Abstract
Efficient in vitro generation of hematopoietic stem cells (HSCs) from embryonic stem cells (ESCs) holds great promise for cell-based therapies to treat hematologic diseases. To date, HoxB4 remains the most effective transcription factor (TF) the overexpression of which in ESCs confers long-term repopulating ability to ESC-derived HSCs. Despite its importance, the components and dynamics of the HoxB4 transcriptional regulatory network is poorly understood, hindering efforts to develop more efficient protocols for in vitro derivation of HSCs. In the present study, we performed global gene-expression profiling and ChIP coupled with deep sequencing at 4 stages of the HoxB4-mediated ESC differentiation toward HSCs. Joint analyses of ChIP/deep sequencing and gene-expression profiling unveiled several global features of the HoxB4 regulatory network. First, it is highly dynamic and gradually expands during the differentiation process. Second, HoxB4 functions as a master regulator of hematopoiesis by regulating multiple hematopoietic TFs and chromatin-modification enzymes. Third, HoxB4 acts in different combinations with 4 other hematopoietic TFs (Fli1, Meis1, Runx1, and Scl) to regulate distinct sets of pathways. Finally, the results of our study suggest that down-regulation of mitochondria and lysosomal genes by HoxB4 plays a role in the impaired lymphoid lineage development from ESC-derived HSCs.
Introduction
The various cells in a tissue are generated by multipotent lineage-specific stem cells, which in turn are derived from pluripotent embryonic stem cells (ESCs). Interest in ESCs has been greatly fuelled by their potential to generate cells for use in cell-replacement therapy for a wide range of diseases. This has also been heightened by the advent of induced pluripotent stem cells, a process in which a somatic cell can be reprogrammed into an ESC-like state.1,2 Despite these great promises, the gene-regulatory pathways controlling lineage-specific differentiation of ESCs are poorly understood, hindering efforts to derive, expand, and manipulate lineage-specific stem cells in vitro for therapeutic purposes.
The spontaneous generation of hematopoietic stem cells (HSCs) by differentiating ESCs has long been documented and characterized.3 In most cases, ESC-derived HSCs show similar clonogenic progenitor capacity and primitive phenotype as somatic sources of hematopoietic progenitors, but possess limited in vivo repopulating capacity when transplanted into immunodeficient mice.4 Therefore, if ESCs are to be used for generating HSCs, we need to know much more about the genetic and epigenetic factors involved in the process that drives undifferentiated ESCs accurately and efficiently down hematopoietic developmental pathways.
HoxB4 is a member of the highly conserved homeodomain transcription factor (TF) family. It was the first TF shown to lead to profound HSC expansion in vitro and in vivo when expressed ectopically in adult BM cells and ESCs.5 These HSCs fully replenish the stem cell pool of lethally irradiated mice and maintain a normal supply of HSCs and mature blood cells for the duration of life.6-8 So far, the data strongly suggest that the HoxB4-mediated ESC differentiation system recapitulates early hematopoiesis observed in vivo during embryonic development.9,10 However, a recent study suggests that the precise control of HoxB4 expression level is important for in vivo HSC development,10 providing a cautionary note for using in vitro models. Nevertheless, the HoxB4 system remains a powerful and convenient in vitro model with which to explore the molecular pathways that specify hematopoietic fate, which otherwise would be difficult to examine in embryos.
The molecular mechanisms behind HoxB4-mediated HSC development remain poorly understood. As a first effort to define the molecular pathways controlled by HoxB4, Schiedlmeier et al performed a genome-wide mRNA profiling of mouse ESCs overexpressing HoxB4.11 Based on differential gene expression, they identified roughly 700 genes that are likely to be direct targets of HoxB4. Those genes are involved in pathways important for controlling the self-renewal, maintenance, and differentiation of stem cells. More recently, using ChIP coupled with a promoter tiling microarray (ChIP-CHIP), 2 studies identified additional HoxB4 target genes in ESC-derived HSCs12 and in a primitive hematopoietic progenitor cell line, EML.13 These 2 pioneering studies expanded the set of HoxB4 targets to roughly 3500. Nevertheless, 2 critical questions remain to be answered with regard to the HoxB4 regulatory network. First, a comprehensive set of HoxB4 direct targets still needs to be established. Because previous studies only used promoter-tiling microarray, they likely missed many gene-distal HoxB4 binding sites. Indeed, based on a HoxB4 DNA motif scan of the mouse genome before this study, we found many thousands more putative HoxB4 binding sites than those identified by the previous ChIP-CHIP studies. Second, the dynamics of the HoxB4 regulatory network during the differentiation process has not been characterized. Like many developmental processes, HoxB4-mediated HSC development involves a precise cascade of regulated pathways. Knowledge about the identities and temporal activities of these pathways will enable us to design more effective strategies for in vitro derivation of HSCs.
In the present study, we report genome-wide distribution of HoxB4 binding sites and global gene-expression profiles at 4 distinct stages of the ESC differentiation to HSCs. Our data demonstrate that the size of HoxB4 transcriptional regulatory network is significantly larger than previously thought, and highlight the role of HoxB4 as a master regulator of hematopoiesis by regulating multiple key hematopoietic TFs and chromatin-modification enzymes. Furthermore, integrated analysis of HoxB4 binding and gene-expression profiles across the time course reveals a dynamic HoxB4 regulatory network with characteristic sets of pathways activated at distinct developmental stages.
Methods
HoxB4 expression vector, ESC line, and culture conditions
Construction of the murine stem cell virus–based expression vector was described previously.14 In this vector, the human HoxB4 gene is under the control of transcriptional regulatory elements within the 5′–long-terminal repeat of the virus, resulting in constitutive expression of HoxB4. We are grateful to Dr Hannes Klump (University of Essen, Essen, Germany) for providing the HoxB4-transduced CCE cells. Transduced cells were grown on gelatinized flasks in feeder cell-free ESC culture medium consisting of DMEM (Invitrogen) supplemented with 15% FCS, 0.1mM l-glutamine, 100 U/mL of penicillin, 100 g/mL of streptomycin, and 1000 U/mL of leukemia inhibitory factor. The culture medium was changed daily, and the cells were passaged every 2-3 days to avoid overgrowth and differentiation.
Differentiation of ESCs to HSCs
We adopted a 2-stage culture strategy, as described previously.6,7 At day 0, HoxB4-transduced CCE ESCs were subjected to embryoid body (EB) formation for 6 days on nonhematopoietic differentiation medium. Briefly, ESCs were plated on an ultra-low-attachment Petri dish at a concentration of 2000 cells/mL in a methylcellulose-based differentiation medium containing IMDM (Invitrogen), 15% FCS, 300 μg/mL of transferrin, 2mM l-glutamine, 100 U/mL of penicillin, 100 μg/mL of streptomycin, 5% protein-free hybridoma medium (Invitrogen), 4 × 10−4M monothioglycerol, and 50 μg/mL of ascorbic acid. At stage II, EBs were dissociated into single-cell suspension using trypsin (2.75%), and 3.5 × 106 cells/mL cells were replated on another ultra-low-attachment Petri dish in a serum-free hematopoietic differentiation medium containing StemPro34 plus nutrient supplement (Invitrogen) and a cocktail of hematopoietic cytokines, including 100 ng/mL of murine SCF (R&D Systems), 2 ng/mL of mIL-3, 5 ng/mL of mIL-6, 10 ng/mL of Flt3-L, 40 ng/mL of insulin-like growth factor-1 (Promega), and 1μM dexamethasone (Sigma-Aldrich). The culture medium was changed every other day and the cell density was maintained below 4 × 106 cells/mL.
ChIP-Seq
We conducted ChIP coupled with deep sequencing (ChIP-Seq) experiments using cells from days 6, 16, and 26 of the differentiation process. For each ChIP experiment, approximately 107 cells were fixed with 1% formaldehyde, lysed, and sonicated to shear chromatin DNA. HoxB4-bound chromatin fragments were enriched by immunoprecipitation with a rabbit mAb raised against human HoxB4 immunogen (Epitomics). After reversal of cross-linking, the enriched DNA fragments were precipitated and purified using a PCR clean-up kit (QIAGEN). Deep sequencing was done per the manufacturer's protocol using an Illumina GA II sequencer. A control ChIP-Seq experiment (ie, without anti-HoxB4 Ab) was run for each time point to ensure that background noise was properly controlled.
Validation of HoxB4 ChIP-Seq peaks using ChIP-qPCR
Cell culture and ChIP were conducted in the same way as in the ChIP-Seq experiments. After the ChIP step, purified DNA from matched IP sample and input control sample were subjected to quantitative PCR (qPCR) analysis using SYBR Green chemistry (Invitrogen). qPCR primers were designed to flank selected HoxB4 peak regions. The fold enrichment was computed as 2−ΔΔCt, where ΔΔCt is the difference in ΔCt values between the IP sample and the input control sample. ΔCt is the cycle number difference between a sample and a GAPDH control.
Gene-expression microarray experiments
The Affymetrix mouse gene ST 1.0 microarray was used to profile mRNA expression levels at days 0, 6, 16, and 26 during the differentiation process. The microarray platform covers 28 853 well-annotated mouse genes. Three biologic replicates were performed for each time point. Microarray data were normalized using the RMA algorithm.15 Differentially expressed genes were detected using the Limma algorithm.16
Results
Differentiation of ESC to hematopoietic cells by overexpression of HoxB4
We developed previously a protocol for deriving HSCs from murine ESCs by the combination of HoxB4 overexpression and incubation with hematopoietic cytokines.7 The differentiation process takes 26 days and the resulting HSCs induce high-level mixed chimerism and long-term engraftment in recipient mice. In the present study, we used this protocol to collect cells at 4 time points during HSC development: days 0, 6, 16, and 26. These 4 time points were chosen based on the expression level of the cell-surface marker CD45, the expression dynamics of which track the developmental maturity of HSCs.6,7,17 Day 0 HoxB4-expressing cells were cultured in ESC medium and represent undifferentiated ESCs, whereas day 6, 16, and 26 cells contained beginning, partially, and fully differentiated HSCs, respectively. Although our differentiation protocol is strongly biased toward hematopoiesis, the cell populations used herein were not sorted and are therefore heterogeneous. The day 6 culture contained cells of 3 germ layers and small numbers of HSCs and progenitors. At later stages, HSCs were gradually enriched and, by the end of the differentiation protocol, the population contained large numbers of HSCs and mature cells (approximately 97% CD45+ cells).7
Genome-wide HoxB4 location maps during ESC differentiation to hematopoietic cells
To understand the mechanisms by which HoxB4 mediates ESC differentiation to HSCs, we used ChIP-Seq to identify direct targets of HoxB4 at days 6, 16, and 26 of the differentiation process. A ChIP-grade rabbit mAb against HoxB4 protein was used and Ab specificity was confirmed by Western blot (supplemental Figure 1A, see the Supplemental Materials link at the top of the article). On average, 8.9 million sequencing reads were obtained for each time point and 67% of all reads were uniquely mapped to the mouse genome (supplemental Table 1). Using a false discovery rate (FDR) of 1%, we identified 3632, 7232, and 29 313 genomic loci bound by HoxB4 at days 6, 16, and 26, respectively (supplemental Methods; Figure 1A; and supplemental Table 2). The median fold enrichment of the read count within identified peaks was 11, 11, and 14, respectively (supplemental Figure 2). Among the set of binding sites, 600 were shared by all 3 time points. Genes near these common sites were enriched for TFs (P = 8.2 × 10−4). A PubMed literature survey revealeds that many of these TFs are involved in hematopoiesis (supplemental Table 3), suggesting that HoxB4 is a master regulator of hematopoiesis.
Summary of HoxB4 ChIP-Seq binding peaks at 3 stages of ESC differentiation to hematopoietic cells. (A) Venn diagram of the HoxB4 ChIP-Seq peaks from day 6 (D6), 16 (D16), and 26 (D26) cells. (B) Distribution of the distance between HoxB4 peak center and nearest RefSeq transcription start site (TSS). Distance is shown in log scale of base 10. Red dashed line shows the median distance. (C) Genomic distribution of HoxB4 peaks. (D) HoxB4 DNA-binding motif identified by a motif search using the top 500 HoxB4 peaks (ranked by ChIP-Seq enrichment ratio) from day 6 cells.
Summary of HoxB4 ChIP-Seq binding peaks at 3 stages of ESC differentiation to hematopoietic cells. (A) Venn diagram of the HoxB4 ChIP-Seq peaks from day 6 (D6), 16 (D16), and 26 (D26) cells. (B) Distribution of the distance between HoxB4 peak center and nearest RefSeq transcription start site (TSS). Distance is shown in log scale of base 10. Red dashed line shows the median distance. (C) Genomic distribution of HoxB4 peaks. (D) HoxB4 DNA-binding motif identified by a motif search using the top 500 HoxB4 peaks (ranked by ChIP-Seq enrichment ratio) from day 6 cells.
We used ChIP-qPCR to assess the quality of our ChIP-Seq data. We randomly selected 30 called peaks (10 peaks/time point) and achieved a validation rate of 83% (supplemental Figure 3), demonstrating excellent corroboration of our ChIP-Seq data. We also examined the overlaps between our peaks and peaks from previous ChIP-CHIP studies. Lee et al used a primitive hematopoietic progenitor cell line (EML) and identified 1910 HoxB4 peaks.13 Oshima et al used an ESC differentiation protocol similar to ours, in which day 6 EB cells were induced by HoxB4 overexpression for 12 days.12 They identified 2292 HoxB4-binding peaks. Target gene overlaps with our day 26 peaks (from cells representing the most mature form of HSCs in our protocol) were 82% and 90% for the Lee et al and Oshima et al studies, respectively. Given the significant amount of differences in the HSC derivation protocols, we conclude that this degree of overlap is excellent.
To further assess the quality of our ChIP-Seq data, we examined the sequence conservation of HoxB4 peaks using precomputed PhastCons conservation scores (see supplemental Methods). Overall, 20.3% of the total length of day 6 HoxB4 peaks was conserved across 30 placental mammalian genomes. The fractions of conserved positions were 18.4% and 14.2% for day 16 and 26 peaks, respectively. In contrast, only 8.14% of the genome was conserved. Most of the conserved positions were located within the central regions of HoxB4 peaks (supplemental Figure 4), which is consistent with the notion that peak centers represent the real protein-DNA binding activity and thus are more conserved than flanking regions.
The median distance between HoxB4 peaks and their nearest genes was 16 529 bp (Figure 1B). Averaging across the 3 time points, 52% of binding peaks were located in intergenic regions, 41% in introns, and 6% in exons (Figure 1C). There was no significant change in the genomic distribution of HoxB4 peaks at the different time points. A de novo motif search using the top 500 HoxB4 peaks from each time point successfully identified the HoxB4 DNA-binding motif (Figure 1D), which matched the HoxB4 binding motif identified by 2 previous studies, 1 using Protein Binding Microarray18 and the other using ChIP-CHIP.13 In summary, extensive quality assessment demonstrated that we have generated a high-quality dataset of HoxB4-binding sites across the time course of ESC-to-HSC differentiation.
Dynamic gene-expression program during ESC differentiation to hematopoietic cells
To examine the dynamics of the transcriptome of the differentiating ESCs, we profiled mRNA expression levels of day 0, 6, 16, and 26 cells. At an FDR of 0.5%, we found 5780 genes that were expressed differentially between 2 adjacent time points, representing roughly 20% of all genes in the mouse genome. For individual time periods, 1665, 3947, and 2068 genes were expressed differentially during the 3 consecutive periods, respectively (supplemental Table 4). Among the differentially expressed genes, 60.6%, 51.4%, and 41.1% were up-regulated during the 3 periods, respectively (supplemental Figure 5). Therefore, it appears that there is a gradual increase in gene silencing during the developmental process.
The time-course gene-expression profile is consistent with the evolving phenotype (ie, loss of self-renewal and pluripotency and concurrent activation of hematopoietic developmental program). For example, the classic ESC genes, Nanog, Oct4, Sox2, and Sall4, were all down-regulated during the time course. Conversely, known regulators involved in early hematopoiesis, such as Fli1, Ikaros, Meis1, Runx1, and Tgfb1, were all up-regulated (Figure 2A).
Gene-expression profiles during ESC differentiation into hematopoietic cells. (A) Expression profiles of classic genes important for ESC and HSC phenotypes. Solid line is ESC genes; dashed line, HSC genes. (B) Overlap between differentially expressed and differentially bound genes between 2 adjacent time points. Differential binding means that the gene is only bound by HoxB4 at 1 of the 2 time points. Top, schematic illustrating the 2 sets of genes and their overlap. Ti and Ti+1 indicate adjacent time points; ●, HoxB4 protein. Bar height represents the expression levels of the target genes; Bottom, for adjacent time points, the second number represents the number of differentially expressed genes and the first number represents the number of differentially expressed genes that overlap with differentially bound genes.
Gene-expression profiles during ESC differentiation into hematopoietic cells. (A) Expression profiles of classic genes important for ESC and HSC phenotypes. Solid line is ESC genes; dashed line, HSC genes. (B) Overlap between differentially expressed and differentially bound genes between 2 adjacent time points. Differential binding means that the gene is only bound by HoxB4 at 1 of the 2 time points. Top, schematic illustrating the 2 sets of genes and their overlap. Ti and Ti+1 indicate adjacent time points; ●, HoxB4 protein. Bar height represents the expression levels of the target genes; Bottom, for adjacent time points, the second number represents the number of differentially expressed genes and the first number represents the number of differentially expressed genes that overlap with differentially bound genes.
To examine the impact of HoxB4 binding on gene expression, we determined the overlap between differential gene expression and differential HoxB4 binding between adjacent time points. Of the 3969 differentially expressed genes between days 6 and 16, 1414 (35.8%) were targeted by HoxB4 sites that were bound differentially between the same 2 time points (Figure 2B). Likewise, 1272 of 2086 (61.5%) differentially expressed genes between days 16 and 26 were targeted by differentially bound HoxB4 sites. By linking differentially bound HoxB4 peaks to neighboring differentially expressed genes, this analysis provides a set of high-confidence HoxB4 target genes during the in vitro differentiation of ESCs to HSCs.
Our data are available through National Center for Biotechnology Information Gene Expression Omnibus using accession number GSE34014. This new dataset provides a powerful resource for the future characterization of HoxB4 target genes and the characterization of regulatory networks in HSCs. For the remainder of this manuscript, we illustrate how this new dataset can provide insights into the HoxB4 transcriptional regulatory network.
Evolving pool of HoxB4 targets during ESC differentiation to hematopoietic cells
A striking result from our time-course ChIP-Seq experiment was the highly dynamic nature of HoxB4 binding during the differentiation process. Specifically, of the 3 sets of HoxB4 peaks, 69.1%, 33.3%, and 83.3%, were unique to each time point (days 6, 16, and 26, respectively; Figure 1A), suggesting a highly dynamic regulatory program controlled by HoxB4 during the developmental process. To reveal the identities of the changing pathways controlled by HoxB4, we first partitioned HoxB4 peaks into nonoverlapping sets unique to each time point or pairs of adjacent time points. We then performed gene ontology term enrichment analysis of the sets of genes associated with time point–specific HoxB4 peaks. Our analysis showed that genes targeted by HoxB4 at different time points are involved in different biologic processes (Figure 3A). For example, genes targeted by HoxB4 at day 6 are enriched (FDR < 0.01) for early embryonic development and patterning. Examples include Dll1, Gli2, Nodal, Sox2, Sim2, Smad2, and Tbx3. In contrast, genes targeted by HoxB4 at later time points are enriched for hematopoietic system–specific functions such as myeloid and lymphocyte proliferation, differentiation, and activation. Examples include Bcl6, CD274, Flt3, Hlx, Ikzf1, Inpp5 days, Spn, and Zap70. To determine whether HoxB4 binding leads to gene-expression changes, we plotted median expression levels of the different sets of genes targeted by HoxB4 at different time points (Figure 3B). This analysis demonstrated that, indeed, different HoxB4-targeted pathways exhibit correct differential expression patterns, suggesting that the pathways are truly regulated by HoxB4. For example, genes targeted by HoxB4 in day 6 cells had the highest expression levels at day 6 compared with the other time points (Figure 3B).
Evolving sets of HoxB4 targets during ESC differentiation to hematopoietic cells. (A) Evolving functional categories enriched among Hoxb4 targets during the differentiation process. Genomic coordinate Hoxb4 binding peaks were used as input to the GREAT algorithm46 for gene ontology term enrichment analysis. D6, D16, and D26 indicate Hoxb4 peaks uniquely observed in day 6, 16, 26 cells, respectively; D16/D26, Hoxb4 peaks shared uniquely by day 16 and 26 cells; red, gene ontology term enriched at FDR = 0.05; black, gene ontology term not enriched. Dendrogram was clustered using hierarchical clustering. (B) Median expression levels of the sets of genes enriched at different time points reported in panel A. Only subsets of genes that were differentially expressed and targeted by HoxB4 at specific time points in panel A were used for the plot.
Evolving sets of HoxB4 targets during ESC differentiation to hematopoietic cells. (A) Evolving functional categories enriched among Hoxb4 targets during the differentiation process. Genomic coordinate Hoxb4 binding peaks were used as input to the GREAT algorithm46 for gene ontology term enrichment analysis. D6, D16, and D26 indicate Hoxb4 peaks uniquely observed in day 6, 16, 26 cells, respectively; D16/D26, Hoxb4 peaks shared uniquely by day 16 and 26 cells; red, gene ontology term enriched at FDR = 0.05; black, gene ontology term not enriched. Dendrogram was clustered using hierarchical clustering. (B) Median expression levels of the sets of genes enriched at different time points reported in panel A. Only subsets of genes that were differentially expressed and targeted by HoxB4 at specific time points in panel A were used for the plot.
Dynamic regulation of TFs by HoxB4 during ESC differentiation to hematopoietic cells
One of the enriched group of genes among HoxB4 targets are TFs, suggesting that HoxB4 functions as a master regulator of hematopoiesis. To examine this issue systematically, we overlapped the set of HoxB4 targets, differentially expressed genes, and the set of 1728 mouse TFs annotated by the FANTOM consortium.19 In total, we identified 31 TF genes that were both targeted differentially by HoxB4 and exhibited differential expression between adjacent time points (Figure 4A). Several of these genes have a documented role in hematopoiesis, including Erg,20 Cited2,21 Ctnnb1,22 Fli1,23 Gata2,24 Id2,25 Id4,26 Ikzf1,27 Lyl1,28 Meis1,29 Pbx1,30,31 Runx1,32,33 Stat5,34 Tcf7,35 and Pml-RARA.36 Direct regulation of multiple hematopoiesis TFs by HoxB4 strongly implies that it is a master regulator positioned high in the hierarchy of the hematopoiesis-regulatory network. Most of the TFs targeted by HoxB4 were either up-regulated or down-regulated; however, several TFs showed more complicated HoxB4 regulation (Figure 4A). For example, Id2, Rb1, Pbx1, Eomes, and Ctnnb1 were up-regulated during the initial phase of ESC differentiation to HSCs and then down-regulated. In contrast, Foxp1 was down-regulated initially and then up-regulated. Such complicated regulatory patterns suggest that the activities of the regulated TFs need to be carefully controlled during the developmental processes.
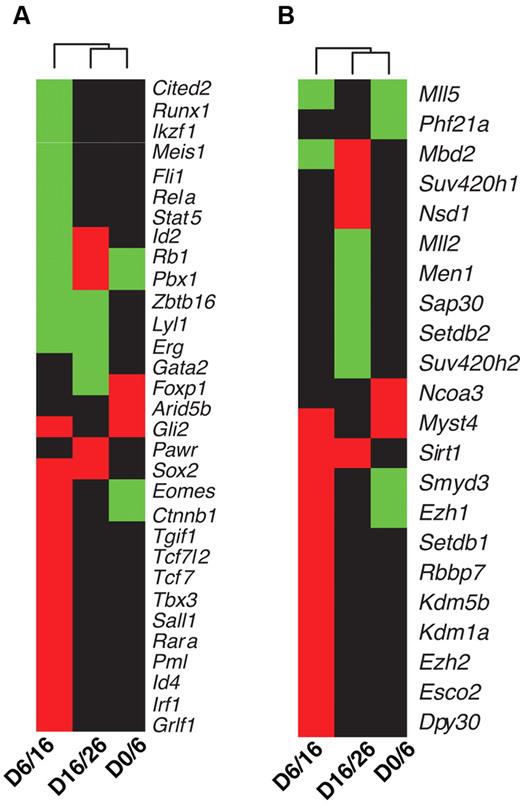
TFs and chromatin-modification enzymes targeted by HoxB4 and differentially expressed. (A) TFs. (B) Chromatin-modification enzymes. Differential gene expression between 2 adjacent time points was detected using the Limma algorithm16 with an FDR cutoff of 0.005. Each column in the dendrogram represents a comparison of data from 2 adjacent time points. For example, D0/D6 represents comparison between day 0 and day 6 data. Each cell represents combined gene-expression and HoxB4-binding information for a given gene; green, up-regulated and targeted by HoxB4; red, down-regulated and targeted by HoxB4; black, not targeted by HoxB4.
TFs and chromatin-modification enzymes targeted by HoxB4 and differentially expressed. (A) TFs. (B) Chromatin-modification enzymes. Differential gene expression between 2 adjacent time points was detected using the Limma algorithm16 with an FDR cutoff of 0.005. Each column in the dendrogram represents a comparison of data from 2 adjacent time points. For example, D0/D6 represents comparison between day 0 and day 6 data. Each cell represents combined gene-expression and HoxB4-binding information for a given gene; green, up-regulated and targeted by HoxB4; red, down-regulated and targeted by HoxB4; black, not targeted by HoxB4.
Dynamic regulation of chromatin-modification enzymes by HoxB4 during ESC differentiation to hematopoietic cells
Chromatin structure plays an important role in modulating gene expression. Therefore, we investigated whether chromatin-modification enzymes are also targeted by HoxB4. We curated a set of 128 genes involved in chromatin modifications based on annotations from the Mouse Genome Database (http://www.informatics.jax.org/). Of these genes, 21 were both differentially targeted by HoxB4 and differentially expressed between adjacent time points (Figure 4B). These genes encode proteins involved in several chromatin-modification activities, including histone methyltransferase activity (Dpy30, Ezh1, Ezh2, Men1, Mll2, Mll5, Nsd1, Setdb1, Setdb2, Smyd3, Suv420h1, and Suv420h2), histone acetyltransferase activity (Myst4 and Ncoa3), histone deacetylase activity (Sirt1, Rbbp7, Sap30, and Phf21a), methyl-CpG binding protein (Mbd2), and histone demethylase activity (Kdm1a and Kdm5b). The expression dynamics of these chromatin-modification enzymes suggest that the epigenome of the developing HSCs also undergoes dynamic changes. Furthermore, our data suggest that, in addition to regulating TF activities, HoxB4 also plays a role in shaping the epigenome of developing HSCs by regulating epigenetic factors.
Combinatorial regulation by HoxB4 and other hematopoietic TFs
Combinatorial interactions of TFs play a critical role in eukaryotic gene regulation and tissue identity. Using a cell line model of HSCs (HPC-70) and ChIP-Seq, Wilson et al recently mapped genome-wide binding sites of 10 key hematopoietic TFs.37 The number of binding peaks ranged from 4349 (Lyl1) to 36 166 (Erg; Figure 5A). In the present study, to examine potential synergistic actions between HoxB4 and these hematopoietic TFs, we determined their binding site overlap. Because our day 26 cells are most similar to HPC-7 cells in terms of developmental maturity as HSCs, we determined the binding peak overlap between the 10 TFs and HoxB4 in our day 26 cells. In total, 26.2% (7690) of HoxB4 peaks colocalized with binding peaks of at least 1 of the 10 hematopoietic TFs. Therefore, these regions represent putative transcriptional enhancers with hallmarks that include combinatorial action of multiple TFs. We also determined whether the frequency of TF pair colocalization in the same regions was greater than would be expected by chance, thus indicating potential coregulatory activities (for details, see supplemental Methods). Our analysis revealed 4 TFs that had a statistically significant binding site overlap (P < .001) with HoxB4. They are, in decreasing order of overlap significance, Runx1, Meis1, Scl, and Fli1 (Figure 5A).
Binding site overlap between HoxB4 and a set of 10 hematopoiesis TFs. ChIP-Seq peaks for the set of hematopoiesis TFs were described previously.37 (A) Number of binding peaks and significance of binding-peak overlap between HoxB4 and the 10 TFs. Significance is shown as the minus logarithm of P value for binding peak overlap. P values were calculated using a permutation test (for details, see supplemental Methods). Overlapping TFs with P < .001 are highlighted in red. (B) Box plot of peak center distance between HoxB4 binding peaks and peaks of other TFs. (C) Breakdown of different classes of overlapping peaks involving the 4 significantly overlapping TFs: H, HoxB4; F, Fli1; M, Meis1; R, Runx1; and S, Scl. Class names are based on the combination of different TFs present in the binding peaks. (D) Gene ontology term enrichment analysis of different classes of combinatorial peaks. Red indicates a gene ontology term enriched at FDR = 0.05; black, gene ontology term not enriched. Dendrogram was clustered using hierarchical clustering.
Binding site overlap between HoxB4 and a set of 10 hematopoiesis TFs. ChIP-Seq peaks for the set of hematopoiesis TFs were described previously.37 (A) Number of binding peaks and significance of binding-peak overlap between HoxB4 and the 10 TFs. Significance is shown as the minus logarithm of P value for binding peak overlap. P values were calculated using a permutation test (for details, see supplemental Methods). Overlapping TFs with P < .001 are highlighted in red. (B) Box plot of peak center distance between HoxB4 binding peaks and peaks of other TFs. (C) Breakdown of different classes of overlapping peaks involving the 4 significantly overlapping TFs: H, HoxB4; F, Fli1; M, Meis1; R, Runx1; and S, Scl. Class names are based on the combination of different TFs present in the binding peaks. (D) Gene ontology term enrichment analysis of different classes of combinatorial peaks. Red indicates a gene ontology term enriched at FDR = 0.05; black, gene ontology term not enriched. Dendrogram was clustered using hierarchical clustering.
Among the 4 TFs, Scl, Lyl1, and Runx1 are known to bind as components of multiprotein complexes to enhancers.37 To further investigate the likelihood of cooperative binding between HoxB4 and other TFs, we determined the relative distances for all pairwise binding events, taking advantage of the fact that ChIP-Seq peak calling algorithms are highly accurate at identifying peak summits.38 This analysis demonstrated that the median peak center distance between HoxB4 and other TFs are 56, 61, 58, and 75 bp for Fli1, Meis1, Runx1, and Scl, respectively (Figure 5B). Statistically significant pairwise cooccupancy, along with the close proximity of the binding sites, strongly suggests that HoxB4 binds DNA cooperatively with each of these 4 TFs. Such cooperative binding would likely be facilitated by protein-protein interactions. Indeed, a recent large-scale mapping of TF protein-protein interactions reported physical interactions between HoxB4 and Meis1 in both humans and mice.19 Validation and characterization of the other newly discovered TF interactions should lead to insights into the combinatorial regulation of hematopoietic pathway genes.
Figure 5C shows a breakdown of the putative enhancer classes based on their participating TFs. The largest class are enhancers containing both HoxB4 and Fli1 (the HF class). This is not surprising, given that Fli1 has the largest number of peaks in the genome. However, the enhancer class that contains all 5 TFs (the HFMRS class) is surprisingly abundant, even more so compared with classes that contain fewer TFs (eg, the HFRS and HFMR classes). The genes targeted by different classes of enhancers are enriched for distinct functions (Figure 5D). For example, HFMS enhancers target genes involved in lymphocyte proliferation, whereas HFMRS and HM enhancers target genes involved in hematopoietic/lymphoid organ development. This set of enhancer targets represents a rich source of potentially new mediators of HSC development.
Many genes down-regulated in lymphoid-biased HSCs are HoxB4 targets
A drawback of the HoxB4-mediated development of HSCs is that the resulting HSCs have impaired lymphoid lineage developmental potential.5,39-41 By comparing the gene-expression profiles of ESC-derived HSCs and HSCs isolated from adult BM, we sought to gain insights into the molecular mechanisms for this deficiency. We compared the genome-wide expression profiles of 4 types of HSCs: (1) ESC-derived HSCs based on HoxB4 overexpression, (2) whole HSCs isolated from adult BM without distinction of subtypes,27 and 2 subtypes of HSCs, (3) myeloid-biased HSCs (My-HSCs), and (4) lymphoid-biased HSCs (Ly-HSCs).42 Furthermore, for ESC-derived HSCs, we used cells from 2 independent sources, our day 26 cells from the present study and those generated by Oshima et al using a similar HoxB4-based protocol.12 Global gene-expression correlation was moderate between adult HSCs and ESC-derived HSCs (supplemental Figure 6), which explains the observed phenotypic differences between in vitro– and in vivo–derived HSCs. Nevertheless, within each group (ESC-derived vs adult HSCs), correlation was high. The high expression correlation with ESC-derived HSCs in the study by Oshima et al suggests that our day 26 cell population was enriched for HSCs even though the population was heterogeneous. Further, there is no distinguishable difference in correlation between ESC-derived HSCs and lineage-biased adult HSCs or whole adult HSCs, suggesting that only a subset of genes are responsible for the bias against lymphoid-lineage development of ESC-derived HSCs. By comparing the expression profiles of the 2 lineage-biased HSCs, Challen et al identified 785 genes that are differentially expressed in one of the HSC subtypes, including 434 genes expressed more highly in My-HSCs and 351 genes expressed more highly in Ly-HSCs.42 We focused on this set of lineage-biased genes to understand the mechanisms underlying the bias against the lymphoid lineage in ESC-derived HSCs.
As shown in Figure 6A, the expression profile of these lineage-biased genes in our ESC-derived HSCs was more similar to that in My-HSCs than that in Ly-HSCs. This observation prompted us to compare the absolute expression levels of these genes in the different cell types. After quantile normalization of the microarray data to account for variations due to nonbiologic factors, we found that genes up-regulated in adult My-HSCs (compared with Ly-HSCs) were expressed at similar levels in day 26 cells (Figure 6B). In stark contrast, genes up-regulated in adult Ly-HSCs were expressed at significantly lower levels in day 26 cells compared with both My-HSCs and Ly-HSCs (Figure 6C). This result suggests that a failure to prime the lymphoid-lineage gene-expression program may be the cause of the developmental defect of the lymphoid lineage from ESC-derived HSCs.
HSCs generated by HoxB4 overexpression have impaired lymphoid-lineage gene-expression pattern. We examined genes up-regulated in 2 subtypes of HSCs, myeloid-biased and lymphoid-biased, described previously by Challen et al referred to herein as lineage-biased genes.42 Normalized expression of lineage-biased genes from our study and Challen et al was compared. (A) Overall expression correlation of lineage-biased genes between various developing and mature HSCs. D0, D6, D16, and D26 refer to day 0, 6, 16, and 26 cells from this study, respectively. (B) Normalized expression levels of myeloid-lineage-biased genes (ie, up-regulated in My-HSCs identified by Challen et al42 ) in ESC-derived HSCs (day 26 cells from the present study), My-HSCs, and Ly-HSCs. (C) Normalized expression level of lymphoid-lineage-biased genes (up-regulated in Ly-HSCs identified by Challen et al42 ) in ESC-derived HSCs (day 26 cells from the present study), My-HSCs, and Ly-HSCs. P values are from 1-tailed t tests.
HSCs generated by HoxB4 overexpression have impaired lymphoid-lineage gene-expression pattern. We examined genes up-regulated in 2 subtypes of HSCs, myeloid-biased and lymphoid-biased, described previously by Challen et al referred to herein as lineage-biased genes.42 Normalized expression of lineage-biased genes from our study and Challen et al was compared. (A) Overall expression correlation of lineage-biased genes between various developing and mature HSCs. D0, D6, D16, and D26 refer to day 0, 6, 16, and 26 cells from this study, respectively. (B) Normalized expression levels of myeloid-lineage-biased genes (ie, up-regulated in My-HSCs identified by Challen et al42 ) in ESC-derived HSCs (day 26 cells from the present study), My-HSCs, and Ly-HSCs. (C) Normalized expression level of lymphoid-lineage-biased genes (up-regulated in Ly-HSCs identified by Challen et al42 ) in ESC-derived HSCs (day 26 cells from the present study), My-HSCs, and Ly-HSCs. P values are from 1-tailed t tests.
A total of 184 Ly-HSC genes were expressed at least 2-fold lower in our day 26 cells than in Ly-HSCs, with an average fold decrease of 7.9 (supplemental Table 5). Strikingly, the majority of these down-regulated genes (157 of 184) were targeted by HoxB4 based on our day 26 ChIP-Seq data (supplemental Table 5). Further, 63.9% of the HoxB4 sites targeting the down-regulated genes overlapped with 1 or more of the 4 TFs, Fli1, Meis1, Runx1, and Scl. In comparison, the fraction of combinatorial sites among all day 26 HoxB4 sites was 26.2% (P = 0 by the one-sample proportion test). Therefore, our data suggest that combinatorial regulation plays an important role in the down-regulation of these Ly-HSC genes.
The down-regulated genes were enriched for lysosomal and mitochondrial functions (Table 1). Most of the lysosome-associated genes are involved in glycolipid and glycoprotein metabolism, including Ids, Gm2a, Cd1d1, Tmem74, Fuca1, Cln5, and Abca5. The mitochondria-associated genes are involved in several functions, including oxidative phosphorylation (D2hgdh and Hk2), response to oxidative stress (Trap1 and Txnrd2), biomolecule synthesis (Pgs1, Ppox, Cyp27a1, Mecr, and Prodh), and apoptosis (Ptrh2, Araf, and Atad3a). Using a mouse model carrying a proofreading-defective mitochondrial DNA polymerase, Norddahl et al established a link between mitochondrial mutations in HSCs and lymphoid-lineage developmental block and myeloid-lineage skewing,43 supporting our finding that down-regulated mitochondrial gene expression is associated with lymphopenia. By comparing the global gene-expression profiles of HSC subtypes and integration with HoxB4 ChIP-Seq data, our results suggest that dysregulation of mitochondrial and lysosomal genes by HoxB4 may underlie the observed impaired lymphoid-lineage development.
Gene ontology (GO) term-enrichment analysis of genes that were at least 2-fold down-regulated in day 26 ESC-derived HSCs compared with Ly-HSCs*
| GO term ID . | Description . | Genes . | Bonferroni-corrected P . |
|---|---|---|---|
| GO:0005764 | Lysosome | Pnpla7, Npc2, Ids, Gm2a, Cd1d1, Tmem74, Fuca1, Cln5, Abca5 | .009 |
| GO:0005739 | Mitochondrion | D2hgdh, Pgs1, Gm2a, 5730469M10RIK, Gimap8, Hk2, Dnhd1, Osgepl1, Ppox, Dpysl2, Ptrh2, Pnpla7, Trap1, Atad3a, Cyp27a1, Hsdl1, Araf, Abhd11, Txnrd2, Mrpl48, Acaa1a, Mecr, Prodh | .03 |
| GO term ID . | Description . | Genes . | Bonferroni-corrected P . |
|---|---|---|---|
| GO:0005764 | Lysosome | Pnpla7, Npc2, Ids, Gm2a, Cd1d1, Tmem74, Fuca1, Cln5, Abca5 | .009 |
| GO:0005739 | Mitochondrion | D2hgdh, Pgs1, Gm2a, 5730469M10RIK, Gimap8, Hk2, Dnhd1, Osgepl1, Ppox, Dpysl2, Ptrh2, Pnpla7, Trap1, Atad3a, Cyp27a1, Hsdl1, Araf, Abhd11, Txnrd2, Mrpl48, Acaa1a, Mecr, Prodh | .03 |
Ly-HSC genes were identified by Challen et al.42
Discussion
Genomic locations of HoxB4 binding sites are critical information for understanding the regulatory role of HoxB4 in hematopoiesis. Two previous studies have reported HoxB4 sites using ChIP coupled with promoter tiling microarray.12,13 Oshima et al used a microarray covering −5.5 kb and +2.5 kb of all RefSeq transcription start sites,12 and Lee et al used a microarray covering −2.5 kb and +0.5 kb of all RefSeq transcription start sites.13 By design, these microarrays cannot detect gene-distal binding sites located either in intergenic and intronic regions. In the present study, we report the first truly global maps of HoxB4 binding sites using ChIP-Seq. Multiple independent lines of evidence, including ChIP-qPCR validation, sequence conservation, DNA motif analysis, and the high degree of overlap with previous results, support that our ChIP-Seq data are of high quality.
Comprehensive bioinformatic analysis revealed several novel features of the HoxB4 transcriptional regulatory network. First, our ChIP-Seq data demonstrate clearly that HoxB4 binding sites are not constrained to gene promoter regions, because the median distance between HoxB4 sites and the nearest genes is 16 529 bp. Therefore, our results greatly expand the repertoire of HoxB4 targets. This comprehensive set of HoxB4 target genes represents a rich source of new mediators of hematopoiesis.
Understanding the dynamics of transcriptional regulatory networks is essential for gaining insights into hematopoiesis. The present study is the first one investigating the dynamics of the HoxB4-regulatory network during ESC differentiation into HSCs. An unexpected finding from our study is that the pool of HoxB4 targets appears to expand during the process. One possible reason could be increased fractions of false-positive sites at later time points. However, the results from quality assessment of our ChIP-Seq data argues against this possibility. Another possibility is that the level of HoxB4 protein increases during the differentiation process, which in turn leads to increased binding. However, our time-course Western blot analysis indicated that this was not the case (supplemental Figure 1B). Hematopoietic multipotential progenitors exhibit low levels of multilineage gene-expression patterns. This phenomenon has been termed transcriptional priming, and it reflects the developmental potency of a multilineage progenitor.44 It is possible that increased HoxB4 binding at later time points reflects the increased demand for multilineage priming of gene expression in the developing HSCs. A recent study of B-cell and myeloid-cell development revealed that a substantial fraction of lymphoid and myeloid transcriptional enhancers are already occupied in HSCs before commitment to the lymphoid or myeloid cell lineages.45 This finding is consistent with the concept of multilineage priming and also suggests that, in our system, many of the expanded HoxB4 sites in the day 26 cells were used for priming the expression of genes involved in downstream developmental pathways.
Consistent with previous ChIP-CHIP studies,12,13 our data suggest that HoxB4 is a master regulator of ESC differentiation into HSCs. By combining gene-expression and ChIP-Seq data, we identified multiple essential hematopoietic TFs and epigenetic factors that are direct targets of HoxB4 (Figure 4). Among these regulators, roles of the epigenetic factors in early hematopoiesis are much less understood compared with the TFs. Because epigenetic regulation of gene expression plays a pivotal role in many developmental systems, future studies of these epigenetic factors will unveil the role of HoxB4 in shaping the epigenomes of HSCs.
In summary, the results of our study demonstrate how comprehensive analysis of genome-wide datasets can provide novel insights into the transcriptional control of hematopoietic stem/progenitor cells. Given that HoxB4 regulates genes with a broad range of biologic functions, the characterization of these novel targets should unveil new aspects of HoxB4 function during hematopoiesis.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank members of the Tan and Zavazava laboratories for helpful discussions.
This study was supported by a March of Dimes Basil O'Connor Starter Scholar Research Award (to K.T.), the National Institutes of Health (grants R01HL073015 and R01NS074987 to N.Z.), and a Veterans Affairs Merit Award (1I01BX001125 to N.Z.).
National Institutes of Health
Authorship
Contribution: R.F., N.Z., and K.T. designed the experiments; all authors acquired the data; R.F., P.G., B.S., C.C., T.M., and K.T. analyzed the data; K.T. wrote the manuscript; and R.F., S.B., P.G., N.Z., and K.T. reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kai Tan, 2294 Carver Biomedical Research Bldg, 285 Newton Rd, Iowa City, IA 52242; e-mail: kai-tan@uiowa.edu.