Abstract
Leukemic transformation (LT) of myeloproliferative neoplasms (MPNs) is associated with a poor prognosis and resistance to therapy. Although previous candidate genetic studies have identified mutations in MPN patients who develop acute leukemia, the complement of genetic abnormalities in MPN patients who undergo LT is not known nor have specific molecular abnormalities been shown to have clinical relevance in this setting. We performed high-throughput resequencing of 22 genes in 53 patients with LT after MPN to characterize the frequency of known myeloid mutations in this entity. In addition to JAK2 and TET2 mutations, which occur commonly in LT after MPN, we identified recurrent mutations in the serine/arginine-rich splicing factor 2 (SRSF2) gene (18.9%) in acute myeloid leukemia (AML) transformed from MPNs. SRSF2 mutations are more common in AML derived from MPNs compared with LT after myelodysplasia (4.8%) or de novo AML (5.6%), respectively (P = .05). Importantly, SRSF2 mutations are associated with worsened overall survival in MPN patients who undergo LT in univariate (P = .03; HR, 2.77; 95% CI, 1.10-7.00) and multivariate analysis (P < .05; HR, 2.11; 95% CI, 1.01-4.42). These data suggest that SRSF2 mutations contribute to the pathogenesis of LT and may guide novel therapeutic approaches for MPN patients who undergo LT.
Introduction
Myeloproliferative neoplasms (MPNs) are chronic, clonal hematopoietic stem cell disorders and include polycythemia vera (PV), essential thrombocytosis (ET), and primary myelofibrosis (PMF).1 These diseases individually carry a risk of morbidity and mortality, including thrombosis, bleeding, and progressive BM fibrosis. Importantly, a substantial proportion of patients with PV, ET, or PMF transform to acute myeloid leukemia (AML). Leukemic transformation (LT) from a pre-existing MPN carries a dismal prognosis.2 Somatic mutations have been identified in most patients with MPN; however, the specific genetic events, which contribute to LT from a pre-existing MPN, are less well understood. Several groups have identified disease alleles in a subset of patients with MPN who undergo LT, including mutations in TET2,3 IDH1/2,4,5 RUNX1,6 WT1, TP53,7 CBL, and NRAS,8 as well as deletions that involve IKZF19 . In addition, high-resolution single nucleotide polymorphism (SNP) analysis has identified several novel regions of gain and loss (chromosome 7q,16q, 19p, and 21q) and focal alterations in new candidate genes (SH2B2, CUTL1, PIN1, ICAM1, CDC37, ERG) in AML cases derived from a pre-existing MPN.10
Although studies have identified individual genes that may predict for increased risk of LT in patients with MPN when mutated,4,11 no studies to date have performed comprehensive sequencing of candidate genes in patients with LT to thoroughly identify the genetic abnormalities present at leukemic state and to identify specific mutations with prognostic significance in this disease entity. Recently, Yoshida et al identified recurrent somatic alterations in multiple genes encoding members of the spliceosome.12 On the basis of this discovery, we performed a comprehensive molecular characterization of a cohort of patients with LT from MPNs by sequencing the commonly mutated spliceosomal genes (SF3B1, U2AF1, SRSF2, and ZRSR2) in addition to genes known to be frequently mutated in patients with chronic phase MPNs and de novo AML. This allowed us to determine the frequency of somatic alterations in known myeloid disease alleles in patients with MPN with LT and to identify recurrent somatic mutations in the serine/arginine-rich splicing factor 2 (SRSF2) gene in LT after MPN. Our data suggest that SRSF2 mutations are associated with adverse outcomes in LT after MPN, implicating spliceosome mutations in the pathogenesis of LT after MPN.
Methods
Patients
DNA was isolated from peripheral blood and/or BM samples from 53 patients with AML transformed from an antecedent MPN, 54 patients with de novo AML, and 42 patients with AML transformed from myelodysplastic syndrome (MDS). Of the 53 patients with LT of an MPN, 17 samples from the chronic phase MPN state were also available for analysis from the same patient who subsequently transformed to AML. We also performed mutational analysis on 38 patients with chronic phase primary myelofibrosis. Approval was obtained from the institutional review boards at the MD Anderson Cancer Center and at Memorial Sloan-Kettering Cancer Center for these studies, and informed consent was provided according to the Declaration of Helsinki. Patient details are in supplemental Tables 1 through 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Cytogenetic classification was performed according to Slovak et al.13
Sequence analysis
DNA resequencing of all coding exons of known mutations in JAK2, MPL, IDH1, IDH2, FLT3, c-KIT, K/N/H-Ras, U2AF1, SF3B1, and SRSF2 and full-length sequencing of GATA2, RUNX1, EZH2, WT-1, DNMT3a, TP53, TET2, ASXL1, PTEN, and ZRSR2 were performed in all secondary AML (sAML) and paired MPN samples. All DNA samples were whole genome amplified with the use of Ø29 polymerase to ensure sufficient material was available for sequence analysis. Primers sequences, target regions, and PCR conditions are listed in supplemental Table 1. PCR amplicons were all smaller than 500 bp, and overlapping PCR amplicons were designed for all exons larger than 400 bp to ensure complete coverage. For each PCR reaction, 20 ng of genomic DNA was used for PCR amplification, followed by magnetic bead purification (SPRI; Agencourt Bioscience) and bidirectional sequencing with the use of ABI 3730 capillary sequencers (Agencourt Bioscience). Bidirectional sequence traces were analyzed for missense and nonsense mutations with the use of Mutation Surveyor (Softgenetics Inc), and all traces were reviewed manually and with Mutation Surveyor for the presence of frameshift mutations. Nonsynonymous alterations not present in dbSNP were annotated as somatic mutations or SNPs based on sequence analysis of matched germline DNA. Nonsynonymous alterations not in dbSNP nor determined to be somatic in paired samples or in published studies were censored with respect to mutational status for that specific gene. All somatic mutations were validated by resequencing nonamplified DNA. Novel frameshift mutations were validated by cloning and sequencing individual isolated colonies (TOPO TA cloning; Invitrogen). Genomic DNA from paired samples was verified to belong to the same patient by mass spectrometry–based genetic fingerprinting assay as described previously.14
Copy number analysis
Copy number status of all genes sequenced in this study was assessed with the use of Affymetrix SNP 6.0 arrays performed on DNA from 5 patients whereby genomic DNA was isolated from paired MPN and AML samples. Data were analyzed using Integrated Genomics Viewer 1.5.64 (www.broadinstitute.org/igv/v1.5).
Statistics
Standard statistical methods were used for parameter comparison, and survival curves were prepared by the Kaplan-Meier method and compared by the log-rank test. Proportional hazards regression was used for multivariate analysis. P values < .05 were considered significant. The MedCalc statistical package (MedCalc Version 12.2.1 software) was used for all computations.
Results
Mutational spectrum of AML derived from MPNs
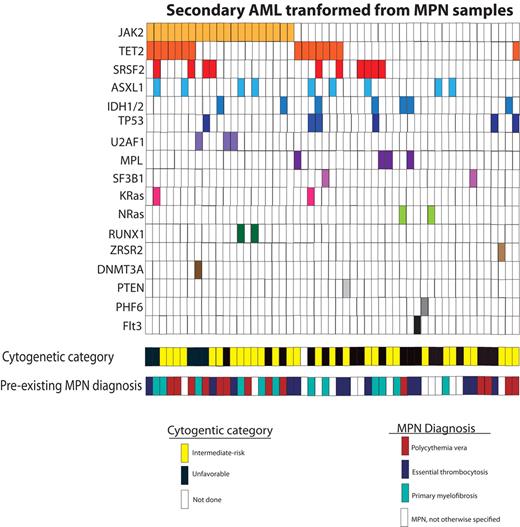
Resequencing of genes known to be mutated in myeloid malignancies in patients with AML secondary to MPNs found that mutations in JAK2, TET2, SRSF2, and ASXL1 were the most commonly mutated genes in this subset of disease (Table 1). These mutations were not exclusive, such that they commonly co-occurred with one another (Figure 1; supplemental Figure 1), although there were no statistically significant co-occurrences among these genes (supplemental Table 5). Notably, several genes commonly mutated in de novo AML, including FLT315 and DNMT3a,16,17 were rarely mutated in AML derived from MPNs, consistent with a divergent mechanism of LT in sAML compared with de novo AML
Frequency of mutations occurring in AML derived from MPNs (n = 53 patients with sAML)
| Gene . | Mutational frequency, % (n/N) . |
|---|---|
| JAK2 | 39.6 (21/53) |
| TET2 | 28.3 (15/53) |
| SRSF2 | 18.9 (10/53) |
| ASXL1 | 17.0 (9/53) |
| IDH1/2 | 13.2 (7/53) |
| TP53 | 11.3 (6/53) |
| MPL | 7.5 (4/53) |
| U2AF1 | 5.7 (3/53) |
| SF3B1 | 3.8 (2/53) |
| KRas | 3.8 (2/53) |
| RUNX1 | 3.8 (2/53) |
| NRas | 3.8 (2/53) |
| DNMT3A | 1.9 (1/53) |
| ZRSR2 | 1.9 (1/53) |
| PTEN | 1.9 (1/53) |
| PHF6 | 1.9 (1/53) |
| FLT3 | 1.9 (1/53) |
| Gene . | Mutational frequency, % (n/N) . |
|---|---|
| JAK2 | 39.6 (21/53) |
| TET2 | 28.3 (15/53) |
| SRSF2 | 18.9 (10/53) |
| ASXL1 | 17.0 (9/53) |
| IDH1/2 | 13.2 (7/53) |
| TP53 | 11.3 (6/53) |
| MPL | 7.5 (4/53) |
| U2AF1 | 5.7 (3/53) |
| SF3B1 | 3.8 (2/53) |
| KRas | 3.8 (2/53) |
| RUNX1 | 3.8 (2/53) |
| NRas | 3.8 (2/53) |
| DNMT3A | 1.9 (1/53) |
| ZRSR2 | 1.9 (1/53) |
| PTEN | 1.9 (1/53) |
| PHF6 | 1.9 (1/53) |
| FLT3 | 1.9 (1/53) |
No mutations were seen in C-Kit, EZH2, WT1, or HRas.
Distribution of mutations among patients with AML derived from a MPN. Pre-exisitng MPN diagnosis and cytogenetic risk category as defined by Slovak et al13 is listed as well.
Distribution of mutations among patients with AML derived from a MPN. Pre-exisitng MPN diagnosis and cytogenetic risk category as defined by Slovak et al13 is listed as well.
Mutations in spliceosomal genes in patients with LT of MPNs and MDS
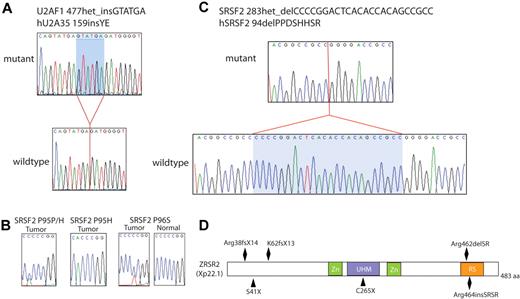
Given that mutations in spliceosomal genes have previously been reported by Yoshida et al12 to be rare in patients with de novo AML and chronic MPNs, we investigated the mutational frequency of mutations in spliceosome components in patients with chronic MPNs, de novo AML, and LT of MPNs and MDS (Table 2). This analysis found that mutations in SRSF2 are more frequent in patients with LT of MPNs compared with patients with de novo AML, chronic MPNs, or LT of MDS (P = .05, 2-tailed, Fisher exact test). Although recent studies have identified recurrent somatic mutations in the spliceosome in myeloid disorders, including SF3B1 in refractory anemia with ringed sideroblasts,12,18 mutations in U2AF1, SF3B1, and ZRSR2 were relatively uncommon in patients with LT of MPNs, occurring in 3.8%, 5.7%, and 1.8% of transformed MPN patients, respectively (Table 2). In addition, our analysis identified previously unreported somatic mutations in U2AF1, SRSF2, and ZRSR2 in patients with MPN, MDS, and sAML or de novo AML (Figure 2). Previous studies identified recurrent heterozygous missense mutations in U2AF1 exclusively at amino acid S34 or Q157 and SRSF2 mutations as heterozygous mutations at amino acid P95 only.12 We also identified rare mutations in U2AF1 occurring as in-frame insertions near one of the regions of recurrent mutation (Figure 2A) as well as a somatic missense mutation in SRSF2 at amino acid 96, adjacent to the previously reported mutated residue, a homozygous mutation in SRSF2 at residue 95 in a de novo AML sample (Figure 2B), a heterozygous in-frame deletion in SRSF2 (Figure 2C), and multiple novel mutations in ZRSR2 (Figure 2D). As noted previously by Yoshida et al, mutations in genes encoding members of the spliceosome were mutually exclusive regardless of diagnosis.12
Frequency of splicing factor mutations in MPNs, AML derived from MPNs, de novo AML, and AML derived from MDSs
| Disease . | Mutational frequency, % . | |||
|---|---|---|---|---|
| SF3B1 . | U2AF1 . | ZRSR2 . | SRSF2 . | |
| Primary myelofibrosis (n = 38) | 5.3 | 2.6 | 5.6 | 2.6 |
| De novo AML (n = 54) | 3.7 | 1.9 | 5.6 | 5.6 |
| Secondary AML from MDS (n = 42) | 4.8 | 9.5 | 2.4 | 4.8 |
| MPNs that later transformed to AML (n = 17) | 0 | 0 | 0 | 11.8 |
| Secondary AML from MPNs (n = 53) | 3.8 | 5.7 | 1.8 | 18.9* |
| Disease . | Mutational frequency, % . | |||
|---|---|---|---|---|
| SF3B1 . | U2AF1 . | ZRSR2 . | SRSF2 . | |
| Primary myelofibrosis (n = 38) | 5.3 | 2.6 | 5.6 | 2.6 |
| De novo AML (n = 54) | 3.7 | 1.9 | 5.6 | 5.6 |
| Secondary AML from MDS (n = 42) | 4.8 | 9.5 | 2.4 | 4.8 |
| MPNs that later transformed to AML (n = 17) | 0 | 0 | 0 | 11.8 |
| Secondary AML from MPNs (n = 53) | 3.8 | 5.7 | 1.8 | 18.9* |
P < .05 compared with frequency in all other disease (2-tailed, Fisher exact test).
Mutations found with TA cloning. (A) TA cloning found a novel, heterozygous in-frame insertion of 2 amino acids in the region of the previously described recurrent mutation in U2AF1. (B) In addition, a single occurrence of a somatic homozygous SRSF2 P95H mutation was seen as well as a novel missense SRSF2 P96S mutation adjacent to the previously described recurrent heterozygous point mutation at codon P95. (C) A novel, heterozygous 24 nucleotide in-frame deletion in SRSF2 was also found. (D) Multiple previously undescribed mutations were also found in ZRSR2, including several nonsense mutations, N-terminal insertions/deletions that resulted in premature stop codons, as well as in-frame insertions/deletions in the Arginine-serine rich domain.
Mutations found with TA cloning. (A) TA cloning found a novel, heterozygous in-frame insertion of 2 amino acids in the region of the previously described recurrent mutation in U2AF1. (B) In addition, a single occurrence of a somatic homozygous SRSF2 P95H mutation was seen as well as a novel missense SRSF2 P96S mutation adjacent to the previously described recurrent heterozygous point mutation at codon P95. (C) A novel, heterozygous 24 nucleotide in-frame deletion in SRSF2 was also found. (D) Multiple previously undescribed mutations were also found in ZRSR2, including several nonsense mutations, N-terminal insertions/deletions that resulted in premature stop codons, as well as in-frame insertions/deletions in the Arginine-serine rich domain.
Sequencing of paired samples
We next performed mutational analysis of paired samples from 17 patients with chronic phase MPN who subsequently transformed to AML. This analysis found multiple genetic differences between the chronic MPN state and the leukemic state within individual patients (Figure 3A). Mutations in genes outside of the JAK-STAT pathway (not in JAK2 or MPL) were significantly enriched in patients at the time of LT compared with the chronic MPN state (average of 1.2 mutations in LT vs 0.6 mutations/sample in chronic phase; P < .02, Mann-Whitney U test). This included acquisition of a variety of different mutations, including mutations in TP53 (28.6% of paired samples) and KRas (14.3% of paired samples). Although clonal analysis could not be performed in these patients, the paired mutational data here suggest the possibility of multiple clones with distinct mutational genotypes within individual patients. Interestingly, mutations in SRSF2, when present in the leukemic state, could always be identified in the chronic MPN sample, suggesting that SRSF2 mutations may either be acquired early in disease pathogenesis and/or are required for LT. Of note, all mutations found in SRSF2 in this study occurred as heterozygous mutations with the exception of a single homozygous SRSF2 P95H mutation in a de novo AML sample (Figure 2B).
multiple genetic abnormalities at LT found with comprehensive sequencing. Comprehensive sequencing of paired MPN samples that later transformed to AML found multiple genetic abnormalities at transformation as shown by (A) paired MPN/AML mutational diagram as well as (B-D) 3 illustrative cases. For instance, gain of TET2 and KRas mutations (B), loss of JAK2 mutation and acquisition of IDH2 and TP53 mutations (C), and loss of the JAK2 mutation but acquisition of activating mutation in FLT3 could all be observed by comparing chronic MPN and leukemic states. SRSF2 mutations when present in the AML state were always present in the paired antecedent MPN sample. Mutations in c-kit, EZH2, WT1, KRas, HRas, IDH1, U2AF1, SF3B1, ZRSR2, PTEN, RUNX1, and DNMT3a were not seen in either state in this set of patients and hence are not displayed here.
multiple genetic abnormalities at LT found with comprehensive sequencing. Comprehensive sequencing of paired MPN samples that later transformed to AML found multiple genetic abnormalities at transformation as shown by (A) paired MPN/AML mutational diagram as well as (B-D) 3 illustrative cases. For instance, gain of TET2 and KRas mutations (B), loss of JAK2 mutation and acquisition of IDH2 and TP53 mutations (C), and loss of the JAK2 mutation but acquisition of activating mutation in FLT3 could all be observed by comparing chronic MPN and leukemic states. SRSF2 mutations when present in the AML state were always present in the paired antecedent MPN sample. Mutations in c-kit, EZH2, WT1, KRas, HRas, IDH1, U2AF1, SF3B1, ZRSR2, PTEN, RUNX1, and DNMT3a were not seen in either state in this set of patients and hence are not displayed here.
In addition to assessing for mutations associated with LT, we assayed for alterations in allelic copy number at LT of MPNs through analysis of SNP arrays in 5 paired MPN/AML samples. Of the 22 genes sequenced in the paired MPN/AML samples, the only gene that differed in copy number in the AML state compared with the MPN state was RUNX1 that was deleted at LT in one patient without a RUNX1 mutation (supplemental Figure 2).
Outcomes associated with common genetic alterations in LT of MPNs
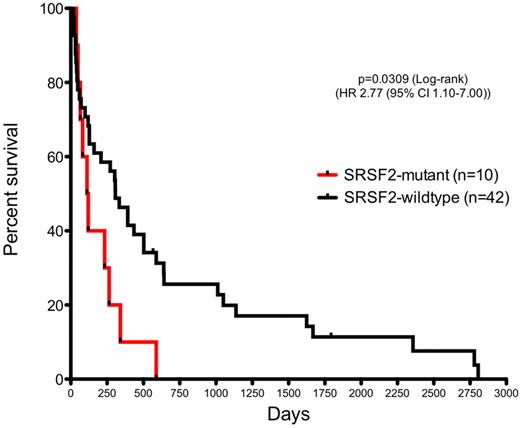
Clinical data were available for 52 of 53 patients with LT of MPNs. Univariate analysis of the 4 most commonly mutated genes found that mutations in SRSF2, but not in JAK2, TET2, or ASXL1, were associated with worsened overall survival (OS) in patients with AML derived from MPNs (P = .03; HR, 2.77; 95% CI, 1.10-7.00; Figure 4). Patients with AML with SRSF2 mutations had a median OS of 116 days (range, 38-588 days) compared with a median survival of 307 days (range, 19-2806 days) in patients wild type for SRSF2 (median, 19-2806 days). The adverse association of SRSF2 mutations to OS was independent of age and cytogenetic risk classification in multivariate analysis (P = .049; HR, 2.11; 95% CI, 1.01-4.42). Other commonly mutated genes (JAK2, TET2, and ASXL1) did not have significant effect on OS (supplemental Figure 3). Only 11 of 52 patients with LT of MPNs included in this analysis achieved complete remission, and only 1 of these 11 patients was SRSF2 mutant. Of note, no difference in the distribution of preceding MPN diagnosis was observed between SRSF2 mutant and SRSF2 wild-type patients who subsequently transformed to AML (supplemental Table 3).
Mutations in SRSF2 are associated with worsened OS in AML derived from MPNs.
Discussion
The recent discovery of a series of genetic events in patients with myeloid malignancies has led to an interest in identifying genetic markers that may be associated with LT. Recently, mutations in genes encoding members of the spliceosome have been identified through whole exome/genome sequencing studies in patients with MDS and AML.12,18,19 Mutational profiling of patients with LT of MPNs here indicate that SRSF2 mutations, but not mutations in other spliceosome factors, are seen at high frequency in patients with MPN who transform to AML. This data, combined with the recent data implicating SF3B1 mutations specifically in refractory anemia with ringed sideroblasts,12,18 suggest that different mutations in the spliceosome are associated with distinct clinical phenotypes and with different roles in myeloid transformation.
Mutations in SRSF2 were found in 18.9% of patients with AML transformed from MPN, which surpassed the frequency of mutations in other genes recently described to be frequently mutated in LT of MPNs, including mutations in TP537 and IDH1/2.4,5 Although TP53 mutations were exclusively seen at LT (Figure 3A), the frequency of TP53 alterations seen in this series of patients (Table 1) was less than that described in one recent candidate gene sequencing series.7 Likewise, although mutations in IDH1/2 were identified in a subset of patients with AML after MPN (13.2%), they were not more common in patients with LT after MPNs than has been previously reported in patients with de novo AML (16%-30%),20-23 suggesting that they probably do not represent a unique transforming event among patients with MPN. In contrast, our data suggest that mutations in SRSF2, which are found in < 5% of patients with de novo AML or chronic phase MPNs,12 are specifically enriched at LT of MPNs. Moreover, genetic profiling of known genes mutated in MPN/AML suggests that there are multiple diverse events present at LT of MPNs (Figures 1 and 3; supplemental Figure 1). In addition, the finding that multiple patients studied here have no identifiable somatic mutations in commonly mutated genes at the MPN or leukemic state (eg, PMF sample no. 6 in Figure 3A), clearly suggests the need for further genomic discovery efforts in patients with this high-risk myeloid neoplasm.
Mutations in SRSF2, as reported previously, occur most commonly as recurrent heterozygous point mutations at codons 95 or 96. The functional implication of this genetic alteration is not clear. Although initial functional studies of recurrent heterozygous missense mutations in U2AF1 suggest that these are dominant-negative alterations that effect splicing,12 the distinct clinical associations of mutations in genes encoding splicing proteins among various myeloid malignancies indicate the need for evaluation of each of these genetic events individually in different oncologic contexts.
The observations that SRSF2 mutations are associated with adverse outcome in AML after MPN and can be detected in patients in the chronic phase of MPN who transform to AML suggest that SRSF2 mutations may predict for disease progression in MPNs and may represent an important event leading to subsequent acute LT. In contrast, other mutations common to LT of MPNs, including mutations in TET2 and ASXL1, did not hold prognostic significance at LT (supplemental Figure 3). Future clinical and biologic studies will need to elucidate the specific role of SRSF2 mutations in MPN/AML pathogenesis and the prognostic significance of SRSF2 mutations in chronic phase PV, ET, and PMF. We predict that subsequent studies will identify additional disease alleles in patients with AML after MPN of biologic, clinical, and therapeutic relevance.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health (grants U54CA143798-01 and 1R01CA138234-01, R.L.L.; and K08CA160647-01, O.A.-W.); the Physical Sciences Oncology Center (R.L.L.); the Peter Solomon Fund at Memorial Sloan-Kettering Cancer Center (R.L.L.); the MPN Research Foundation (R.L.L.); the Starr Cancer Consortium (R.L.L.); the Howard Hughes Medical Institute (R.L.L.); the National Natural Science Foundation of China (grant 81170490, S.-J.Z.); and the Charles A. Dana Foundation Clinical Scholars Program at Memorial Sloan-Kettering Cancer Center (R.R.). O.A.-W. is a Basic Research Fellow of the American Society of Hematology.
National Institutes of Health
Authorship
Contribution: S.-J.Z., R.R., T.M., R.L.L., O.A.-W., and S.V. designed the study; T.M., N.M., C.H., and S.V. collected samples and provided genetic and clinical annotation; S.-J.Z., R.R., J.P., A.K., A.H., and O.A-W. performed sequence analysis and validated mutations; T.H. and M.G. helped to perform statistical analysis of the data; H.K. collected samples and provided genetic and clinical annotations; and R.R., R.L.L, O.A.-W., and S.V. wrote the manuscript with assistance from J.P. and T.M.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Srdan Verstovsek, Department of Leukemia, MD Anderson Cancer Center, Unit 428, 1515 Holcombe Blvd, Houston, TX 77030; e-mail: sverstov@mdanderson.org; or Omar Abdel-Wahab, 1275 York Ave, Box 20, Leukemia Service, Department of Medicine, Human Oncology and Pathogenesis Program, Memorial-Sloan Kettering Cancer Center, New York, NY 10065; e-mail: abdelwao@mskcc.org.
References
Author notes
S.-J.Z. and R.R. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal