16 (51.6) | 21 (60.0) | |
| < 10 × 109/Lt | 18 (58.1) | 21 (60.0) | NS |
| ≥ 10 × 109/Lt | 13 (41.9) | 14 (40.0) | |
| Platelets | |||
| ≤ 20 × 109/Lt | 18 (58.1) | 23 (65.7) | NS |
| > 20 × 109/Lt | 13 (41.9) | 12 (34.3) | |
| Good-risk group* | 6 (19.4) | 8 (22.9) | NS |
| High-risk group | 25 (80.6) | 27 (77.1) | |
| RT-PCR | |||
| bcr1 | 20 (64.5) | 21 (60) | |
| bcr2 | 2 (6.5) | 4 (11.4) | NS |
| bcr3 | 9 (29.0) | 10 (28.6) | |
| Lactate dehydrogenase, IU/Lt | 527 (280-2385) | 564 (226-2200) | NS |
| Serum creatinine, mg/100 mL | 0.9 (0.5-1.2) | 0.8 (0.5-1.6) | NS |
| Additional CTG finding | |||
| Yes | 15 (60.0) | 9 (29.0) | .030 |
| No | 10 (40.0) | 22 (71.0) | |
| FLT3 mutation (ITD/TKD) | |||
| Yes | 10 (35.7) | 11 (34.4) | NS |
| No | 18 (64.3) | 21 (65.6) |
NS indicates not significant.
Defined as WBC < 5 × 109/Lt and platelets > 20 × 109/Lt.
Comparison of PML-RARA clearance by RT-PCR and RQ-PCR after initiation of therapy.
Comparison of PML-RARA clearance by RT-PCR and RQ-PCR after initiation of therapy.
From cases and controls mentioned in the preceding paragraph, at the end of induction, 46 of 54 (85.2%) were still positive and the sensitivity, specificity, and positive predictive value of a positive RQ-PCR at this time point to predict relapse would be 89.3%, 19.2%, and 54.3%, respectively.
Role of sequential RQ-PCR monitoring in predicting molecular relapse before hematologic relapse
Retrospectively, RQ-PCR was done, based on availability of banked cDNA, on most hematologic relapse cases (n = 25) on a sample collected in the 3 to 4 months before a documented hematologic relapse. RQ-PCR was also done on most of the set of age and WBC at presentation-matched controls (Table 3) who remained in continuous complete remission for greater than 2 years from completion of maintenance (n = 35).
Compared with RT-PCR, which picked up MRD in 15 of 31 cases before frank relapse, the RQ-PCR would have picked up 21 of 31 relapses; this included all the cases that were positive by RT-PCR. A total of 21 of 31 molecular relapse as detected by RQ-PCR was specific to hematologic relapse, and relapse occurred within a median of 2.5 months (range, 0.9-7.3) months from documentation of a positive RQ-PCR. The median RQ-PCR level for this group was 0.04 (range, 0.001-3.9). The remaining 10 RQ-PCR–positive cases were in the control group, and they all remain in continuous CRm at a median follow-up of 52 months (range, 24-69 months) from the time of a positive reading, and the median RQ-PCR level in this group was 0.072 (range, 0.04-5.69). The median RQ-PCR values between these 2 groups were not significantly different. Based on this analysis, the sensitivity, specificity, and positive predictive value of a positive RQ-PCR to predict a hematologic relapse after achievement of CRm was 84%, 71.4%, and 67.7%, respectively. Among the cases that had hematologic relapses, the median log increase per month was 0.34 (range, 0.12-1.17) as shown in supplemental Figure A5.
Discussion
MRD monitoring of patients with APL with a conventional RT-PCR or RQ-PCR is a well-accepted standard of care and an important component of international guidelines for the management APL.19,20 Most of the available data that go toward making such recommendations come from studies that have used a conventional ATRA plus chemotherapy regimen.5,6,21
In a conventional ATRA plus chemotherapy regimen, the majority of patients remain RT-PCR positive at the end of induction therapy, and a positive RT-PCR at this time point does not predict relapse.5,6 In the MRC study, it was noted that detection of PML-RARA transcripts at the end of course 2 (after induction and one consolidation course) and above predicted risk of subsequent relapse. In this study, using single-agent ATO, it is noted that detection of PML-RARA at the end of the initial induction therapy correlates best with subsequent risk of relapse. This increased risk persisted even on a multivariate analysis after adjusting for all conventional risk factors. Detection of PML-RARA transcripts at subsequent time points during therapy, such as preconsolidation and premaintenance therapy, did not predict an increased risk of relapse. A positive RT-PCR reading after induction, in this study, was more significant than conventional risk factors in predicting relapse. Moving forward with this low-intensity, low-cost regimen, the presence of a positive RT-PCR reading at the end of induction could be used to stratify patients into a more intensive consolidation and maintenance regimen to reduce cumulative incidence of relapse. However, it must be recognized that such an approach would result in more than 60% of patients who are RT-PCR positive after induction being overtreated.
From the data reported here, it would be reasonable in future to consider stopping MRD monitoring for all good-risk group patient who were in CRm at the end of induction therapy, even with this low intensity regimen, although it is important to note that in this series less than 10% of the entire cohort would fall into this group. For those who are RT-PCR positive at the end of induction and for all high risk cases, MRD monitoring should probably be continued for at least 3 years after completion of maintenance therapy.
Disappointingly, MRD monitoring by RT-PCR after achieving CRm at the frequency done in this study had a low sensitivity and positive predictive value. Even after considering the limitations of the retrospective study design to address the role of serial MRD detection by RQ-PCR to predict relapse in this series, it would appear that this strategy would not have significantly improved our ability to predict relapse. As suggested and demonstrated in an earlier study,6 the use of a bone marrow sample and the RARA-PML transcript detection could improve the sensitivity, although the absolute number of patients who would benefit over and above having done a qualitative RT-PCR for PML-RARA transcripts alone is not clear. In our setting (developing country) and we suspect in many other resource-constrained environments and also from a patient's perspective, these are not practical options. In our own analysis, an additional 6 cases would have had a hematologic relapse predicted by the use of an RQ-PCR over that achieved by RT-PCR alone, which would take successful prediction from 10% to 13.9% of the entire cohort. This degree of increment in sensitivity would not make this a cost-effective option for us. Further, we are reassured by our own experience that, even in the event of a frank hematologic relapse, treatment with an ATO-based induction and consolidation followed by an autologous SCT in CRm can achieve a cure in up to 70% of cases.22
In conclusion, MRD detection with an ATO-based regimen demonstrates significant differences in kinetics of leukemia clearance and ability to predict relapse. To improve the sensitivity of MRD detection by peripheral blood monitoring, a practical option that could potentially be considered is to increase the frequency of testing to once in 2 months for the first 2 years after completion of therapy when the majority of hematologic relapses happen. This remains to be validated prospectively.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Department of Biotechnology, New Delhi, India (projects BT/PR4460/MED/14/531/2003 and BT/PR7513/MED/12/287/2006).
Authorship
Contribution: E.C. performed research and molecular tests, analyzed data, and wrote the paper; P.B. performed and standardized molecular tests and analyzed data; B.G., A.V., A.A., and R.A. performed research and clinical data accrual and analyzed data; A.A.A. performed research and flow cytometric analysis for diagnosis and analyzed data; S.G. performed research and molecular tests and analyzed data; K.M.L. performed research and statistical analysis; U.S. performed morphologic data and clinical data accrual; S.C.N. performed research, clinical data accrual, and morphologic data and analyzed data; M.C. designed the study, performed research and clinical data accrual, analyzed data, and provided administrative assistance; N.B.J. and V.M.S. performed research and cytogenetic analysis and analyzed data; A.S. performed research and clinical data accrual, analyzed data, provided administrative assistance, and wrote the paper; and V.M. designed the study, performed research and clinical data accrual, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vikram Mathews, Department of Haematology, Christian Medical College and Hospital, Vellore 632004, India; e-mail: vikram@cmcvellore.ac.in.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal