Abstract
Human natural killer (NK) cell development is a step-by-step process characterized by phenotypically identified stages. CD161 is a marker informative of the NK cell lineage commitment, whereas CD56, CD117, and CD94/NKG2A contribute to define discrete differentiation stages. In cells undergoing in vitro differentiation from CD34+ umbilical cord blood (UCB) progenitors, LFA-1 expression allowed to discriminate between immature noncytolytic CD161+CD56+LFA-1− and more differentiated cytolytic CD161+CD56+LFA-1+ NK cells. CD161+CD56+LFA-1− NK cells produce large amounts of CXCL8 after phorbol myristate acetate (PMA) or cytokine treatment. Remarkably, CXCL8 mRNA expression was also detected in fresh stage III immature NK cells isolated from tonsils and these cells expressed CXCL8 protein on PMA stimulation. Within in vitro UCB-derived CD161+CD56+LFA-1− NK cells, CXCL8 release was also induced on antibody-mediated cross-linking of NKp44 and CD161. Such unexpected activating function of CD161 was confined to the CD161+CD56+LFA-1− subset, because it did not induce cytokine release or CD107a expression in CD161+CD56+LFA-1+ cells or in mature peripheral blood NK cells. Anti-CXCL8 neutralizing antibody induced a partial inhibition of NK cell differentiation, which suggests a regulatory role of CXCL8 during early NK cell differentiation. Altogether, these data provide novel information that may offer clues to optimize NK cell maturation in hematopoietic stem cell transplantation.
Introduction
Natural killer (NK) cells play a key role in the host defense because of their ability to mediate cytolytic activity and to release cytokines during the early phases of immune responses against tumor cells, viruses, and other pathogens.1-3 In humans, two main cell subsets have been identified: CD3−CD56dimCD16+ cells (CD56dim), which represent the majority of NK cells circulating in the peripheral blood (PB) and CD3−CD56brightCD16− cells (CD56bright), that are a minority in PB but are largely represented in peripheral tissues. These subsets differ for the expression and the surface density of the main inhibitory receptors. Thus, killer immunoglobulin (Ig)–like receptors (KIR), specific for HLA-class I allotypes are confined to CD56dim cells (that may also express the HLA-E-specific CD94/NKG2A receptor), whereas CD56bright cells express CD94/NKG2A but not KIRs.2,4,5 Cytolytic activity is mostly associated with the CD56dim subset, whereas CD56bright NK cells are poorly cytotoxic but secrete cytokines playing an immunoregulatory role in innate and in adaptive immunity.2,5,6 Recent data demonstrated that the “cytolytic” CD56dim subset also releases large amounts of cytokines within a short time interval after receptor-mediated activation. Thus, CD56dim cells may provide a prompt intervention mediating not only rapid killing, but also production of proinflammatory cytokines and chemokines.7,8 On the other hand, CD56bright cells would guarantee a late, but prolonged cytokine production ensuring an efficient inflammatory response also at late intervals.7 There is now evidence that CD56bright may give rise to CD56dim cells representing two different stages of NK cell differentiation.9-11
A number of in vitro studies had focalized on the human NK cell differentiation process from CD34+ hematopoietic stem cell (HSC) precursors. These studies revealed that NK cell maturation is associated with the sequential acquisition of specific receptors and functions. CD161, CD56, 2B4 (CD244), and the activating receptor NKp44 are the first cell-surface markers to be expressed during early NK cell development. Subsequently, the NKp46, NKp30, NKG2D, and DNAM-1 activating receptors,12 the CD94/NKG2A inhibitory receptor, and the LFA-1 adhesion molecule are gradually acquired. The expression of CD16 and KIRs occurs at later stages and is hardly detectable in vitro.13-16 NK cell progenitors are noncytolytic but secrete a set of cytokines including interleukin (IL)–13, GM-CSF, and IL-22.17,18 On the other hand, mature NK cells are characterized by the production of IFN-γ, tumor necrosis factor (TNF)–α, and the acquisition of cytolytic activity.2,17 Therefore, human NK cells undergo differentiation through phenotypically defined stages of development also characterized by different functional capabilities.17 During the past two decades, experimental evidences have indicated that NK-committed human CD34+ precursors are present in the thymus, secondary lymphoid organs, gut mucosa, and decidua, thus implying that NK cell differentiation may also occur in other sites beside the bone marrow (BM).17,19,20 These data suggest that at least a fraction of NK-committed precursors may migrate from BM to peripheral tissues, representing an important reservoir of cells able to rapidly differentiate toward NK cells on microenvironment stimulation.
Understanding how and where NK cells undergo differentiation from CD34+ HSCs has become an important issue because of the role exerted by these cells in the positive clinical outcome of patients undergoing allogeneic HSC transplantation (HSCT), particularly haploidentical-HSCT, to treat high-risk leukemias.21,22
Studies performed both in classic allogeneic human leukocyte Ag (HLA)–matched HSCT and in haplo-HSCT, revealed that the first NK cell population recovered from PB at early time intervals (2-3 weeks) after HSCT is characterized by the CD56brightCD16−CD94/NKG2A+ surface phenotype, whereas the appearance of CD56+CD16+KIR+ NK cells occurs after 6 to 8 weeks. A delay in NK cell differentiation and perturbations in the acquisition of the NK cell repertoire have been observed in vivo, in some patients, as a possible result of radiotherapy, pharmacologic treatment, infections, inflammation or graft-versus-host disease (GVHD).23-26 Interestingly, we have recently shown that methylprednisolone (MePDN), usually used as first-line treatment for GVHD, accelerated in vitro NK cell differentiation and favored switching of myeloid cell precursors toward the NK cell lineage.27 Furthermore, we detected high levels of the proinflammatory CXCL8 chemokine in cultures of CD34+ cells undergoing in vitro differentiation toward NK cells, suggesting that immature NK cells themselves could produce CXCL8.27 Notably, CXCL8 has been show to play an important role not only in inflammatory responses, but also in hematopoiesis, angiogenesis, and tissue remodeling.28-31
In this study, in view of the pleiotropic function of CXCL8, we investigated whether it could play a role also in the process of NK cell differentiation. We show that high CXCL8 expression and release is confined to the in vitro derived immature CD117+CD161+CD56+CD94/NKG2A−LFA-1− NK cell subset and to the corresponding stage III immature NK cells freshly isolated from tonsils. On the other hand, more differentiated CD161+CD56+CD94/NKG2A+LFA-1+ NK cells, displaying cytolytic activity and IFN-γ release, did not produce CXCL8. Importantly, we provide evidence that CXCL8 may contribute to the preferential expansion of cells of the NK cell lineage. Abundant CXCL8 was released also in response to cross-linking of CD161 molecule. This finding revealed an unexpected activating receptor function associated with CD161. Because this function is confined to immature NK cells, this suggests a possible role of this receptor during early stages of NK cell development.
Methods
Cell isolation and in vitro culture
Umbilical cord blood (UCB) samples were provided by Liguria Cord Blood Bank (AOUSM-IST, Genova, Italy). Peripheral blood samples were obtained by AOUSM-IST. Tonsils were obtained from Giannina Gaslini Institute, Genova, Italy, and from Charité University Hospital, Berlin, Germany. The relevant institutional review boards approved the study and all patients gave their written informed consent according to the Declaration of Helsinki. Mononuclear cell suspension from UCB and PB were obtained as previously described.27 For detailed description of cell isolation and culture, please see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Monoclonal antibodies and flow cytometry
Cells were preincubated with human IgG (Baxter). To know the complete list of mAbs used in this study, please refer to supplemental Methods. Flow cytometric analyses were performed on FACSCalibur (BD Biosciences) and Gallios flow cytometer (Beckman Coulter). Analyses were performed on PI-negative or 7AAD-negative or Fixable viability dye (eBioscence)–negative gated cells. Data were analyzed by FlowJo Version 8.8.6 software (TreeStar). FACS sorting was performed on FACSAria (BD Biosciences). Postsort analysis showed > 99% purity.
Cytotoxicity and CD107a mobilization assays
Cell cytotoxicity was analyzed in a 4-hour 51Cr-release assay against the human melanoma FO1 cell line as previously described.32 Experiments were performed in duplicates; data are expressed as percentage of target cell lysis. CD107a mobilization was analyzed in a 3-hour redirected-killing assay against FcRγ+P815 mastocytoma murine cell line. Cell precursors and FcRγ+P815 cells were incubated in the presence of anti-CD107a mAb in the absence or presence of stimulating mAbs. After 1 hour, monensin (GolgiStop; BD) was added to the cells. At the end of incubation, cell-surface immunofluorescence was performed as previously described.
Analysis of cytokine production and receptor signaling
Cytokine production was evaluated by mean of intracellular staining or by culture supernatants analyses. Supernatants derived from culture and/or stimulation of sorted cell fractions were collected and analyzed by enzyme-linked immunosorbent assay (ELISA) multiplex assay (Bioclarma). Detailed description of intracellular staining assay is available in supplemental Methods.
RT-PCR and real-time PCR
For detailed description of RT-PCR and real-time RT-PCR analysis, please refer to supplemental Methods.
Statistical analysis
We performed Wilcoxon signed ranked test, 2-tailed, or Mann-Whitney test as indicated in figure legends. We considered significant P values ≤ .05.
Results
Analyses of informative surface markers expressed by CD34+ cells undergoing in vitro differentiation toward NK cells
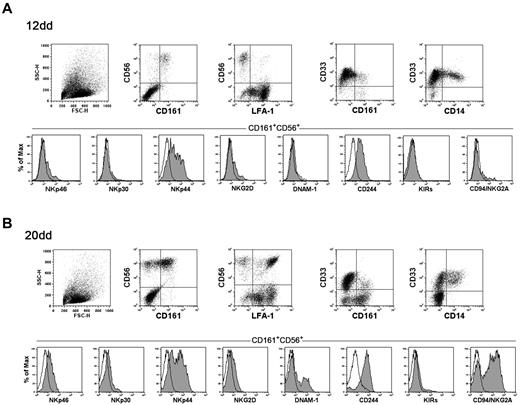
As previously described, on culture in the presence of SCF, Flt3-L, IL-7, IL-15, and IL-21 (cytokine-mix supplemented medium), CD34+ cell precursors undergo differentiation toward NK cells. We analyzed UCB CD34+ cells that had been cultured for different time intervals with the cytokine-mix. Figure 1 shows informative markers analyzed at day 12 (Figure 1A) and day 20 (Figure 1B), corresponding to early stages of in vitro NK cell differentiation. At day 12, a fraction of cells expressed both CD161 and CD56 but not CD33, thus indicating their commitment toward the NK cell lineage. In addition, this subset contained low proportion of LFA-1+ cells. Gated CD161+CD56+ cells did not express significant amounts of the main activating NK receptors with the remarkable exception of NKp44. Regarding inhibitory NK receptors, both CD94/NKG2A and KIRs were absent, whereas CD244 was homogenously expressed by CD161+CD56+ cells. At day 20 larger proportions of CD161+CD56+ cells could be detected. Of note, most of these cells expressed LFA-1. Higher proportions of CD161+CD56+ cells expressed NKp44 and NKp46 compared with cells at day 12. DNAM-1 receptor was detectable on a sizable fraction of CD161+CD56+ cells and the majority of cells expressed CD94/NKG2A. At a later culture interval (day 30), the majority of CD161+CD56+ cells, in addition to LFA-1, up-regulated NKp46 and DNAM-1, whereas both NKp30 and NKG2D displayed a dull expression (not shown).
Flow cytometric analysis of cell surface markers of NK and myeloid cell differentiation. UCB CD34+ cells were purified and cultured in the presence of SCF, Flt3-L, IL-7, IL-15, and IL-21 (cytokine-mix medium). After 12 days (A) and 20 days (B) of culture cells were analyzed for the expression of the indicated markers. NK activating and inhibitory receptors expression was analyzed gating on CD161+CD56+ cells. Black line with empty profile in histograms indicates isotype control.
Flow cytometric analysis of cell surface markers of NK and myeloid cell differentiation. UCB CD34+ cells were purified and cultured in the presence of SCF, Flt3-L, IL-7, IL-15, and IL-21 (cytokine-mix medium). After 12 days (A) and 20 days (B) of culture cells were analyzed for the expression of the indicated markers. NK activating and inhibitory receptors expression was analyzed gating on CD161+CD56+ cells. Black line with empty profile in histograms indicates isotype control.
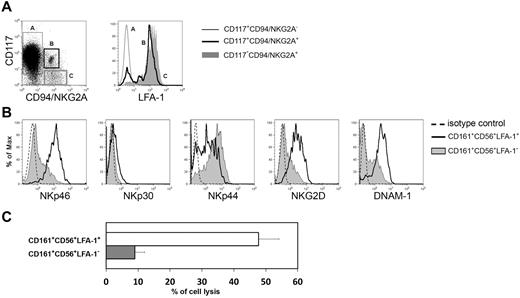
An important stage of NK cell maturation corresponds to the expression of CD94/NKG2A and to down-regulation of CD117. As previously suggested also the acquisition of LFA-1 helps to identify a stage of NK cell differentiation.27,33 Thus, we analyzed CD161+CD56+ cells undergoing in vitro differentiation for the simultaneous surface expression of CD117, CD94/NKG2A, and LFA-1. The expression of LFA-1 was detectable on CD56brightCD117−CD94/NKG2A+ cells. This surface phenotype corresponds to the so called stage IV of NK cell differentiation,2 whereas CD56brightCD117+CD94/NKG2A− NK cells corresponds to stage III of NK cells differentiation. These cells were LFA-1 negative (Figure 2A). In agreement with these data, the CD56brightLFA-1+ NK cells were characterized by higher percentages of cells expressing activating NK receptors and were cytolytic against the NK-susceptible human M-FO1 melanoma cell line (Figure 2B and C, respectively).
LFA-1 molecule is a marker of NK cell differentiation. (A) Flow cytometry analysis of LFA-1 expression on CD161+CD56+CD117+CD94/NKG2A− (light gray line, empty profile), CD161+CD56+CD117lowCD94/NKG2A+ (bold black line, empty profile), CD161+CD56+CD117−CD94/NKG2A+ (gray profile) cells. (B) After 20 days of culture, in vitro derived NK cells were analyzed for the expression of activating receptors gating separately on CD161+CD56+LFA-1− cells (gray line, gray profile) or CD161+CD56+LFA-1+ cells (black line, empty profile). Dashed line in histograms indicates isotype control. (C) CD161+CD56+LFA-1− immNK cells and CD161+CD56+LFA-1+ difNK cells were isolated by cell sorting and analyzed for their cytotoxic activity in a 4-hour 51Cr release assay against melanoma FO1 cell line. The E/T ratio was 5/1.
LFA-1 molecule is a marker of NK cell differentiation. (A) Flow cytometry analysis of LFA-1 expression on CD161+CD56+CD117+CD94/NKG2A− (light gray line, empty profile), CD161+CD56+CD117lowCD94/NKG2A+ (bold black line, empty profile), CD161+CD56+CD117−CD94/NKG2A+ (gray profile) cells. (B) After 20 days of culture, in vitro derived NK cells were analyzed for the expression of activating receptors gating separately on CD161+CD56+LFA-1− cells (gray line, gray profile) or CD161+CD56+LFA-1+ cells (black line, empty profile). Dashed line in histograms indicates isotype control. (C) CD161+CD56+LFA-1− immNK cells and CD161+CD56+LFA-1+ difNK cells were isolated by cell sorting and analyzed for their cytotoxic activity in a 4-hour 51Cr release assay against melanoma FO1 cell line. The E/T ratio was 5/1.
Immature NK cells produce large amounts of CXCL8
In a previous study, assessing the effect of MePDN on NK cell differentiation from CD34+ precursors, we detected the production of CXCL8 in control cultures performed in the absence of steroids.27 Given the potential interest of this finding, we further analyzed the pattern of chemokines and cytokines expression and release in NK cells undergoing in vitro differentiation from UCB-derived CD34+ HSCs. In particular, we compared the expression of CXCL8, IL-22, and IFN-γ, that are cytokines that have been proposed to correlate with different stages of NK cell differentiation.17,18
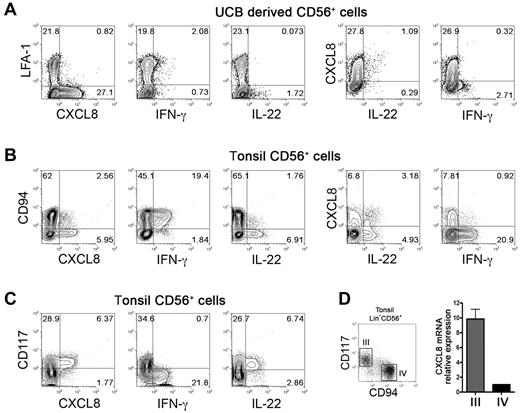
Cells cultured for 20 days with cytokine-mix, were stimulated with phorbol myristate acetate plus IL-23, and analyzed for the expression of CXCL8, IFN-γ and IL-22 by intracytoplasmic staining. CD56+LFA-1− cells selectively expressed CXCL8, whereas small percentages of CD56+LFA-1+ cells expressed IFN-γ (Figure 3A; supplemental Figure 1C). Analysis of IL-22 expression revealed that low amounts of this cytokine were detected only in CD56+LFA-1−CXCL8+ cells. As shown in supplemental Figure 1A, these cells expressed RORC mRNA. Our data indicate that the surface expression of LFA-1 on CD56+ cells can discriminate not only between cytolytic and noncytolytic NK cells, but also identifies NK cells with different patterns of cytokine expression.
Differential expression of CXCL8, IL-22, and IFN-γ in human “stage III” NK cells both in vitro and in vivo. (A) Cytokine expression in in vitro derived NK cells. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix. At day 20 of culture cells were harvested, washed and stimulated 4 hours in the presence of PMA 25ng/mL plus Ionomycin 1μg/mL and IL-23 50ng/mL in the presence of monensin. We assessed CXCL8, IL-22 and IFN-γ expression by flow cytometric gating on CD56+ cells. Representative experiment of 4 performed. (B-C): Cytokine expression in tonsil-derived stage III and stage IV NK cells. CD56+ cell were enriched by positive selection from fresh tonsil-derived cell suspensions and were stimulated 4 hours as described above. CXCL8, IL-22 and IFN-γ expression was analyzed by flow cytometry gating on Lin−(CD3−CD19−CD14−CD34−)CD56+ cells. Representative experiments of 6 performed. (D) Real-time RT-PCR analysis of CXCL8 expression in fresh tonsils-derived Lin−CD56+CD117−CD94+ and Lin−CD56+CD117+CD94− cell subsets, purified by cell sorting. We calculated CXCL8 relative expression in Lin−CD56+CD117+CD94− on the basis of CXCL8 expression level detected in Lin−CD56+CD117−CD94+, arbitrarily normalized to one. Data are expressed as mean ± SEM of relative expression obtained in 3 independent experiments. Samples were run in triplicate, we normalized gene expression levels to GAPDH mRNA, and performed relative quantification using the ΔΔCT method.
Differential expression of CXCL8, IL-22, and IFN-γ in human “stage III” NK cells both in vitro and in vivo. (A) Cytokine expression in in vitro derived NK cells. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix. At day 20 of culture cells were harvested, washed and stimulated 4 hours in the presence of PMA 25ng/mL plus Ionomycin 1μg/mL and IL-23 50ng/mL in the presence of monensin. We assessed CXCL8, IL-22 and IFN-γ expression by flow cytometric gating on CD56+ cells. Representative experiment of 4 performed. (B-C): Cytokine expression in tonsil-derived stage III and stage IV NK cells. CD56+ cell were enriched by positive selection from fresh tonsil-derived cell suspensions and were stimulated 4 hours as described above. CXCL8, IL-22 and IFN-γ expression was analyzed by flow cytometry gating on Lin−(CD3−CD19−CD14−CD34−)CD56+ cells. Representative experiments of 6 performed. (D) Real-time RT-PCR analysis of CXCL8 expression in fresh tonsils-derived Lin−CD56+CD117−CD94+ and Lin−CD56+CD117+CD94− cell subsets, purified by cell sorting. We calculated CXCL8 relative expression in Lin−CD56+CD117+CD94− on the basis of CXCL8 expression level detected in Lin−CD56+CD117−CD94+, arbitrarily normalized to one. Data are expressed as mean ± SEM of relative expression obtained in 3 independent experiments. Samples were run in triplicate, we normalized gene expression levels to GAPDH mRNA, and performed relative quantification using the ΔΔCT method.
To analyze whether immature NK cells, potentially capable of CXCL8 expression, were present also in vivo, we analyzed tonsil-derived Lin−CD56+ cells that are known to contain phenotypically defined subsets of immature NK cells.17 Purified tonsil CD56+ cells were stimulated with PMA plus IL-23 and analyzed for the expression of CXCL8, IFN-γ, and IL-22. As shown in Figure 3B-C, this NK-enriched cell population produced all the above cytokines after 4 hours incubation with PMA plus IL-23. Notably, CXCL8 was mainly confined to Lin−CD56+CD94−CD117+ cell fraction, corresponding to stage III immature NK cells. Interestingly, CXCL8+ cells did not coexpress IFN-γ but display a partial IL-22 coexpression (Figure 3B right dot plots). Analysis of tonsil-derived NK cell subsets after 12 hours incubation with PMA plus IL-23, revealed that Lin−CD56+CD94−(CD117+) increased CXCL8 expression, comparable with that observed in in vitro–derived CD161+CD56+LFA-1− immature NK cells (supplemental Figure 1B-C). Moreover, a more prolonged stimulation (12 hours), revealed and increase of the proportions of IL22+ cells that also expressed CXCL8 (supplemental Figure 1B).
To further confirm that the ability to produce CXCL8 was a feature of stage III CD56+CD117+CD94− NK cells, fresh tonsil-derived CD56+ cells were sorted into Lin−CD56+CD117+CD94− (stage III) and Lin−CD56+CD117−CD94+ (stage IV) cell fractions and analyzed for CXCL8 mRNA. As shown in Figure 3D, CXCL8 mRNA content was higher in stage III with respect to stage IV cell fraction.
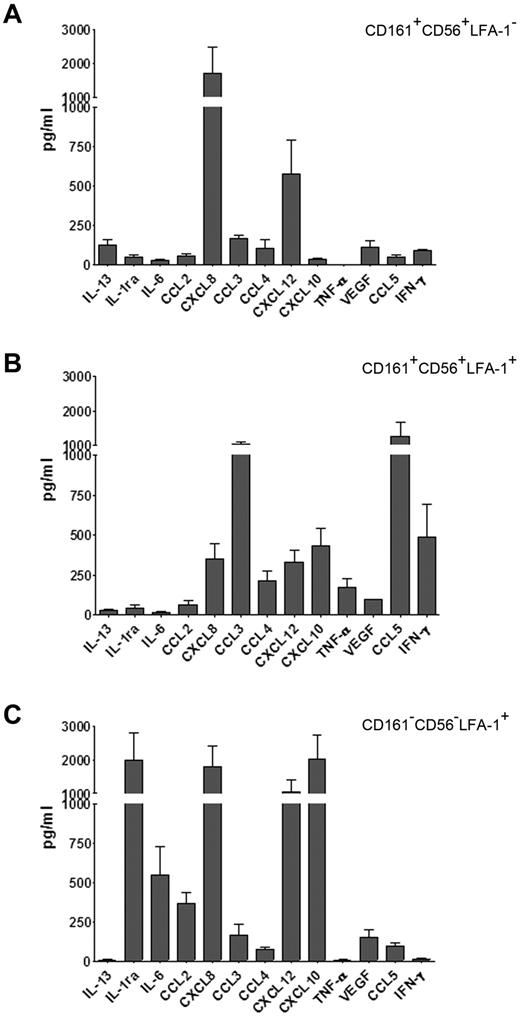
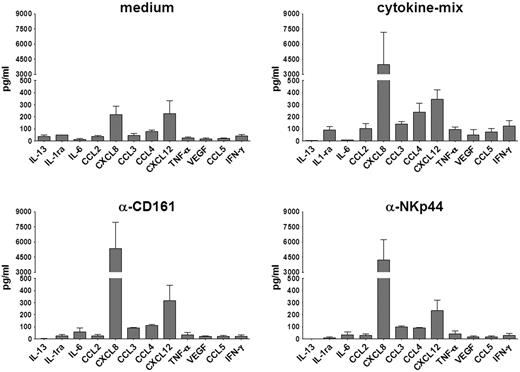
We further analyzed the repertoire of cytokines secreted by phenotypically defined cell subsets detectable at day 20, representing different stages of in vitro NK cell differentiation. These subsets were purified by cell sorting on the basis of CD56, CD161, and LFA-1 surface expression as previously described.27 Three distinct cell subsets were obtained, namely: CD161+CD56+LFA-1− (corresponding to immature NK cells, immNK), CD161+CD56+LFA-1+ (ie, more differentiated NK cells, difNK), and CD161−CD56−LFA-1+ (corresponding to CD33+CD14± myelomonocytic cell precursors). These subsets were further cultured for 72 hours in the presence of cytokine-mix. Supernatants were then analyzed for cytokine production. Figure 4 shows the release of various informative cytokines and chemokines. In agreement with data obtained with intracytoplasmic staining, the immNK cell fraction (CD161+CD56+LFA-1−) released high amounts of CXCL8 (Figure 4A). In addition, they released CXCL12 (SDF-1α) and low amounts of IL-13, CCL3 (MIP-1α), and VEGF. CD161+CD56+LFA-1+ difNK cells displayed a different pattern of cytokine secretion. Indeed a marked down-regulation of CXCL8 and CXCL12 secretion occurred, whereas IL-13 was undetectable (Figure 4B). On the other hand, difNK cells released chemokines and cytokines typical of mature NK cells including CCL3, CCL4 (MIP-1β), CCL5 (RANTES), and IFN-γ. Note that, in these experiments, IFN-γ was measured after a much longer time interval than experiments shown in Figure 3. Figure 4C shows that the myeloid cell fraction (CD161−CD56−LFA-1+) produced large amounts of IL-1 receptor antagonist (IL-1ra) and different chemokines including CXCL8, CXCL12, and CXCL10 (IP-10).
immNK cells display an unique pattern of chemokine and cytokine release. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix for 20 days. At this time interval, we purified by cell sorting: (A) immNK cells (CD161+CD56+LFA-1−), (B) difNK cells (CD161+CD56+LFA-1+), and (C) myelomonocytic cell precursors (CD33+CD161−CD56−LFA-1+). Purified cells were further cultured in the presence of cytokine-mix and after 72 hours of culture supernatants were collected and analyzed by ELISA multiplex assay for the indicated cytokines and chemokines. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments.
immNK cells display an unique pattern of chemokine and cytokine release. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix for 20 days. At this time interval, we purified by cell sorting: (A) immNK cells (CD161+CD56+LFA-1−), (B) difNK cells (CD161+CD56+LFA-1+), and (C) myelomonocytic cell precursors (CD33+CD161−CD56−LFA-1+). Purified cells were further cultured in the presence of cytokine-mix and after 72 hours of culture supernatants were collected and analyzed by ELISA multiplex assay for the indicated cytokines and chemokines. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments.
In immNK cells CD161 functions as triggering receptor inducing CXCL8 release but not cytotoxicity
Because mature NK cells respond to different stimuli by producing different cytokines and chemokines,7 we further investigated the responsiveness of the immNK cell subset to stimulation via NK receptors that were already expressed at early stages of differentiation, including NKp44, CD244, and CD161. Notably, although NKp44 has been shown to exert an activating function at different stages of NK cell maturation, CD244 has a dichotomous function in immature versus mature NK cells.16,27 On the other hand, CD161 has been reported to function as an inhibitory receptor in mature NK cells.34,35 As shown in Figure 5, anti-NKp44 mAbs induced release of large amounts of CXCL8. This data indicate that immNK cells can secrete cytokines, not only on exposure to stimulating cytokines, but also on surface receptor-mediated cell triggering. Importantly, strong CXCL8 release was also induced by cross-linking of CD161, and similar results were obtained by intracytoplasmic staining (supplemental Figure 2A). These data clearly indicate that, in immNK cells, CD161 exerts an unexpected activating receptor function. On the other hand, cross-linking of CD244 did not result in cytokine release by both immNK and difNK cells (not shown). Analysis of CXCL8 mRNA synthesis revealed that NKp44 induced higher expression of CXCL8 mRNA compared with CD161 (supplemental Figure 2B).
CXCL8 release is induced by CD161 and NKp44 cross-linking. Purified CD161+CD56+LFA-1− immNK cells were incubated in GAM-coated plates in the presence of RPMI+15% serum (medium), cytokine-mix, anti-CD161, or anti-NKp44 mAbs. After 24 hours, supernatants were collected and analyzed for the indicated cytokines and chemokines by ELISA multiplex assay. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments.
CXCL8 release is induced by CD161 and NKp44 cross-linking. Purified CD161+CD56+LFA-1− immNK cells were incubated in GAM-coated plates in the presence of RPMI+15% serum (medium), cytokine-mix, anti-CD161, or anti-NKp44 mAbs. After 24 hours, supernatants were collected and analyzed for the indicated cytokines and chemokines by ELISA multiplex assay. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments.
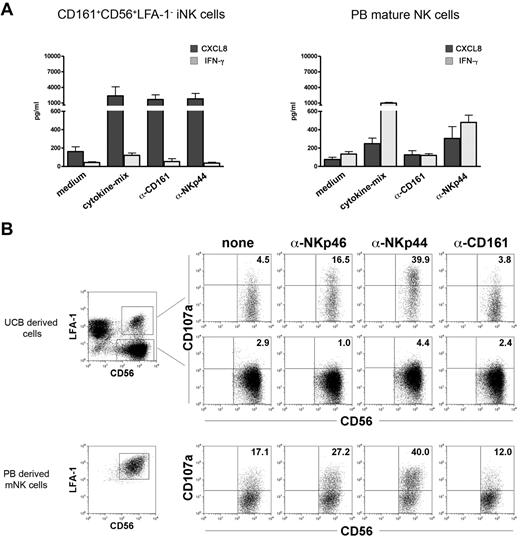
We further analyzed whether CD161 could induce cytokine release in PB-derived mature NK cells. PB NK cells cultured in IL-15 were assessed for their ability to secrete CXCL8 and/or IFN-γ in response to anti-CD161 or anti-NKp44 mAbs or IL-15 (Figure 6A). Cell triggering via CD161 induced CXCL8 release in immNK cells, but not in mature NK cells. Cross-linking of CD161 did not lead to IFN-γ release in either immNK cells or in mature NK cells.
CD161 triggers CXCL8 release, but not cytotoxicity in UCB-derived NK cells. (A) Purified CD161+CD56+LFA-1− immNK and PB NK cells were incubated in GAM-coated plates in the presence of RPMI+15% serum (medium), cytokine-mix, anti-CD161 and anti-NKp44 mAbs. After 24 hours, supernatants were collected and analyzed for the presence of CXCL8 (dark gray bars) and IFN-γ (light gray bars) by ELISA multiplex assay. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments. (B) UCB-derived NK cells and PB-mature NK cells were analyzed for CD107a expression in a 3-hour redirected killing assay against the FcγR+ P815 murine mastocytoma cell line. The E/T ratio was 2/1 and was calculated on the percentages of NK cells present in each culture. Top dot plots shows the analyses of CD107a expression in UCB derived CD56+LFA-1+ difNK cells and CD56+LFA-1− immNK cells. Bottom dot plots represent CD107a expression in PB-derived NK cells. Representative experiments of 5 performed.
CD161 triggers CXCL8 release, but not cytotoxicity in UCB-derived NK cells. (A) Purified CD161+CD56+LFA-1− immNK and PB NK cells were incubated in GAM-coated plates in the presence of RPMI+15% serum (medium), cytokine-mix, anti-CD161 and anti-NKp44 mAbs. After 24 hours, supernatants were collected and analyzed for the presence of CXCL8 (dark gray bars) and IFN-γ (light gray bars) by ELISA multiplex assay. Data are expressed as mean (± SEM) of pg/mL obtained in 3 independent experiments. (B) UCB-derived NK cells and PB-mature NK cells were analyzed for CD107a expression in a 3-hour redirected killing assay against the FcγR+ P815 murine mastocytoma cell line. The E/T ratio was 2/1 and was calculated on the percentages of NK cells present in each culture. Top dot plots shows the analyses of CD107a expression in UCB derived CD56+LFA-1+ difNK cells and CD56+LFA-1− immNK cells. Bottom dot plots represent CD107a expression in PB-derived NK cells. Representative experiments of 5 performed.
Given the capability of CD161 to induce cell triggering resulting in CXCL8 expression and release, we further investigated whether it could mediate cytolytic activity. To this end, cytolytic difNK cells and noncytolytic immNK cells (Figure 2) were analyzed in comparison with PB-derived mature NK cells for their responsiveness to anti-CD161 mAbs. In these experiments, we analyzed the CD107a surface expression in a redirected killing assay against the FcγR+P815 murine mastocytoma cell line. Figure 6B shows that difNK cells expressed CD107a on triggering via NKp44 and NKp46 receptors, but not on cross-linking of CD161. As expected, immNK cells did not express CD107a in the presence of any stimulus. Statistical analysis confirmed that, whereas NKp46 and NKp44 induced significant CD107a expression compared with control, CD161 cross-linking did not (supplemental Figure 2C).
Previous studies reported that cross-linking of CD161 in CD4+ T cells results in the activation of the PI3K/PKB-Akt pathway and subsequent cytokine release.36 Thus, we investigated whether activation of the same pathway also occurred in immNK cells. As shown in supplemental Figure 3A, mAb-mediated cross-linking of CD161 induced an increase of Akt-pS473 expression in immNK cells. This increase was statistically significant (P = .015) compared with isotype-matched control mAb (supplemental Figure 3B). On the other hand, Akt-pS473 phosphorylation did not increase in PB NK cells or in immNK cells stimulated with anti-NKp44 mAb (P = .015; supplemental Figure 3B). Moreover, CD161-induced CXCL8 expression was significantly inhibited by the presence of Akt inhibitor XI (P = .03, supplemental Figure 3C). As shown in supplemental Figure 3D and E, CD161 cross-linking induced also a significant increase of ERK1/2 pT202-pY204 expression in immNK and PBNK cells (P = .008 and P = .03, respectively). The NKp44 cross-linking-mediated effect on ERK1/2 pT202-pY204 phosphorylation was instead more evident in PB-derived NK cells, as previously described (supplemental Figure 3F).12,37
Effect of CXCL8 on NK cell differentiation
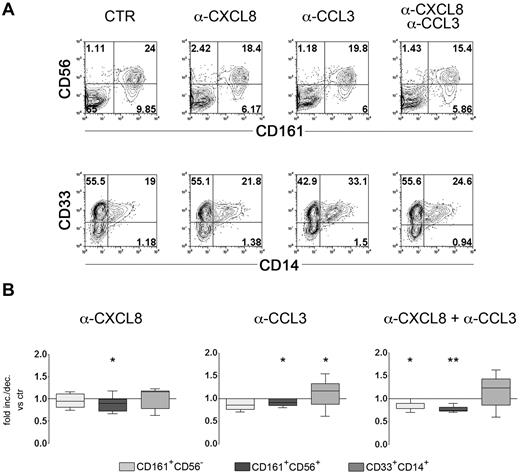
The ability of immNK cells and myeloid cells to produce high amounts of CXCL8 prompted us to investigate whether this chemokine could exert any functional effect on cell precursors present in the culture. In this context, CXCL8 and CCL3 have been shown to sharply inhibit human myelopoiesis both in vitro and in vivo.29,30,38 To define the possible role of CXCL8 secreted during in vitro NK cell differentiation, CD34+ precursors were cultured for 12 days in the presence of a cytokine-mix. As illustrated in Figure 1, at this time interval, myelomonocytic cell precursors (CD33+CD14±) were present in culture together with immNK cells and difNK cells. Neutralizing mAbs specific for CXCL8 and CCL3, alone or in combination, were added to cultures to verify whether they affect monocyte differentiation. Control cultures were performed in the absence of any mAbs or in the presence of anti-CCL4 mAbs, because differentiating NK cells can secrete CCL4 on cytokine stimulation (Figures 4–5). Cells were cultured for additional 5 days, and then analyzed by flow cytometry for informative surface markers, namely CD161, CD56, CD33, and CD14. Figure 7A, shows a representative experiment. The simultaneous presence of anti-CXCL8 and anti-CCL3 neutralizing mAbs resulted in a reduction of both CD161+CD56− and CD161+CD56+ cell subsets. On the other hand, increases of CD33+CD14+ cells were detectable, in particular in the presence of anti-CCL3 mAb. Statistical analyses revealed that reduction of the percentages of CD161+CD56+ NK cells in the presence of anti-CXCL8 or anti-CCL3 mAbs was significant (P = .047 and P = .031, respectively, Figure 7B). A more pronounced reduction was obtained by the combined use of these mAbs (P = .007). Under these conditions, there was a significant reduction also of the percentages of CD161+CD56− cells (P = .015). On the other hand, neutralization of CCL3 resulted in increases in the percentages of CD33+CD14+ cells (Figure 7B). Addition of control anti-CCL4 mAbs did not result in any modification of the proportions of various cell subsets compared with control cultures (not shown). In parallel experiments, we further investigated the effect of addition of recombinant CXCL8 and CCL3. Chemokines were added at day 12 and cells analyzed after additional 5 days of culture. A significant decrease of CD33+CD14+ cells was observed only in the presence of CXCL8 (P = .019) and an increase of CD161+CD56+ NK cells only in the presence of CCL3 (P = .011; supplemental Figure 4A). Of note, analysis of the chemokine receptor expression revealed that CXCR1 and CXCR2 (ie, the CXCL8 receptors) and CCR1 (ie the CCL3 receptor) were mainly expressed by the CD161−CD56−CD33+ myeloid precursor subset (supplemental Figure 4B). Our data would suggest that CXCL8 and CCL3 could positively influence NK cell maturation, possibly by inhibiting the immature myeloid precursors present at these early culture intervals.
CXCL8 favors NK cell differentiation. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix. At day 12 of culture anti-CXCL8 and anti-CCL3 blocking antibodies were added to the culture alone or in combination. Cells were further cultured for 5 days and then analyzed for the expression of CD56, CD161, CD33, and CD14. (A) Flow cytometric analysis for the surface expression of the indicated markers. Representative experiments of 8 performed. (B) Statistical analysis of the effect of anti-CXCL8 and anti-CCL3 neutralizing mAbs on NK and monocyte cell differentiation. Data are expressed as median values of fold increase/decrease of percentages of cell positive for the indicated surface markers versus control (arbitrarily normalized to 1) obtained in 8 independent experiments. Data were analyzed by Wilcoxon signed rank test (**P ≤ .001; *P ≤ .05).
CXCL8 favors NK cell differentiation. UCB CD34+ cells were purified and cultured in the presence of cytokine-mix. At day 12 of culture anti-CXCL8 and anti-CCL3 blocking antibodies were added to the culture alone or in combination. Cells were further cultured for 5 days and then analyzed for the expression of CD56, CD161, CD33, and CD14. (A) Flow cytometric analysis for the surface expression of the indicated markers. Representative experiments of 8 performed. (B) Statistical analysis of the effect of anti-CXCL8 and anti-CCL3 neutralizing mAbs on NK and monocyte cell differentiation. Data are expressed as median values of fold increase/decrease of percentages of cell positive for the indicated surface markers versus control (arbitrarily normalized to 1) obtained in 8 independent experiments. Data were analyzed by Wilcoxon signed rank test (**P ≤ .001; *P ≤ .05).
Discussion
In this study, we analyzed different stages of NK cell differentiation from CD34+ HSCs isolated from UCB. Different NK cell subsets, defined by the expression of surface markers including CD161, CD56, and LFA-1, were analyzed for their functional capabilities. We show that in vitro-derived immNK cells express and secrete large amounts of CXCL8. Remarkably, similar data were also obtained by ex vivo analysis of stage III immature NK cells isolated from tonsils. Importantly, we show that CD161, a major marker of early NK cell differentiation, functions as a triggering receptor inducing cytokine release in immNK cells.
CD34+ precursors, in the presence of appropriate cytokines, progressively acquired NK cell markers. Thus at day 12, a sizable fraction of cells expressed CD161 and CD56, whereas only a minority of these cells expressed LFA-1. Notably, the majority of CD161+CD56+ immNK cells expressed NKp44. NKp44 was originally described as an activating NK receptor expressed by mature NK cells on culture in IL-2 (or IL-15).32 Our data indicate that NKp44 expression can be induced by cytokines also in immature NK cells that do not express yet the major activating NK receptors. Indeed, the acquisition of these receptors (including NKp46, NKp30, DNAM-1, and NKG2D) occurs later during differentiation (at day 20 in our experimental setting). Notably, at this stage, a sizable fraction of differentiating NK cells acquire the expression of the HLA-E–specific CD94/NKG2A inhibitory receptor, which is paralleled by the loss of CD117, the appearance of LFA-1, and the acquisition of cytolytic activity.
NK cell differentiation appears to proceed through several steps, characterized by the sequential acquisition not only of different surface markers, but also of functional capabilities including cytolytic activity and secretion of different cytokines.17 In this context, it has been shown that the CD56+CD117+CD94− NK cells (“stage-III” NK cells) identified in secondary lymphoid organs contain a subset of cells expressing IL-22.39 Recently, IL-22–producing NK cells were also derived in vitro from UCB CD34+ cells cultured in the presence of murine stromal cells and cytokines.18 In the present study, IL-22 positive cells were also identified in a small fraction of UCB-derived immNK cells. However, in our experimental setting, the prominent cytokine released by immNK cells was CXCL8. Importantly, CXCL8 expression could also be demonstrated in vivo, in immature NK subsets isolated from tonsils. Indeed, analysis of ex vivo-isolated NK cells from human tonsils revealed that stage III fresh iNK cells expressed higher CXCL8 mRNA amounts compared with stage IV fresh NK cells. Moreover, analysis of intracytoplasmic protein expression indicated that, on stimulation, CXCL8 was mainly detectable in stage III NK cells. It is of note that CXCL8 was not coexpressed with IFN-γ, whereas IL-22 and CXCL8 coexpression could be detected. In addition tonsil stage IV NK cells expressed limited amounts of CXCL8 only on stimulation. The role of CXCL8 or other proinflammatory cytokines, produced by tonsil NK cells at different stages of differentiation, could be an interesting issue for further investigations. In this context, CXCL8 production occurs also in decidua-associated NK cells characterized by a peculiar mature phenotype.31,40,41 Taken together these findings suggest that CXCL8 production/release by NK cells may depend on given maturational stages and/or on peculiar microenvironments. The best-defined function of CXCL8 is the chemotactic activity primarily exerted on neutrophils, thus playing an important role in acute inflammatory responses.28 However, more recently, CXCL8 has been shown to function also as a potent inducer of neoangiogenesis. In particular, during pregnancy, NK-derived CXCL8 has been proposed to play a pivotal role in building of new vessels and placental tissues.31
One may ask which could be the functional meaning of CXCL8 production during the early stages of NK cell differentiation. We show that CXCL8 secreted by immNK cells favored the in vitro expansion of the NK cell lineage. This effect on NK cell differentiation could be amplified by CCL3, produced by more differentiated NK cells. Moreover, CCL3 displayed an inhibitory effect on myeloid cell differentiation. These data are in agreement with previous reports showing an inhibitory effect of CCL3 and CXCL8 on monocyte development from early myeloid progenitors.29,30,38 Taken together, our findings suggest that the combined effect of CCL3 and CXCL8 may favor the preferential growth of cells belonging to the NK cell lineage, in part as a consequence of the inhibitory effect on myelopoiesis. Release of CXCL8 was characteristic not only of immNK cells, but also of CD161−CD56−LFA-1+CD33+ cells, corresponding to myelo-monocyte precursors. It has been suggested that CXCL8 and/or CCL3 secretion by different cell types may contribute to the control of stem/progenitor cell proliferation in different organ sites in late phases of inflammatory responses.30 On the other hand, our data support the existence of a relationship between myelomonocytic precursors and NK precursors by the finding that both cell types secrete CXCL8 and CXCL12. The present results are in line with our previous study showing that purified CD161−CD56−LFA-1+CD33+ cells, on culture with the cytokine-mix and MePDN, acquire the CD161+CD56+LFA-1−NKp44+(CD33−) phenotype typical of immNK cells, suggesting that NK cells may also derive from myeloid precursors.27 These data have recently been confirmed and extended by Grzywacz et al by a limiting dilution approach.42,43
Similar to mature NK cells, immNK cells could secrete cytokines not only in response to cytokine-mediated stimulation, but also in response to receptor cross-linking. Our results suggest that, NKp44 cross-linking in immNK cells induced the synthesis of CXCL8 and the release of CXCL8 and CXCL12. More importantly, also cross-linking of CD161 led to CXCL8 and CXCL12 release, thus revealing an activating role of this receptor. Although it was conceivable that, in immNK cells, NKp44 could exert a role by inducing cytokine production and secretion that favor the cross-talk with other cells, an activating function of CD161 was unexpected. Moreover, the ability of CD161 to trigger cytokine release was restricted to immNK cells, because PB NK cells were unable to release cytokines on CD161 cross-linking. Of note, CD161 cross-linking did not lead to cytokine secretion or CD107a expression in (potentially cytotoxic) UCB-derived CD161+CD56+LFA-1+ difNK cells.
In mature PB NK cells, CD161 receptor has been reported to exert a minor inhibitory effect on NK cell-mediated cytolytic activity.34,35 On the other hand, it has also been suggested that CD161 may be involved in γδ (Vδ2+) T lymphocytes migration to peripheral tissues, through the activation of the calcium calmodulin kinase II (CAMKII) signaling pathway.44 Of note, CD161 cross-linking has been shown to induce proliferation of immature thymocytes.45 Further analysis of CD161 signaling pathway in T lymphocytes indicated that CD161 triggers the activation of the PI3K-PKB/Akt-1 pathway.36 This study shows that the PI3K-PKB/Akt-1 pathway is activated by CD161 cross-linking in immNK cells. The ability of CD161 to function either as inhibitory or activating receptor is still a matter of debate.46 However, our data suggest that at least in early stages of NK development, CD161, which represents one of the first NK cell marker detectable on NK cell precursors,47 may play an activating function in the regulation of cytokine release. Remarkably, lectin-like transcript 1, the CD161 physiologic ligand, is expressed not only by PB lymphocytes and monocytes, but also by endothelial cells and osteoblasts.48-50 Thus, our results suggest that CD161 may play a role in the cross-talk of NK cell precursors with other cells present in the BM microenvironment or in other peripheral tissues where NK cell differentiation occurs.17,18,20
In conclusion, our study provides relevant novel information on the early stages of NK cell differentiation, revealing that immNK cells express high proportions of CXCL8 both in vitro and in vivo. Perhaps more importantly, we provide evidence of a new unexpected function of CD161, that is the ability to function as an activating NK receptor, contributing to CXCL8 release by NK cells at early stages of development. In turn, CXCL8 appears to play a role in favoring preferential expansion of the NK cell lineage. Our data also provide further information on the relationship existing between myeloid and NK cell precursors, and together with previous findings, suggest that the first wave of NK cells appearing in PB at early time intervals after HSCT22,24,25 may derive, at least in part, from CD34+CD33+ precursors committed toward the myeloid cell lineages. Taken together, these data may help to better define different stages of NK cell differentiation and may be relevant in haplo-HSCT, in which NK cells play a central role in the successful clinical outcome.21,22
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Poggi (AOUSM-IST, Genova) for providing anti-CD161 antibody; D. Reverberi and F. Loiacono (AOUSM-IST, Genova) for technical support in cell sorting purification; and A. Bo (Liguria Cord Blood Bank) for providing selected UCB samples.
This work was supported by grants awarded by the Associazione Italiana per la Ricerca sul Cancro (AIRC; IG 2010 project no. 4725, L.M.), and “Special Program Molecular Clinical Oncology 5 × 1000” (no. 9962, L.M.); Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR)–FIRB 2003 project RBLA039LSF-001 (L.M.), MIUR-PRIN 2007 project 20077NFBH8_005 (M.C.M), and MIUR-PRIN 2008 project 2008PTB3HC_005 (L.M.); Ministero della Salute RF2006–Ricerca Oncologica–Project of Integrated Program 2006-08, agreement RO strategici 3/07 (L.M.) and RFPS-2007-4-633146 agreement RO strategici 8/07 (M.C.M.); Progetto Ricerca Ateneo 2010 020302005059, University of Genova (C.V.); and the European Federation of Immunological Societies–Immunology Letters (EFIS-IL) fellowship (E.M.). E.M. is now the recipient of a fellowship awarded by Fondazione Italiana per la Ricerca sul Cancro (FIRC).
Authorship
Contribution: E.M. and C.V. designed and performed experiments and wrote the paper; F.C., R.C., T.G., and P.A. performed experiments; and L.M. and M.C.M. supervised the project and wrote the paper.
Conflict-of-interest-disclosure: The authors declare no competing financial interests.
Correspondence: Lorenzo Moretta, Giannina Gaslini Institute, Largo G. Gaslini 5, 16147 Genova, Italy; e-mail: lorenzomoretta@ospedale-gaslini.ge.it.
References
Author notes
E.M. and C.V. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal