Abstract
Th17 cells represent a subset of CD4+ T helper cells that secrete the proinflammatory cytokine IL-17. Th17 cells have been ascribed both a beneficial role in promoting clearance of pathogenic fungi and bacteria, and a pathogenic role in autoimmune diseases. Here we identify the tyrosine phosphatase SHP-1 as a critical regulator of Th17 development, using 3 complementary approaches. Impaired SHP-1 activity through genetic deletion of SHP-1, transgenic expression of an inducible dominant negative SHP-1, or pharmacologic inhibition of SHP-1 strongly promotes the development of Th17. Ex vivo Th17 skewing assays demonstrate that genetic or pharmacologic disruption of SHP-1 activity in T cells results in a hyper-response to stimulation via IL-6 and IL-21, 2 cytokines that promote Th17 development. Mechanistically, we find that SHP-1 decreases the overall cytokine-induced phosphorylation of STAT3 in primary CD4+ T cells. These data identify SHP-1 as a key modifier of IL-6–and IL-21–driven Th17 development via regulation of STAT3 signaling and suggest SHP-1 as a potential new therapeutic target for manipulating Th17 differentiation in vivo.
Introduction
Th17 cells are a subset of CD4+ T helper cells defined by their ability to secrete IL-17A and IL-17F.1,2 IL-17 is an inflammatory cytokine important in mediating host defense against bacterial and fungal pathogens.3,4 Under physiologic conditions, Th17 cells are found in the intestinal lamina propria and Peyer patches, where they are regulated by the local cytokine milieu and support responses against pathogenic bacteria and fungi.5-8 However, unregulated Th17 development and IL-17 production have been shown to contribute to the development of allergic and autoimmune diseases.1,9-12 Recently, Th17 cells have also been linked to cancer, but their involvement toward cancer ablation or progression varies widely depending on the type of cancer.13-16 Therefore, characterizing the intracellular signaling within CD4+ T cells that modifies Th17 development will have important clinical implications for a broad range of diseases. To date, few studies have addressed how modifying early signaling events in CD4+ T cells affects Th17 differentiation.
Stimulation of naive T cells with either IL-6 plus TGF-β or IL-21 plus TGF-β leads to the activation and induction of several key transcription factors essential for Th17 differentiation, including STAT3, RORγt, and RORα.2,9,12,17,18 The signaling cascade via the IL-6 receptor leads to the downstream activation of Jak kinases and, in turn, Jak-mediated phosphorylation of STAT3 proteins. This leads to homodimerization and translocation of STAT3 into the nucleus, where STAT3 directly binds to the il17a promoter and is required for the induction of RORγt.17,19 Consistent with this, STAT3−/− mice completely lack Th17 cells and are resistant to experimental autoimmune encephalitis. To date, a network of transcription factors has been linked to Th17 differentiation, yet modifiers of the signaling cascade from cytokine stimulation to transcription, and in turn Th17 development, are not well understood.20
The Src homology region 2 domain-containing tyrosine phosphatase-1 (SHP-1) is a cytoplasmic protein tyrosine phosphatase expressed in all hematopoietic cell lineages. Motheaten (me/me) mice are homozygous for a mutation that abrogates SHP-1 protein expression and display hematopoietic abnormalities resulting in death approximately 2 to 3 weeks after birth. SHP-1 is a negative regulator of signaling via cytokines, chemokines, growth factors, and antigens.21 Specifically, SHP-1–deficient T cells display increased responses to TCR stimulation and concomitant hyperproliferation in ex vivo cultures.22 However, the role of SHP-1 during Th17 cell differentiation is not known. Here, using mice in the motheaten background, as well as a new tissue-specific transgenic mouse line expressing a dominant negative mutant of SHP-1 in T cells, we demonstrate that SHP-1 naturally dampens Th17 cell development in vivo. SHP-1–deficient mice have increased percentages of Th17 cells in their Peyer patches and intestinal lamina propria, and T cells with decreased SHP-1 activity hyper-respond to IL-6 or IL-21 stimulation, in turn generating higher numbers of Th17 cells. As an independent nongenetic approach, we used sodium stibogluconate (SSG), a small molecule inhibitor of SHP-1 activity23,24 that is currently tested in clinical trials as treatment option of patients with advanced solid tumors.25-27 SSG-mediated inhibition of SHP-1 again demonstrated the regulatory role of SHP-1 in Th17 differentiation. Mechanistically, SHP-1 decreases the tyrosine phosphorylation of STAT3 after IL-6 or IL-21 stimulation, thereby directly dampening a transcription factor critical for Th17 development. Collectively, these data identify SHP-1 as a new player that naturally regulates Th17 cell differentiation in vivo.
Methods
Mice
me/+ (C57BL/6) mice were bred to generate +/+, me/+, and me/me mice. me/+ DO11.10 TCR-Tg+ (BALB/c) mice were bred to generate +/+ DO11.10 TCR-Tg+, me/+:DO11.10 TCR-Tg+, and me/me:DO11.10 TCR-Tg+. Genotyping for all mice was performed as previously described.23 C57BL/6 and rag1−/− (C57BL/6) were obtained from The Jackson Laboratory. CD4-Cre (C57BL/6) mice were obtained from Taconic Farms.28 For all experiments including me/me mice, 15- to 19-day-old mice were used. For all other studies, 4- to 6-week-old mice were used.
The DN-SHP-1 construct (SHP-1-D419A) was subcloned into the modified pLITMUS28 plasmid, in which the EF-1α promoter and DN-SHP-1 cDNA were separated by a transcription-translation STOP cassette with flanking loxP sites (see Figure 4A).29 DN-SHP-1 mice were generated by the UVA Transgenic Core Facility and bred onto the C57BL/6 background for more than 12 generations. DN-SHP-1 Tg+ C57BL/6 mice were crossed with CD4-Cre (C57BL/6) expressing mice to drive T cell–specific expression of DN-SHP-1. Genotyping and confirmation of stop cassette deletion (supplemental Figure 3A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were performed using the following primers from Integrated DNA Technologies: 5′ loxP 3040, 5′-GGG GCT CTA GTG AAC CTC TCC G-3′; 3′ loxP 3506, 5′-GAT AGG TGG CAA GTG GTA TTC CG-3′; and 3′ SHP1 674, 5′-CAC CCT CGT GGC ATA GTA CGG C-3′.
All mice were bred and maintained in accordance with the policies of the Institutional Animal Care and Use Committee at the University of Virginia. All experiments involving mice were conducted with the approval of the Institutional Animal Care and Use Committee.
Isolation of primary cells from Peyer patches and intestinal lamina propria
Intestines were isolated from mice and placed in complete media (RPMI 1640 from Invitrogen, supplemented with 10% FBS, 5 × 10−5M 2-β-mercaptoethanol, 2mM l-glutamine, 10mM HEPES, 20mM sodium bicarbonate, and antibiotics. Peyer patches were isolated manually from the intestine surface with forceps, and cells were dispersed by straining through a 40-mm cell strainer. The intestines were cut longitudinally and washed in PBS followed by 20-minute incubation at 37°C in HBSS supplemented with 5mM EDTA. The intestines were washed again in PBS, cut into 1.5-cm pieces, and digested in 50 mL of digestion solution (RPMI 1640 supplemented with 10% FBS, 5mM 2-β-mercaptoethanol, 1 mg/mL collagenase type II, Worthington Biochemical; 1 mg/mL Dispase, Invitrogen; and 40 μg/mL DNase I, Roche Diagnostics) for 1 hour at 37°C. After digestion, intestinal pieces were strained through a 40-mm cell strainer to collect released lamina propria cells, which were then stimulated and stained for flow cytometeric analyses.
Cell staining and flow cytometry
For intracellular cytokine staining, cells were seeded at 2 × 106 cells/mL in complete media and incubated for 4 to 5 hours with 60 ng/mL phorbol-12-myristate-13 acetate (Calbiochem), 750 ng/mL Mixed Calcium Magnesium Salt (Calbiochem), and 1.0 mL/mL BD GolgiStop (BD Biosciences). Cells were fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences) and stained with the appropriate antibodies. Staining antibodies are as follows: CD4-FITC, CD8-FITC, CD4-PE, IL-17-PE, IL-17A-PE, IgG1-PE, IFN-γ-allophycocyanin (APC), IgG1-APC, CD4-Alexa647, CD4-PE, CD4-Percp, TCRβ chain-APC, CD45RB-FITC, CD45RB-PE, CD25-APC (BD Biosciences); IL-17F–Alexa647, IgG2-Alexa647, IL-6Rα–PE, IL-21R–PE, IL-21–PE (eBioscience). Stained cells were collected on the FACSCalibur instrument using the CellQuest Pro Version 6.0 software (BD Biosciences) and analyzed with FlowJo 9.4.11 software (TreeStar). Analyses were conducted on live cells (> 95%) as defined by forward and side-angle scatter. Gates were set based on isotype-matched controls.
IL-6 ELISA
Orbital eye bleeds were performed to collect sera. Concentrations of IL-6 in sera were measured using the BD OptEIA mouse IL-6 ELISA Set with BD OptEIA buffers (BD Biosciences).
Colitis transfer model
Naive splenic CD4+ T cells were isolated (> 98%) from +/+ or me/+ C57BL/6 mice via negative selection as described in “Th17 culture conditions,” and stained with 4,6-diamidino-2-phenylindole (Sigma-Aldrich), CD4-FITC, CD25-APC, CD45RB-PE (BD Biosciences). CD4+CD25− CD45RBhi T cells were sorted in the UVA Flow Cytometry Core facility to more than or equal to 95% purity. A total of 5 × 105 CD4+CD25− CD45RBhi T cells were intraperitoneally injected into rag1−/− mice. Mice were observed daily and weighed weekly. Mice developed clinical signs of colitis by 7 to 8 weeks after transfer and were killed for analysis. Colon segments were fixed in 10% formalin, embedded in paraffin, and 5-μm sections were stained with H&E for pathologic scoring. The slides were studied with an Olympus BH-2 light microscope with a 20× objective and a 10× eyepiece; they are photographed with an Olympus DP12 digital camera and the image adjusted with the Picasa software. Colitis severity was scored semiquantitatively from 0 to 4 in a blinded fashion as follows: grade 1 indicates focal monocytic inflammation at the base of the glands; grades 2 and 3, incremental inflammation and epithelial changes with severity above grade 1, but below grade 4; and grade 4, loss of all goblet cells, hyperplastic and dysplastic glandular epithelium, and massive inflammation through all bowel layers with giant cells and crypt abscesses.
Th17 culture conditions
CD4+ T-cell purification.
Splenocytes were dispersed followed by red blood cell lysis using BD Pharm lyse buffer (BD Biosciences PharMingen). CD4+ T cells were isolated from the spleen by negative selection using the CD4+ T cell isolation kit (Miltenyi Biotec) according to the manufacturer's protocol.
Th17 culture.
The 1.5 × 105 CD4+ T cell together with 1.5 × 105 T cell–depleted splenocytes serving as antigen presenting cells (irradiated at 2000 cGy) were seeded per well into 24-well plates in 500 μL complete RPMI 1640 media supplemented with: 1.25 μg/mL anti-CD3 Ab (145-2c11; Cedarland Laboratories), or 5 μg/mL OVA 323-339 (ISQAVHAAHAEINEAGR) peptide (Bimolecular Research Facility, University of Virginia), 1 μg/mL anti-CD28 Ab (Southern Biotechnology), 5 ng/mL TGF-β (R&D Systems; and eBioscience), 40 ng/mL IL-23, 5 μg/mL anti–IL-4 Ab, 5 μg/mL anti–IFN-γ Ab, and IL-6 (0-75 ng/mL), or IL-21 (0-200 ng/mL) purchased from eBioscience. Cells were harvested 3 to 5 days later and restimulated as described.
SSG treatment during Th17 culture.
EC50 determination for IL-6 and IL-21 dose-responses.
To determine the EC50 of IL-6 and IL-21 for optimal TH17 differentiation, the maximal percentage of differentiated Th17 cells per experiment was set to 100%, and all other percentages were calculated in relation to this highest Th17 population. These values were then plotted against the IL-6 or IL-21 concentrations to determine the EC50 values.
Immunoblotting
Splenic CD4+ T cells were isolated as described in “Th17 culture conditions.” A total of 0.7 × 106 CD4+ T cells were stimulated with 50 ng/mL IL-6 or 50 ng/mL IL-21 at 37°C for 0 to 60 minutes before lysis in SDS buffer (2% SDS, 50mM Tris, pH 7.6, 10% glycerol, 5% 2-β-mercaptoethanol, and protease inhibitors as described previously). Lysates were separated via 10% SDS-PAGE followed by transfer and immunoblotting using the following antibodies: anti–pSTAT3-Tyr705 (clone D3A7, 1:500 dilution), anti-STAT3 (clone 124H6, 1:500 dilution; Cell Signaling), and anti–β-actin (clone AC-15 peroxidase-labeled, 1:40 000 dilution, Sigma-Aldrich). Relative protein levels were calculated based on densitometry measurements of immunoblots at linear range using the ImageJ program (National Institutes of Health).
Quantitative RT-PCR
Splenic CD4+ T cells were purified as described, and total RNA was extracted using the RNeasy mini-kit followed by DNase digestion (RNase Free DNase Set; QIAGEN). cDNA was generated with the Superscript III First-strand kit (Invitrogen). Quantitative RT-PCRs for STAT3 and HPRT1 were performed using the TaqMan Fast Universal PCR Master mix and commercially available TaqMan Gene expression assays for STAT3 and HPRT1 (Applied Biosystems). Using HPRT1 expression for normalization, relative STAT3 expression was calculated with the 2−DCt method.
Statistical analysis
P values were calculated using the unpaired Student t test. P values less than .05 were considered significant.
Results
SHP-1 limits natural intestinal Th17 development
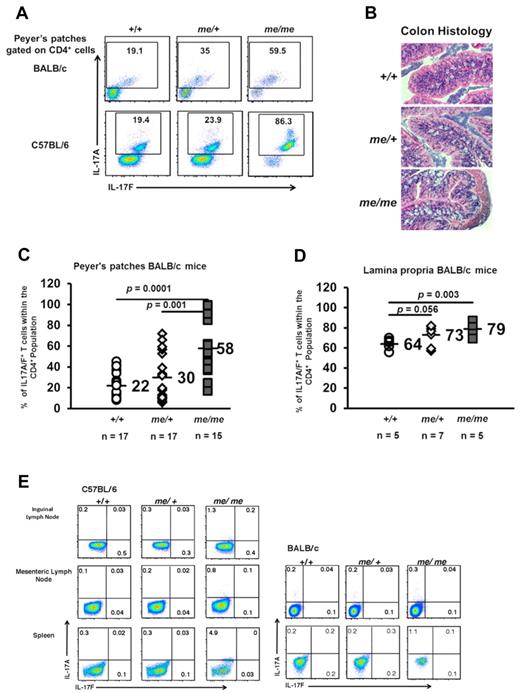
Th17 cells are naturally present in the Peyer patches and the lamina propria of the small intestines in mice where they are thought to be induced by local cytokine responses that occur in response to bacteria and fungi. We first asked whether SHP-1 affected intestinal Th17 development in vivo using the motheaten DO11.10 mouse model.23,30 DO11.10 mice express a transgenic (Tg) TCR derived from the DO11.10 T cell hybridoma line, which specifically recognizes chicken OVA peptide, amino acids 323 to 339 in the context of I-Ad.30 SHP-1–deficient me/me mice and me/+ BALB/c DO11.10 mice had higher percentages of CD4+ IL-17A/F producing cells in their Peyer patches and intestinal lamina propria compared with +/+ littermates (Figure 1A top, C-D). This was also seen in a non-TCR transgenic background; me/me and me/+ C57BL/6 mice also displayed increased levels of intestinal Th17 cells in their Peyer patches (Figure 1A bottom). Because Th17 cells have been demonstrated to contribute to colitis, we examined the intestines for pathologic signs of disease or damage. However, basally, the intestines of me/+ and me/me mice showed no signs of intestinal damage or inflammation despite having higher percentages of Th17 cells (Figure 1B).
SHP-1 limits natural intestinal Th17 development. (A) IL-17 expression of CD4+ gated T-cell population isolated from the Peyer patches of +/+, me/+, and me/me BALB/c DO11.10 and C57BL/6 mice. +/+ and me/+ mice have comparable numbers of CD4+ cells, whereas me/me mice have 50% reduced CD4+ cell numbers in their Peyer patches correlating with their decreased size (data not shown). (B) Representative colon segments from +/+ (n = 8), me/+ (n = 7), and me/me (n = 8) BALB/c DO11.10 mice (H&E, original magnification ×200). (C-D) Percentages of IL-17A/F+ cells within the CD4+ compartment isolated from the Peyer patches (C) and intestinal lamina propria (D) of mice with the indicated genotypes. (E) IL-17 expression of CD4+ gated T-cell population isolated from inguinal lymph nodes, mesenteric lymph nodes, and spleen of +/+, me/+, and me/me C57BL/6 and BALB/c DO11.10 mice. Data are representative of 2 to 4 independent experiments with multiple mice for each genotype per experiment.
SHP-1 limits natural intestinal Th17 development. (A) IL-17 expression of CD4+ gated T-cell population isolated from the Peyer patches of +/+, me/+, and me/me BALB/c DO11.10 and C57BL/6 mice. +/+ and me/+ mice have comparable numbers of CD4+ cells, whereas me/me mice have 50% reduced CD4+ cell numbers in their Peyer patches correlating with their decreased size (data not shown). (B) Representative colon segments from +/+ (n = 8), me/+ (n = 7), and me/me (n = 8) BALB/c DO11.10 mice (H&E, original magnification ×200). (C-D) Percentages of IL-17A/F+ cells within the CD4+ compartment isolated from the Peyer patches (C) and intestinal lamina propria (D) of mice with the indicated genotypes. (E) IL-17 expression of CD4+ gated T-cell population isolated from inguinal lymph nodes, mesenteric lymph nodes, and spleen of +/+, me/+, and me/me C57BL/6 and BALB/c DO11.10 mice. Data are representative of 2 to 4 independent experiments with multiple mice for each genotype per experiment.
We then asked whether the Th17 representation in other lymphoid tissues was altered in the absence of SHP-1. Comparable Th17 cell populations were seen in the inguinal and mesenteric lymph nodes of me/+ mice and +/+ mice (Figure 1E), but a small increase in the population of Th17 cells was observed in the spleens of me/me BALB/c/DO11.10 and me/me C57BL/6 mice (Figure 1E). This suggested that the effect of SHP-1 deficiency on Th17 development is not widespread and largely limited to the physiologic localization of Th17 cells.
Because IL-6 is a critical cytokine for Th17 differentiation, we next examined IL-6 levels in +/+, me/+, and me/me mice. Although comparable low levels of IL-6 were found in the sera of wild-type (+/+) and heterozygous me/+ mice, significantly higher levels of IL-6 were observed in the sera of homozygous me/me mice (supplemental Figure 1A-B). On analyzing many homozygous me/me mice, we consistently found a correlation between higher sera levels of IL-6 and an increase in splenic Th17 cells. We also observed that me/me mice in the C57BL/6 background had higher sera levels of IL-6 compared with me/me mice on the BALB/c background, which again correlated with larger percentages of splenic Th17 cells. Because increased IL-6 sera levels in me/me mice appear to contribute to higher percentages of splenic Th17 cells and to avoid the potentially complicating effects that continuous exposure to IL-6 creates, we focused on comparing heterozygous me/+ with +/+ mice for all of the following studies addressing Th17 differentiation. Although +/+ and me/+ mice have comparable sera IL-6 levels and comparable populations of Th17 cells in their spleens, me/+ mice still have an increased Th17 cell population in their lamina propria and Peyer patches, suggesting that SHP-1 directly regulates Th17 differentiation.
T cells from SHP-1–deficient mice induce colitis
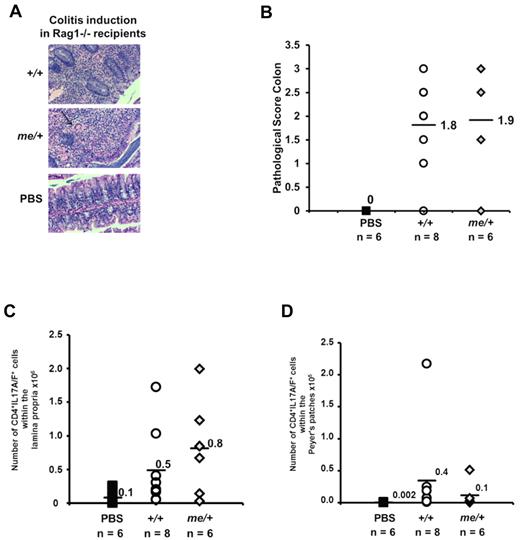
Th17 cells have been causally associated in the pathogenesis of colitis.1,12,31 Because me/+ and me/me mice showed no pathologic signs of intestinal disease at steady state, we asked whether SHP-1–deficient cells are capable of inducing colitis. In the colitis transfer model, adoptive transfer of CD4+CD25−CD45RBhigh T cells into rag1−/− recipients induces colitis within 7 to 8 weeks,31,32 and the Th17 cell–associated cytokines impact colitis development and progression. Therefore, we transferred CD4+CD25−CD45RBhigh T cells purified from +/+ and me/+ C57BL/6 mice into rag1−/− recipients, and assessed colitis severity and Th17 cell populations. Over a period of 7 to 8 weeks, the recipients of me/+ as well as +/+ cells developed diarrhea with associated weight loss. Histologic examination of the intestines confirmed that both +/+ and me/+ diseased mice exhibited colon thickening, comparable degree of monocytic and neutrophilic infiltration, a reduction in goblet cells, and moderate epithelial hyperplasia (Figure 2A-B). Furthermore, we observed comparable percentages and absolute numbers of the IL-17A/F producing cells in the Peyer patches and intestinal lamina propria in recipients of cells from SHP-1–deficient or control mice (Figure 2C-D). Thus, the Th17 cells derived from SHP-1–deficient mice are functional and capable of inducing pathogenesis in this model of colitis.
SHP-1–deficient T cells are pathogenic and able to induce colitis. CD4+CD25− CD45RBhi T cells (5 × 105) isolated from +/+ or me/+ mice were transferred into rag1−/− recipients. (A) Representative colon segments of recipient mice 7 to 8 weeks after adoptive T-cell transfer (+/+, me/+, and PBS, H&E, original magnification ×200). The arrow points to a giant cell among inflammatory cells. (B) Pathologic scoring of colon segments from recipient mice 7 to 8 weeks after adoptive T-cell transfer (+/+ vs me/+, P = .5, not significant; +/+ vs PBS, P = .001; me/+ vs PBS, P = .02). (C-D) Numbers of IL-17A/F+ T cells within the CD4+ compartment isolated from the lamina propria (C, +/+ vs me/+, P = .5, not significant) and Peyer patches (D, +/+ vs me/+, P = .7, not significant) of the indicated donor-derived populations 7 to 8 weeks after transfer. Data are representative of 3 independent experiments with multiple mice for each genotype per experiment.
SHP-1–deficient T cells are pathogenic and able to induce colitis. CD4+CD25− CD45RBhi T cells (5 × 105) isolated from +/+ or me/+ mice were transferred into rag1−/− recipients. (A) Representative colon segments of recipient mice 7 to 8 weeks after adoptive T-cell transfer (+/+, me/+, and PBS, H&E, original magnification ×200). The arrow points to a giant cell among inflammatory cells. (B) Pathologic scoring of colon segments from recipient mice 7 to 8 weeks after adoptive T-cell transfer (+/+ vs me/+, P = .5, not significant; +/+ vs PBS, P = .001; me/+ vs PBS, P = .02). (C-D) Numbers of IL-17A/F+ T cells within the CD4+ compartment isolated from the lamina propria (C, +/+ vs me/+, P = .5, not significant) and Peyer patches (D, +/+ vs me/+, P = .7, not significant) of the indicated donor-derived populations 7 to 8 weeks after transfer. Data are representative of 3 independent experiments with multiple mice for each genotype per experiment.
SHP-1 negatively regulates IL-6–mediated signaling in a T cell–intrinsic manner
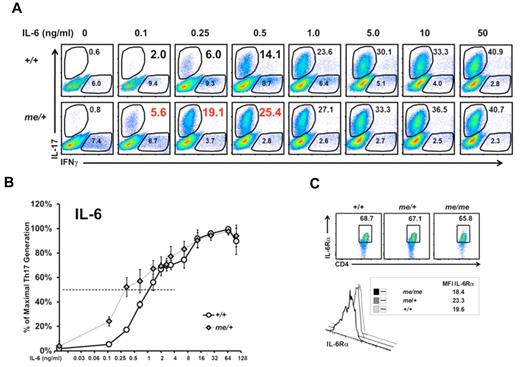
We observed enhanced Th17 development in the intestines of me/+ mice compared with +/+ mice, suggesting the possibility that SHP-1 might regulate Th17 development, and that the absence of SHP-1 could then manifest as higher numbers of Th17 cells. Both IL-6 and IL-21 promote Th17 cell differentiation by inducing STAT3 activation, which in turn regulates RORγt and RORα activation.2,9,12,17,18 To address the possible link between SHP-1, IL-6 signaling, and Th17 development, we performed Th17 lineage differentiation assays using an IL-6 dose-response (while maintaining a constant optimal level of TGF-β; supplemental Figure 1C). T cells derived from me/+ mice developed increased percentages and numbers of Th17 cells, especially at the lower range of IL-6 concentrations compared with +/+ T cells (Figure 3A-B; supplemental Figure 1D). Furthermore, T cells from me/+ mice reached their EC50 at 0.25 ng IL-6 compared 1.0 ng for +/+ T cells. This 4-fold reduction in EC50 for me/+ cells suggests that SHP-1–deficient T cells are hyper-responsive to IL-6 stimulation. Both +/+ and me/+ T cells respond similarly at higher concentrations of IL-6, indicating that the regulatory effect of SHP-1 on IL-6 signaling and Th17 differentiation is most pronounced at suboptimal IL-6 concentrations. We have previously demonstrated that SHP-1 negatively regulates TCR-mediated signaling.22 Therefore, to exclude TCR-mediated effects, we used optimal levels of TCR stimulation where effects of SHP-1 on TCR-mediated signaling are not detectable. Consistently, we observed higher percentages of Th17 cells in cultures of me/+ T cells compared with +/+ T cells at all antigen concentrations tested (supplemental Figure 1E). Finally, comparable numbers and percentages of IFN-γ+ T cells were generated from +/+ and me/+ T cells, suggesting a preferential influence of SHP-1 on Th17 differentiation (Figure 3A; supplemental Figure 1F).
SHP-1 negatively regulates IL-6–mediated Th17 development. (A) Representative IL-6 dose-response assay. CD4+ T cells from +/+ and me/+ C57BL/6 mice that were cultured under Th17-inducing with the indicated IL-6 concentrations followed by intracellular staining for IFN-γ and IL-17. Profiles show CD4+-gated population. (B) Relative Th17 T-cell generation from IL-6 dose-response assays presented in panel A. Percentages of maximal Th17 cells generated in cultures (maximum % Th17 cells: 37% +/+, 40% me/+) were set to 100% to determine EC50. EC50 values were calculated as: +/+ cells 1.0 ng/mL IL-6 (n = 4) and me/+ cells 0.25 ng/mL IL-6 (n = 7; P = .001). (C) IL-6R surface expression of splenic +/+, me/+, or me/me CD4+ T cells. Data are representative of 2 or 3 independent experiments with multiple mice for each genotype per experiment.
SHP-1 negatively regulates IL-6–mediated Th17 development. (A) Representative IL-6 dose-response assay. CD4+ T cells from +/+ and me/+ C57BL/6 mice that were cultured under Th17-inducing with the indicated IL-6 concentrations followed by intracellular staining for IFN-γ and IL-17. Profiles show CD4+-gated population. (B) Relative Th17 T-cell generation from IL-6 dose-response assays presented in panel A. Percentages of maximal Th17 cells generated in cultures (maximum % Th17 cells: 37% +/+, 40% me/+) were set to 100% to determine EC50. EC50 values were calculated as: +/+ cells 1.0 ng/mL IL-6 (n = 4) and me/+ cells 0.25 ng/mL IL-6 (n = 7; P = .001). (C) IL-6R surface expression of splenic +/+, me/+, or me/me CD4+ T cells. Data are representative of 2 or 3 independent experiments with multiple mice for each genotype per experiment.
Because T cells from me/+ mice are hyper-responsive to IL-6 stimulation, we asked whether SHP-1–deficient T cells expressed higher levels of IL-6R, which might have been advantageous. However, T cells isolated from SHP-1–deficient and +/+ mice express comparable expression levels of IL6Rα (Figure 3C), suggesting that differences in intracellular signaling downstream of the IL-6R are responsible for the hyper-responsiveness of the SHP-1–deficient T cells.
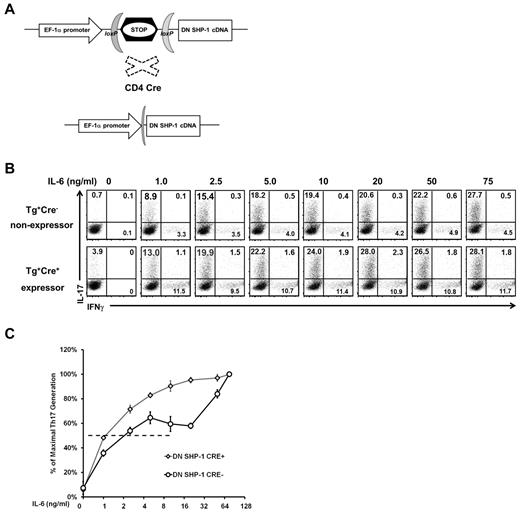
In the context of the in vivo me/+ mouse studies, the phenotypes observed in me/+ mice could be complicated by secondary effects by other lineages because the me mutation is present in every hematopoietic cell lineage.21 Therefore, to test whether SHP-1 can influence Th17 differentiation in a T cell–intrinsic manner, we generated transgenic mice that can conditionally express a dominant negative mutant of SHP-1 (DN-SHP-1). Because of the loxP-flanked STOP cassette in front of the coding sequence for DN-SHP-1, its expression is conditional and can be induced in a tissue specific manner (Figure 4A; supplemental Figure 3A). The DN-SHP-1 mutant contains an aspartic acid to alanine point mutation (D419A) in the WPD loop of the phosphatase, which renders it catalytically inactive and enables it to act as a dominant negative mutant.33 DN-SHP-1 transgenic mice were crossed with CD4-Cre transgenic mice, where the Cre expression is under the CD4 promoter and Cre-mediated deletion occurs at the late DN4 and DP stages.28 Thymic cellularity and CD4/CD8 profiles were comparable between DN-SHP-1 expressing and nonexpressing mice, indicating that T cell–specific DN-SHP-1 expression does not affect overall thymic development (supplemental Figure 3B). Next, we assessed whether DN-SHP-1 expression was functional. The DN-SHP-1–expressing T cells hyper-respond to TCR stimulation at lower concentrations of OVA peptide, similar to me/+ mice, confirming that the transgenically expressed DN-SHP-1 protein can act in a dominant negative fashion (supplemental Figure 3C). To test the effect of DN-SHP-1 on Th17 differentiation, T cells were purified and exposed to a range of IL-6 concentrations under Th17 skewing conditions. DN-SHP-1–expressing T cells were also hyper-responsive to IL-6 stimulation, manifested by increased generation of Th17 cells at suboptimal IL-6 concentrations (Figure 4B-C). Furthermore, T cells from DN-SHP-1–expressing mice reach their EC50 at approximately 2-fold to 3-fold lower IL-6 concentration than nonexpressing T cells (1.0 ng/mL IL-6 vs 2.5 ng/mL IL-6, Figure 4C). To focus on the responsiveness to IL-6 in these assays, the T cells were cultured in the presence of optimal TGF-β and TCR stimulation. Collectively, based on 2 different in vivo models (the me mutation and transgenic expression of dominant negative SHP-1), these data suggest that SHP-1 negatively regulates IL-6–driven Th17 development in a T cell–intrinsic manner.
SHP-1 negatively regulates IL-6 mediated Th17 development in a T cell–intrinsic manner. (A) Graphic map of the construct used to generate DN-SHP-1 mice. (B) Representative IL-6 dose-response assay of DN-SHP-1–expressing and –nonexpressing T cells that were cultured under Th17-inducing, with the indicated IL-6 concentrations, followed by intracellular staining for IFN-γ and IL-17. Profiles show CD4+-gated population. (C) Relative Th17 T-cell generation from IL-6 dose-response assays as described in panel B (maximum % Th17 cells: 35% nonexpressers, 34% DN-SHP-1–expressers). EC50: DN-SHP-1–expressers 1.0 ng/mL IL-6 (n = 3) and nonexpressers 2.5 ng/mL IL-6 (n = 4; P = .13). Data are representative of 2 independent experiments with multiple mice for each genotype.
SHP-1 negatively regulates IL-6 mediated Th17 development in a T cell–intrinsic manner. (A) Graphic map of the construct used to generate DN-SHP-1 mice. (B) Representative IL-6 dose-response assay of DN-SHP-1–expressing and –nonexpressing T cells that were cultured under Th17-inducing, with the indicated IL-6 concentrations, followed by intracellular staining for IFN-γ and IL-17. Profiles show CD4+-gated population. (C) Relative Th17 T-cell generation from IL-6 dose-response assays as described in panel B (maximum % Th17 cells: 35% nonexpressers, 34% DN-SHP-1–expressers). EC50: DN-SHP-1–expressers 1.0 ng/mL IL-6 (n = 3) and nonexpressers 2.5 ng/mL IL-6 (n = 4; P = .13). Data are representative of 2 independent experiments with multiple mice for each genotype.
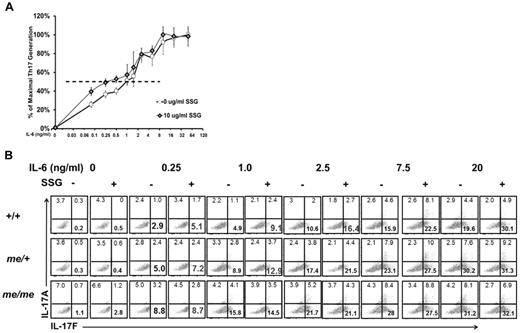
We next asked whether disturbing SHP-1 function in the context of wild-type T cells would also favor their development to Th17 lineage. For these studies, we took advantage of the drug SSG, which specifically inhibits SHP-1.24 Purified CD4+ T cells from wild-type C57BL/6 mice were cultured under Th17 promoting conditions in the presence or absence of SSG, and in a dose-response with IL-6. T cells in the SSG-treated conditions were hyper-responsive to suboptimal IL-6, leading to increased numbers of Th17 cells (Figure 5A). SSG-treated T cells reached their EC50 at an approximately 4-fold lower concentration of IL-6 compared with untreated T cells (0.25 ng/mL IL-6 vs 1.0 ng/mL IL-6). Both SSG-treated and -untreated T cells responded similarly at higher concentrations of IL-6, again supporting our finding that SHP-1 is a negative regulator of IL-6 signaling at suboptimal IL-6 concentrations and thereby influences Th17 differentiation. To confirm that under our conditions SSG is a specific inhibitor of SHP-1, we skewed T cells purified from +/+, me/+, and me/me mice toward the Th17 lineage. SSG treatment of +/+ and me/+ T cells, but not me/me T cells, causes a hyper-response to IL-6 stimulation, as evidenced by the generation of increased percentages (Figure 5B) and absolute numbers (data not shown) of Th17 cells compared with untreated T cells. Thus, the observed effect of SSG on IL-6–driven Th17 differentiation appears to be directly the result of changes in SHP-1 activity (Figure 5B). Remarkably, the effect of SSG on Th17 differentiation essentially reproduced our data in the genetic models with me/+ and DN-SHP-1 T cells. Collectively, based on the 3 different approaches, SHP-1 negatively regulates IL-6–mediated Th17 differentiation in a T cell–intrinsic manner.
SSG-mediated SHP-1 inhibition increases Th17 development. (A) Relative Th17 T-cell generation in IL-6 dose-response assay comparing SSG-treated and -untreated +/+ T cells (maximum % Th17 cells: 46% untreated, 47% SSG treated). EC50: untreated +/+ cells 1.0 ng/mL IL-6 (n = 2) and SSG-treated cells 0.25 ng/mL IL-6 (n = 2; P = .04). (B) IL-17A/F expression of CD4+ T cells from +/+, me/+, and me/me BALB/c DO11.10 mice that were cultured with or without SSG under Th17-inducing conditions with the indicated concentrations of IL-6. Profiles show CD4+-gated population. Data are representative of 2 independent experiments with multiple mice for each genotype.
SSG-mediated SHP-1 inhibition increases Th17 development. (A) Relative Th17 T-cell generation in IL-6 dose-response assay comparing SSG-treated and -untreated +/+ T cells (maximum % Th17 cells: 46% untreated, 47% SSG treated). EC50: untreated +/+ cells 1.0 ng/mL IL-6 (n = 2) and SSG-treated cells 0.25 ng/mL IL-6 (n = 2; P = .04). (B) IL-17A/F expression of CD4+ T cells from +/+, me/+, and me/me BALB/c DO11.10 mice that were cultured with or without SSG under Th17-inducing conditions with the indicated concentrations of IL-6. Profiles show CD4+-gated population. Data are representative of 2 independent experiments with multiple mice for each genotype.
SHP-1 regulates IL-6–dependent STAT3 phosphorylation
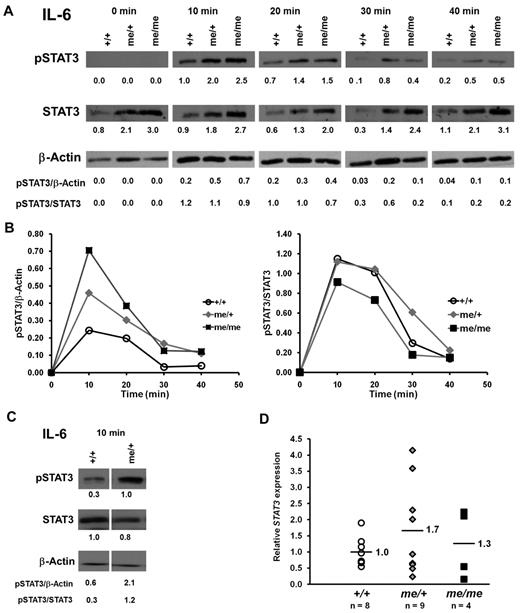
We next sought to understand the molecular mechanism, by which SHP-1 might regulate IL-6–dependent Th17 differentiation. IL-6 stimulation has been shown to induce the activation of the Jak/STAT3 pathway, which in turn directly regulates RORγt and RORα activation and Th17 cell differentiation.2,9,12,17,18 To address whether SHP-1 is a negative regulator of Jak/STAT3 signaling, purified CD4+ T cells from +/+, me/+, and me/me mice were stimulated with IL-6 and examined for levels of induced STAT3 phosphorylation as an indicator of STAT3 activation.19 We made 2 key observations. First, SHP-1–deficient T cells had overall higher levels of phosphorylated STAT3 in response to IL-6 stimulation compared with +/+ T cells, whereas the kinetics of STAT3 phosphorylation were unaffected by SHP-1 (Figure 6A-B). Second, surprisingly, we also observed higher levels of STAT3 protein in T cells from me/me mice and some of the me/+ mice (Figure 6A). As noted earlier, there is some heterogeneity among the me/+ mice and the subset of me/+ mice that had STAT3 protein levels comparable with +/+ mice still had higher levels of phosphorylated STAT3 in response to IL-6 stimulation (Figure 6C).
SHP-1 negatively regulates IL-6–induced STAT3 activation. (A) CD4+ T cells (0.7 × 106) purified from +/+, me/+, and me/me C57BL/6 mice were stimulated with IL-6 for the indicated times. Cells were assessed for STAT3 phosphorylation (Tyr705), STAT3, and β-actin protein expression by immunoblotting. Numbers represent relative band densities in arbitrary units. (B) Kinetics of IL-6–induced STAT3 phosphorylation presented in panel A. pSTAT3 band densities normalized to β-actin (left panel) and pSTAT3 band densities normalized to STAT3 (right panel). (C) CD4+ T cells (0.7 × 106) purified from +/+ and me/+ mice were stimulated with IL-6 for 10 minutes and analyzed as described in panel A. (D) Quantitative RT-PCR analysis of STAT3 mRNA isolated from splenic CD4+ T cells of +/+, me/+, and me/me C57BL/6 mice. STAT3 mRNA expression levels were normalized to HPRT1 and levels for +/+ set to 1. Data are representative of 2 or 3 independent experiments with multiple mice for each genotype.
SHP-1 negatively regulates IL-6–induced STAT3 activation. (A) CD4+ T cells (0.7 × 106) purified from +/+, me/+, and me/me C57BL/6 mice were stimulated with IL-6 for the indicated times. Cells were assessed for STAT3 phosphorylation (Tyr705), STAT3, and β-actin protein expression by immunoblotting. Numbers represent relative band densities in arbitrary units. (B) Kinetics of IL-6–induced STAT3 phosphorylation presented in panel A. pSTAT3 band densities normalized to β-actin (left panel) and pSTAT3 band densities normalized to STAT3 (right panel). (C) CD4+ T cells (0.7 × 106) purified from +/+ and me/+ mice were stimulated with IL-6 for 10 minutes and analyzed as described in panel A. (D) Quantitative RT-PCR analysis of STAT3 mRNA isolated from splenic CD4+ T cells of +/+, me/+, and me/me C57BL/6 mice. STAT3 mRNA expression levels were normalized to HPRT1 and levels for +/+ set to 1. Data are representative of 2 or 3 independent experiments with multiple mice for each genotype.
We also asked whether SHP-1 is affecting the generation of STAT3 protein at the transcriptional level. Quantitative RT-PCR for Stat3 message on T cells purified from +/+, me/+, and me/me mice found comparable levels of Stat3 mRNA between +/+, me/+, and me/me T cells (Figure 6D). This indicates that SHP-1 does not affect STAT3 transcription, rather suggesting a regulatory mechanism at the protein level. Thus, SHP-1 appears to negatively regulate STAT3 signaling at 2 levels, by affecting STAT3 phosphorylation (as seen in the me/+ mice) and by altering STAT3 protein levels. Collectively, our data support the hypothesis that SHP-1 negatively regulates IL-6–driven Th17 development via limiting STAT3 activation.
SHP-1 negatively regulates IL-21–mediated signaling in a T cell–intrinsic manner
IL-21 has been identified as a second cytokine that can induce STAT3 activation and thereby mediate Th17 cell differentiation along with TGF-β.2,34,35 Therefore, we next addressed whether SHP-1 also regulates IL-21–driven Th17 differentiation. We performed in vitro Th17 differentiation assays using purified SHP-1–deficient and wild-type T cells in response to a range of IL-21 concentrations under conditions of optimal TGF-β and TCR stimulation. me/+ T cells develop increased percentages and absolute numbers of Th17 cells in response to IL-21 compared with +/+ T cells (Figure 7A-C). Furthermore, me/+ T cells have an approximately 2-fold lower EC50 than +/+ T cells (5 ng/mL IL-21 vs 10 ng/mL IL-21), indicating that SHP-1 negatively regulates IL-21–mediated signaling and IL-21–driven Th17 differentiation. In contrast to what was observed in response to IL-6, me/+ cells continue to hyper-respond, even at higher IL-21 concentrations. Surface IL-21R levels and MFI values are comparable between +/+ and me/+ splenic T cells (Figure 7D), confirming that SHP-1 affects IL-21–mediated signaling downstream of the IL-21R and that SHP-1–deficient T cells have an intrinsic ability to hyper-respond to IL-21 stimulation. Collectively, these data demonstrate that SHP-1 negatively regulates IL-6– as well as IL-21–driven Th17 development.
SHP-1 negatively regulates IL-21–mediated Th17 development. (A) Representative IL-21 dose-response assay. CD4+ T cells from +/+ and me/+ C57BL/6 mice that were cultured under Th17-inducing with the indicated IL-21 concentrations followed by intracellular staining for IL-17A/F. Profiles show CD4+-gated population. (B) Relative Th17 T-cell generation in IL-21 dose-response assays comparing +/+ and me/+ T cells (maximum % Th17 cells: 23% +/+, 30% me/+). EC50: +/+ cells 10 ng/mL IL-21 (n = 6); and me/+ cells 5 ng/mL IL-21 (n = 6; P = .01). (C) Representative graph of absolute numbers of IL-17+CD4+ T cells generated in IL-21 dose-response assays, as shown in panel A. (D) IL-21R surface expression of splenic CD4+ T cells. (E) CD4+ T cells (0.7 × 106) purified from +/+, me/+, and me/me C57BL/6 mice were stimulated with IL-21 for the indicated times and assessed for STAT3 phosphorylation (Tyr705), STAT3, and β-actin protein expression by immunoblotting. Numbers represent relative band densities in arbitrary units. (F) Kinetics of IL-21–induced STAT3 phosphorylation presented in panel E. pSTAT3 band densities normalized to β-actin (left panel) and pSTAT3 band densities normalized to STAT3 (right panel). Data represent 2 or 3 independent experiments with multiple mice per genotype.
SHP-1 negatively regulates IL-21–mediated Th17 development. (A) Representative IL-21 dose-response assay. CD4+ T cells from +/+ and me/+ C57BL/6 mice that were cultured under Th17-inducing with the indicated IL-21 concentrations followed by intracellular staining for IL-17A/F. Profiles show CD4+-gated population. (B) Relative Th17 T-cell generation in IL-21 dose-response assays comparing +/+ and me/+ T cells (maximum % Th17 cells: 23% +/+, 30% me/+). EC50: +/+ cells 10 ng/mL IL-21 (n = 6); and me/+ cells 5 ng/mL IL-21 (n = 6; P = .01). (C) Representative graph of absolute numbers of IL-17+CD4+ T cells generated in IL-21 dose-response assays, as shown in panel A. (D) IL-21R surface expression of splenic CD4+ T cells. (E) CD4+ T cells (0.7 × 106) purified from +/+, me/+, and me/me C57BL/6 mice were stimulated with IL-21 for the indicated times and assessed for STAT3 phosphorylation (Tyr705), STAT3, and β-actin protein expression by immunoblotting. Numbers represent relative band densities in arbitrary units. (F) Kinetics of IL-21–induced STAT3 phosphorylation presented in panel E. pSTAT3 band densities normalized to β-actin (left panel) and pSTAT3 band densities normalized to STAT3 (right panel). Data represent 2 or 3 independent experiments with multiple mice per genotype.
SHP-1 regulates STAT3 phosphorylation in response to IL-21 stimulation
Because SHP-1–deficient T cells hyper-respond to IL-21–driven Th17 differentiation, we asked whether IL-21–mediated STAT3 signaling is also regulated by SHP-1. Purified CD4+ T cells from +/+ and me/+ mice were stimulated with IL-21, and the levels of induced phosphorylated STAT3 were measured. me/+ T cells showed overall increased levels of phosphorylated STAT3 compared with +/+ T cells but unchanged kinetics of phosphorylation (Figure 7E-F left panel). When me/+ mice that expressed STAT3 protein levels comparable to +/+ mice were analyzed, a considerable increase in relative STAT3 phosphorylation was observed (Figure 7E-F right panel). This correlated well with the significant difference in IL-21–driven Th17 development between me/+ and +/+ T cells (Figure 7A-C). These data indicate that SHP-1 regulates STAT3 activity by negatively regulating STAT3 phosphorylation in response to IL-21 stimulation. Collectively, our data support a model where SHP-1 limits IL-6– and IL–21–driven Th17 development, via decreasing STAT3 activation.
Discussion
To date, numerous studies have identified transcription factors that are critical for Th17 differentiation, but the upstream signaling events that modulate these transcription factors are less well understood. STAT3 is one of the key transcription factors known to promote Th17 differentiation. Here, we identify SHP-1 as a critical negative regulator of STAT3 signaling and IL-6– and IL-21–driven Th17 cell development. Our data suggest that SHP-1 can regulate Th17 differentiation via 2 mechanisms: (1) by negatively regulating STAT3 phosphorylation in response to IL-6 and IL-21 stimulation; and (2) by limiting the levels of STAT3 protein available for activation. Because we observed that there are no differences in stat3 transcription between wild-type and SHP-1–deficient T cells, these data suggest that SHP-1 limits STAT3 at the protein level, perhaps by indirectly regulating its degradation.
Our in vitro Th17 skewing studies identified SHP-1 as a negative regulator of both IL-6– and IL-21–driven Th17 development. To address whether SHP-1 might also be enhancing Th17 cell survival or expansion, we assessed these parameters in Th17 cultures and observed comparable levels of cell survival and proliferation between +/+ and me/+ T cells (data not shown). This finding is consistent with our hypothesis that SHP-1 regulates Th17 differentiation and that differences in Th17 differentiation potential between +/+ and me/+ T cells are not the result of selective expansion or survival of the SHP-1–deficient T cells.
Recent studies have demonstrated that the hypoxia-inducible factor 1 (HIF-1α) is induced by STAT3 and promotes Th17 development through direct transcriptional activation of RORγt, an essential transcription factor for Th17 differentiation.36,37 Therefore, we tested whether SHP-1 affects the generation of HIF-1α at the transcriptional level. Quantitative RT-PCR for HIF-1α mRNA indicated that HIF-1α induction was comparable between Th17 skewed +/+ and me/+ cells, suggesting that this pathway is not being affected by SHP-1 deficiency (supplemental Figure 2A-B). In addition, we examined the mRNA levels of Th17 family cytokines il17a, il17f, il22, and the lineage-specific transcription factor RORγt in Th17 skewed +/+ and me/+ cells by quantitative RT-PCR. Whereas il-22 and Rorc expression levels were comparable between Th17 skewed +/+ and me/+ cells, il17a and il17f induction was elevated in Th17 skewed me/+ cells consistent with an up-regulation of Th17 cytokine expression in SHP-1–deficient cells (supplemental Figure 2A-B).
Colitis is thought to be a Th17-mediated disease. Despite the increase in resident colonic Th17 cells, we did not detect any signs of colitis in any of the SHP-1–deficient mice. Although there are several potential explanations for the lack of any pathogenesis, the following is perhaps the most likely one. Recent studies suggest that “nonpathogenic” Th17 cells can develop under specific cytokine and TCR stimuli.38-42 Although the precise components of “nonpathogenic” Th17 development are not fully defined, lowered SHP-1 levels could promote the generation of nonpathogenic Th17 cells in the intestines of these mice.
SHP-1–deficient cells are capable of being pathogenic and inducing colitis mediated through T-cell transfer. SHP-1–deficient and wild-type T cells were equally pathogenic, and both cell types generated comparable Th17 populations on transfer. This suggests that, in this model using a lymphopenic host, different factors, such as homeostatic T cell expansion, may be the main force in driving T-cell proliferation and differentiation, thereby overriding any differences in the response to the Th17-promoting cytokine milieu found in a nonlymphopenic mouse. To assess whether SHP-1 deficiency affects the kinetics of colitis development, we also examined mice at earlier time points and found no differences indicating that SHP-1 does not influence the timing of Th17 development (data not shown). At earlier time points before the appearance of colitis or Th17 T cells (4-6 weeks after T-cell transfer), we observed the development of CD4+IFN-γ+ T cell populations, which has been described for other autoimmune models, such as experimental autoimmune encephalitis and orchitis.43,44 However, these populations were comparable between recipients of +/+ or me/+ T cells, consistent with what we had observed in vitro.
SSG treatment of T cells results in hyper-responsiveness to IL-6 with increased in vitro Th17 development. Interestingly, SSG is currently being tested in phase 1 and 2 clinical trials as a possible treatment option for patients with advanced solid tumors, as well as hematopoietic malignancies, such as lymphomas, or myelomas.25-27 Current studies suggest that the presence of Th17 cells can be either beneficial or detrimental for the cancer patient, depending on the type of cancer.13-16 Therefore, our findings that SHP-1 deficiency/SSG treatment promotes IL-6– and IL-21–driven Th17 development could be of high clinical significance and suggest SHP-1 as a potential new therapeutic target for manipulating Th17 differentiation in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Joanne Lannigan and Mr Michael Solga (UVA Flow Cytometry Core) for their help with cell sorting, the UVA histology core for all histology work presented in this paper, Dr Kodi Ravichandran for critical reading of the manuscript, Ms Kaitie Farenholtz for technical assistance, and Mr Josh Mauldin for help and suggestions during the course of these studies.
This work was supported by the National Institutes of Health (grant RO1 AI48672, U.M.L.; and grants AI 41236 and AI 51420, K.S.T.).
National Institutes of Health
Authorship
Contribution: I.S.M. designed the research, performed the experiments, analyzed the results, created the figures, and wrote the manuscript; K.S.T. analyzed histologic slides and scored pathology of colitis experiments; and U.M.L. designed the research, supervised planning of experiments, and helped with data analysis, figure preparation, and manuscript writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ulrike M. Lorenz, Beirne Carter Center of Immunology, MR-6, Rm 2708, University of Virginia Health System, PO Box 801386, Charlottesville, VA 22908; e-mail: ulorenz@virginia.edu.