Abstract
The interactions between hematopoietic cells and the bone marrow (BM) microenvironment play a critical role in normal and malignant hematopoiesis and drug resistance. These interactions within the BM niche are unique and could be important for developing new therapies. Here, we describe the development of extramedullary bone and bone marrow using human mesenchymal stromal cells and endothelial colony-forming cells implanted subcutaneously into immunodeficient mice. We demonstrate the engraftment of human normal and leukemic cells engraft into the human extramedullary bone marrow. When normal hematopoietic cells are engrafted into the model, only discrete areas of the BM are hypoxic, whereas leukemia engraftment results in widespread severe hypoxia, just as recently reported by us in human leukemias. Importantly, the hematopoietic cell engraftment could be altered by genetical manipulation of the bone marrow microenvironment: Extramedullary bone marrow in which hypoxia-inducible factor 1α was knocked down in mesenchymal stromal cells by lentiviral transfer of short hairpin RNA showed significant reduction (50% ± 6%; P = .0006) in human leukemic cell engraftment. These results highlight the potential of a novel in vivo model of human BM microenvironment that can be genetically modified. The model could be useful for the study of leukemia biology and for the development of novel therapeutic modalities aimed at modifying the hematopoietic microenvironment.
Introduction
The relevance of the bone marrow (BM) microenvironment in regulating hematopoietic stem cell (HSC) behavior has only recently been established.1-5 The maintenance of HSC quiescence and normal hematopoiesis requires complex bidirectional interactions between the BM niche and HSCs.6-8 Moreover, much evidence supports the idea that the BM microenvironment also plays a pivotal role in the initiation and propagation of leukemia.9-11 Leukemic cells have been shown to hijack the homeostatic mechanisms of normal HSCs and take refuge within the BM niche. This mechanism is pivotal during chemotherapy and contributes to disease relapse.12,13 Although human HSCs can be genetically modified and transplanted into immunodeficient mice, the BM micro-environment is not transplantable. Numerous studies in patients and mice undergoing bone marrow transplantation have failed to demonstrate consistent engraftment of donor BM stroma cells.14-16 A better understanding of the BM niche will not only improve our understanding of HSC self-renewal and hematopoiesis in general but also accelerate the development of new therapeutic modalities and targeted agents for the therapy of hematopoietic malignancies. Although the concept of a BM “niche” was formulated in 1978, it remains largely unidentified owing to technical limitations.13,17-19 Currently, the xenotransplant NOD/SCID and NOD/SCID/IL-2rγnull mouse repopulation assays are the most widely used and relevant readout systems for studying human normal and malignant hematopoiesis. However, one of the major restrictions of these models is that the human cells engraft in a murine BM environment that may not reflect the interactions between the HSCs and the BM microenvironment in humans. The inadequacy of the presently used murine systems has recently been highlighted by the finding of distinct genetic abnormalities in the stromal cells from patient with myelodysplatic syndromes and acute myeloid leukemias.20,21 Mesenchymal stromal cells (MSCs) have been shown to serve as scaffolds for the formation of stem cell niches in BM and exert both positive and negative regulatory effects on the self-renewal, proliferation, and differentiation of HSCs22,23 However, transplanted human MSCs do not efficiently or consistently engraft at murine sites of hematopoiesis even after intra-BM injection, perhaps because they are competing with a functional murine microenvironment. In the past 2 decades, researchers have attempted to mimic the native BM environment in 2-dimensional culture systems by adding the proper cytokines and growth factors to cell cultures, coculturing HSCs with stromal cells, or both.24-26 However, this method does not recapitulate the development and self-renewal of hematopoietic or leukemic stem cells. Recently, several 3-dimensional bone-tissue–like models have been developed with scaffolds that better mimic the physiologic in vivo situation27-29 ; however, assembly of such bone-tissue analogs is challenging owing to the complexity of the BM microenvironment. The BM environment comprises various kinds of nonhematopoietic cells, including adipocytes, osteocytes, chondrocytes, fibroblasts, and macrophages. MSCs have been shown to serve as precursors of most types of these cells.23 Previously, our group reported that endothelial colony-forming cells (ECFCs), which are blood- or vasculature-derived endothelial progenitor cells, characterized by robust proliferative potential, also can form perfused long-lasting blood vessels in vivo.30 Thus, we hypothesized that we could develop ectopic, extramedullary bones and BM using MSCs and ECFCs.
In this study, we developed a novel in vivo extramedullary bone model in NOD/SCID/IL-2rγnull mice that establishes a human BM microenvironment using human BM-derived MSCs and human peripheral blood–derived ECFCs. Because the MSCs and ECFCs can be genetically modified, this model also allows the identification and modification of genes critical for leukemia development and maintenance of normal and leukemic HSCs.
Methods
Mice
NOD/SCID/IL-2rγnull mice were purchased from The Jackson Laboratory. All animal work was done in accordance with a protocol approved by the institutional animal care and use committee at The University of Texas MD Anderson Cancer Center.
Isolation and expansion of MSCs
Human BM was diluted immediately after harvest in αMEM (Cellgro) supplemented with 10% pooled human platelet lysate31 (pHPL; supplied by D.S.), 2mM l-glutamine, 100 U/mL penicillin, and 100 mg/mL streptomycin (Sigma-Aldrich). The BM mononuclear cell content was analyzed by automated blood count (Beckman Coulter), and cells were seeded at a density of 5 × 104 mononuclear cells/cm2 in tissue culture flasks and cultured at 37°C in an atmosphere of 5% CO2 at 95% humidity. Nonadherent cells were removed by completely changing the medium after 3 days, and the adherent cells were continuously cultured. The cultures were fed twice weekly by replacing 30% of the medium with fresh supplemented medium. Cells were harvested before reaching confluence by applying 0.25% trypsin and 1mM EDTA (Invitrogen). MSC aliquots were frozen after primary culture and stored in liquid nitrogen. For large-scale expansion, the MSCs derived from primary culture were seeded in αMEM/10% pHPL at a density of 300 cells/cm2 in 4-layered cell factories. The culture medium was changed (30% replaced) twice weekly, and MSCs were harvested on days 11 to 15 by trypsinization.
Induction of MSC differentiation into the mesenchymal lineage
MSCs were grown to 90% confluence in αMEM/10% pHPL and then moved to NH OsteoDiff medium or NH Adipo-Diff medium (Miltenyi Biotec) to induce differentiation. Each differentiation medium was changed every 3 days. To confirm that the MSCs differentiated into osteocytes and adipocytes, they were stained with alizarin red (to detect calcium deposits indicative of osteocytes) and oil red O (to detect lipids indicative of adipocytes). To induce chondrogenic differentiation, MSC pellets were cultured in 1 mL of NH Chodro-Diff medium (Miltenyi Biotec) in 15-mL Falcon tubes for 21 days. Then, the pellets were fixed in 4% paraformaldehyde and embedded in paraffin. Sections (5 μm) were stained with Alcian blue and analyzed by light microscopy.23 MSCs before and after the inductions also were harvested for RNA extraction. Real-time RT-PCR was performed on these samples to demine the expression of lipoprotein lipase, collagen type II α 1, and osteocalcin during the differentiation. Primer information was described in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Knockdown of HIF1α in MSCs
Lentiviral infections were carried out according to the standard procedures for silencing experiments. In brief, 293T cells were cotransfected with pMD2.G and psPAX2 (Addgene), along with either a specific hypoxia-inducible factor 1α (HIF1α)-short hairpin (sh)RNA (pGIPZ-HIF1α-shRNA) or a nonsilencing (NS) shRNA control (pGIPZ-NS-shRNA) lentiviral construct (catalog no. RHS4531, source ID V2LHS_132150, Open Biosystems), using JetPrime transfection reagent (Polyplus) according to the manufacturer's protocol. The transfection medium was replaced after 12 hours with fresh Dulbecco modified Eagle medium (Cellgro)/10% fetal bovine serum (FBS), and 48 hours later the viral supernatants were collected and concentrated using Centricon Plus-70 filter units (Millipore). The MSCs were infected in the presence of 8 μg/mL polybrene (Sigma-Aldrich) to promote the lentiviral infections. Two days after infection, stably transduced MSCs were selected with 2 μg/mL puromycin during a 2-week period, resulting in a homogeneous population of 100% turbo GFP-positive (GFP+) cells.
Isolation and expansion of endothelial colony-forming cells
Endothelial growth medium (EGM; Lonza) was prepared as suggested by the manufacturer, except that FBS was replaced with 10% pHPL. Human ECFCs were isolated and expanded as described previously.30 In brief, heparinized blood (6 mL) from a healthy donor was directly diluted in EGM/10% pHPL without additional cell separation and seeded into a 75-cm2 culture flask. Nonadherent cells were removed by washing with prewarmed phosphate-buffered saline (PBS) after culturing for 24 hours. Cultures were maintained until we observed the outgrowth of cobblestone-type colonies, defined as ECFCs. The primary culture-derived ECFCs were then expanded in EGM/10% pHPL in two 4-layered cell factories for 2 to 3 weeks.
Flow cytometry
The expression of surface markers on MSCs and ECFCs was analyzed with a flow cytometer (Becton Dickinson) as described previously.32 In brief, cells were trypsinized and washed with PBS. Then, they were stained for 30 minutes at room temperature with fluorochrome-labeled monoclonal antibodies against CD14, CD31, CD34, CD44, CD45, CD73, CD90, CD105, CD114, and CD146 (Becton Dickinson). The appropriate isotype-matched antibodies were used as negative controls. To determine the cell engraftment, extramedullary bones and femurs were cut into pieces and flushed with 2 mL of PBS/5% FBS. Cells were incubated in 1 × red cell lysis buffer at room temperature for 20 minutes, washed with PBS, and stained with fluorochrome-labeled antibodies. Antibodies included anti–mouse CD45, Gr-1, B220 (all eBioscience). and anti–human CD45. DAPI (4,6-diamidino-2-phenylindole) staining was used to exclude the dead cells.
Western blot analysis
Cell lysates were separated on 10% polyacrylamide gels, transferred to nitrocellulose membrane, immunoblotted with HIF1α antibody (Cell Signaling Technology) followed by infrared secondary antibodies (LI-COR Biosciences), and then detected by the Odyssey imaging system (LI-COR Biosciences).
ELISA
SDF-1α levels in culture media were quantified using ELISA (Quantikine Human CXCL12/SDF-1α Immunoassay; R&D Systems) according to the manufacturer's protocol.
In vivo extramedullary bone formation
Human BM-derived MSCs (1.5 × 106) were mixed with 1.5 × 106 human ECFCs in 0.2 mL Matrigel (Millipore) immediately before being subcutaneously injected into the flanks of NOD/SCID/IL-2rγnull mice. Both the MSCs and ECFCs were obtained from the large-scale expansions described in the preceding paragraph, with low passages (1-3). To monitor bone formation in mice, we used animal microcomputed tomography weekly, starting on the fourth week after injection. At each time point, mice were anesthetized and detailed 3-dimensional images of the soft tissue and bone structure were obtained with microcomputed tomography. When a positive signal was observed from the implants by microcomputed tomography, the mice were injected with OsteoSense 750 (PerkinElmer), a fluorescent agent that targets hydroxylapatite, and further scanned with a VisEn FMT 2500 imaging system (PerkinElmer) to generate a tomographic database consisting of the bone structure and fluorescence signal.
Human cord blood mononuclear cell transplantation
The mononuclear cells were separated from human whole cord blood (MD Anderson Cord Blood Bank) by Ficoll-Hypaque (Cellgro) density-gradient centrifugation. After the viability and density of the mononuclear cells were determined, cells were centrifuged for 5 minutes at 1500 rpm, and PBS was added before these cells were injected into the mice (2 × 106 cells/100 μL). Mice were irradiated with 2 Gy under specific pathogen-free conditions 1 day before cell transplantation. The engraftment of human-derived cells in mouse peripheral blood, femurs, and extramedullary bones was determined by flow cytometry, immunofluorescence staining, or both.
Generation of the acute myeloid leukemia model
Human acute myeloid leukemia cell line MOLM13 was purchased from ATCC, infected with lentivirus expressing Luc-GFP, and maintained in RPMI-1640 medium containing 10% FBS. Mice with extramedullary bones were intravenously injected with MOLM13/Luc/GFP acute myeloid leukemia cells at a concentration of 2 × 106 cells/100 μL. Bioluminescence imaging was used to monitor the tumor burden. In brief, mice were anesthetized and imaged noninvasively with an in vivo imaging system (IVIS-200; Xenogen) after injection of the luciferase substrate colenterazine (native; Biotium). Two weeks after the transplantation of MOLM13/Luc/GFP cells, mice were humanely killed by CO2 asphyxiation, and the extent of leukemic infiltration was assessed by both hematoxylin and eosin (H&E; Sigma-Aldrich) staining and staining for GFP+ leukemic cells with an anti-GFP antibody.
Immunohistochemistry, immunofluorescence, and FISH analysis
Fresh tissues collected from mice were fixed in 4% paraformaldehyde and embedded in paraffin. The sections (5 μm) were stained with H&E and analyzed by light microscopy. For immunohistochemical staining, the tissue sections were first incubated with sodium citrate buffer (pH 6.0) for antigen retrieval and incubated for 30 minutes in blocking solution (1 × PBS, 0.5% Tween 20, 0.1% bovine serum albumin, and 10% FBS), followed by an overnight incubation with the primary antibody against GFP (Abcam), pimonidazole (Hypoxyprobe-1; Chemicon), HIF1α (Abcam), or appropriate negative control antibody. The tissue sections were then sequentially incubated with a biotinylated antibody and peroxidase-labeled streptavidin (Dako). The staining was completed by a 5-minute incubation with 3,3′ diaminobenzidine tetrahydrochloride/hydrogen peroxide, resulting in a brown precipitate at the antigen site. For fluorescence staining, slides were stained overnight at 4°C with anti–human collagen type I (Abcam), α smooth muscle actin (αSMA; Abcam), CD31 (Dako), CD19 (Abcam), CD33 (eBioscience), or appropriate isotype controls; then, a secondary Alexa Fluor antibody was added at a 1:500 dilution in diluted blocking buffer and placed on the slide for 1 hour at room temperature. The slides were washed 3 times in PBS-Tween before being stained with the nuclear marker DAPI. The slides were washed 3 more times in PBS-Tween, covered with a glass coverslip over Dako Fluorescent Mounting Media, and left to dry in the dark before the edges were sealed with nail hardener. Spectral images were obtained using a CRi attachment (CRi) on an Ix81 microscope (Olympus) with a disc-scanning unit confocal attachment using Nuance Version 2.8 software (Nuance Communications), and images were analyzed using InForm Version 1.0 software (INFORM). Five images per slide were quantified and averaged at 3 different focal depths within the tissue section. FISH for mouse and human DNA also was performed using Spectrum Red–conjugated mouse X-chromosome probe (DXMit190; ID LABS) and Spectrum Green–conjugated human X-chromosome probe (CEP X DXZ1 Spectrum Green; Abbott Molecular) according to the manufacturers' instructions.
Statistical analysis
Results are shown as the mean ± SEM from 5× independent experiments. Student paired t test was used for statistical comparisons between groups. P values less than .05 were considered statistically significant.
Results
Extramedullary bones developed from human MSCs and ECFCs
To develop an ectopic extramedullary in vivo model of human BM (Figure 1), we first isolated MSCs from normal human BM as described in “In vivo extramedullary bone formation.” After expansion, these MSCs in culture expressed CD44, CD73, CD90, and CD105, but they were negative for macrophage and hematopoietic lineage markers, including CD14, CD31, CD34, and CD45 (supplemental Figure 1Aii). To determine the “stemness” of these MSCs, the cells were induced to differentiate into different mesenchymal lineages with special media (supplemental Figure 1Aiii-iv). These studies confirmed that the human BM-derived MSCs used in our study met the criteria postulated by the Mesenchymal and Tissue Stem Cell Committee of the International Society of Cellular Therapy for MSCs.33
Schema of extramedullary bone and marrow generation. MSCs and ECFCs were isolated from heparinized human BM or peripheral blood through an initial adhesion step and subsequently allowed to proliferate in specific media. Cells mixed with Matrigel were subcutaneously injected into the flanks of NOD/SCID/IL-2rγnull mice, and these cells developed into bonelike tissues with high osteoblastic activity in 8 weeks.
Schema of extramedullary bone and marrow generation. MSCs and ECFCs were isolated from heparinized human BM or peripheral blood through an initial adhesion step and subsequently allowed to proliferate in specific media. Cells mixed with Matrigel were subcutaneously injected into the flanks of NOD/SCID/IL-2rγnull mice, and these cells developed into bonelike tissues with high osteoblastic activity in 8 weeks.
Second, proliferating human ECFCs isolated from normal peripheral blood showed the typical cobblestone morphology of endothelial lineage cells in vitro (supplemental Figure 1Bi). More than 95% of these cells were positive for human EC lineage markers, including CD31 (platelet-endothelial cell adhesion molecule-1), CD144 (vascular endothelial cadherin), and CD146 (melanoma and endothelial cell adhesion molecule). Cultures contained less than 0.1% contamination with CD45+ hematopoietic cells (supplemental Figure 1Bii). As we reported previously, the immune phenotype was unchanged after freezing, thawing, and further culture.30
Next, we mixed human MSCs and ECFCs with Matrigel and injected the mixture subcutaneously into NOD/SCID/IL-2rγnull mice (n > 10). Within 8 weeks after implantation, well-defined vascularized bone-like tissues developed in the flank. Mice were intravenously injected with OsteoSense 750 and scanned in a VISEN FMT 2500 imaging system (PerkinElmer) 24 hours later. After data generation, normalization, and fluorescence tomographic reconstruction, tomographic databases were generated that allowed us to quantify hydroxyapatite in vivo and in real time (supplemental Video 1). Strong fluorescent signals, representing high osteoblast activity, were observed from the MSC-ECFC-Matrigel–derived tissues (Figure 2A). Histologic analysis further revealed that these human cell–derived tissues had a trabecular bone structure similar to normal long bones (Figure 2B). Circulating murine HSCs engrafted into the extramedullary bone marrow. Different types of hematopoietic cells, including erythroid and myeloid cells, lymphocytes, and megakaryocytes, were seen in the bone marrow cavities on the morphologic appearances (Figure 2Ci). Cells also were harvested from the extramedullary bone and analyzed by flow cytometry after staining. Results showed ∼ 39% cells were positive for the myeloid lineage marker Gr-1 and 28% of them were CD45R (B220)+, which belong to lymphoid lineage (Figure 2Cii). Using the hypoxia probe pimonidazole and HIF1α staining,17,34,35 we demonstrated that the extramedullary bones contained discrete hypoxic areas similarly to normal femurs (Figure 2D). Extramedullary bone slides stained positive with anti–human collagen type I, an important matrix protein produced in normal bones, confirming that they were human-derived (Figure 2E). Human vasculatures were found in the extramedullary bones that displayed similar composition as the vessels in normal human bone biopsy. As shown in Figure 2F, human-CD31+ endothelial cells were surrounded by αSMA+ pericytes. Moreover, FISH for mouse and human X chromosome was performed. CD31/αSMA/DAPI and H&E staining were first carried out to define the typical blood vasculature and bone structure area, and then the adjacent slides were used for FISH. Spectral images were obtained using the Cri system (Cambridge Research & Instrumentation), and images were analyzed using InForm Version 1.0 software. Positive signals for human X chromosome were observed in more than 95% cells that composed vasculatures inside extramedullary bone and the bone structure; these results further confirmed the extramedullary bone structures were dominantly derived from human cells (Figure 2G). Taken together, these findings suggest that the MSC-ECFC-Matrigel mixture developed into humanized ectopic bones with a robust hematopoietic environment in vivo.
Establishment a human bone marrow microenvironment in NOD/SCID/IL-2rγnull mice. Human BM-derived MSCs and ECFCs mixed with Matrigel were subcutaneously injected into the flanks of mice to induce the development of extramedullary bones. (A) Eight weeks after implantation, mice were scanned in a VISEN FMT 2500 imaging system to measure osteoblastic activity using OsteoSene 750, and the fluorescence throughout the extramedullary bones was measured. (B) Representative H&E staining (shown at low magnification) shows an overview of the extramedullary bone with the typical bone structures (i). Representative imagines from mouse femur (ii) and human bone biopsy (iii) were displayed as normal bone control. Scale bar represents 1 mm. (C) Higher magnification of the H&E staining shows different kinds of hematopoietic cells with variety of morphologic appearances in the extramedullary bone cavities (i). Cells were flushed from the extramedullary bone, stained with mouse lineage antibodies, and analyzed by flow cytometry (ii). (D) Staining with the hypoxia probe pimonidazole (i-ii) and HIF1α (iii-iv) was performed to determine the presence of hypoxic conditions. Pimonidazole staining was converted to fluorescent (green) signal using the CRi multispectral system. (E) Phase-contrast (i) and confocal fluorescent (ii) images of extramedullary bone stained positive with human collagen type I (COL-1) antibody. (F) Human vasculatures were observed in the extramedullary bones (i) that displayed similar composition as the vessels in normal human bone biopsy (ii). Human CD31+ endothelial cells (red) were surrounded by αSMA+ pericytes (green). (G) CD31/αSMA/DAPI (i) and H&E (ii) staining displayed typical blood vasculature and bone structure, the adjacent slides were used for FISH (iii,iv); green spots indicate positive hybridization on human X chromosome (white arrows). The figures show 1 representative result of at least 5 experiments.
Establishment a human bone marrow microenvironment in NOD/SCID/IL-2rγnull mice. Human BM-derived MSCs and ECFCs mixed with Matrigel were subcutaneously injected into the flanks of mice to induce the development of extramedullary bones. (A) Eight weeks after implantation, mice were scanned in a VISEN FMT 2500 imaging system to measure osteoblastic activity using OsteoSene 750, and the fluorescence throughout the extramedullary bones was measured. (B) Representative H&E staining (shown at low magnification) shows an overview of the extramedullary bone with the typical bone structures (i). Representative imagines from mouse femur (ii) and human bone biopsy (iii) were displayed as normal bone control. Scale bar represents 1 mm. (C) Higher magnification of the H&E staining shows different kinds of hematopoietic cells with variety of morphologic appearances in the extramedullary bone cavities (i). Cells were flushed from the extramedullary bone, stained with mouse lineage antibodies, and analyzed by flow cytometry (ii). (D) Staining with the hypoxia probe pimonidazole (i-ii) and HIF1α (iii-iv) was performed to determine the presence of hypoxic conditions. Pimonidazole staining was converted to fluorescent (green) signal using the CRi multispectral system. (E) Phase-contrast (i) and confocal fluorescent (ii) images of extramedullary bone stained positive with human collagen type I (COL-1) antibody. (F) Human vasculatures were observed in the extramedullary bones (i) that displayed similar composition as the vessels in normal human bone biopsy (ii). Human CD31+ endothelial cells (red) were surrounded by αSMA+ pericytes (green). (G) CD31/αSMA/DAPI (i) and H&E (ii) staining displayed typical blood vasculature and bone structure, the adjacent slides were used for FISH (iii,iv); green spots indicate positive hybridization on human X chromosome (white arrows). The figures show 1 representative result of at least 5 experiments.
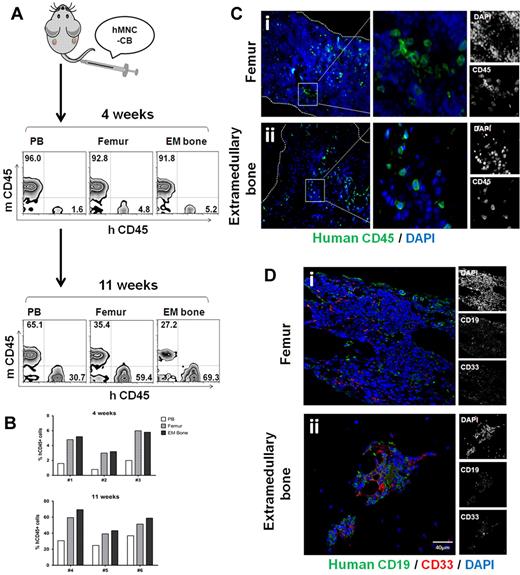
HSC engraft into the extramedullary bones
Next, we investigated whether normal human hematopoietic cells could engraft into the extramedullary bones. As shown in Figure 3A, mononuclear cells isolated from fresh human cord blood, which served as a rich source of hematopoietic stem cells and progenitor cells, were injected into mice with established extramedullary bones (n = 13). Four weeks later, 5 of the mice were killed, and mononuclear cells were harvested from peripheral blood, femurs, and extramedullary bones. Using flow cytometry, 1.5% ± 0.4% of peripheral blood mononuclear cells, 4.6% ± 0.9% of femur mononuclear cells, and 4.8% ± 0.8% of extramedullary bone mononuclear cells were positive for human CD45 (n = 3). Surviving mice were killed after 11 weeks, and 57.1% ± 7.6% of the extramedullary bone mononuclear cells were human CD45+, which was a little higher than 49.7% ± 5.8% in the femur (n = 3). However, no statistical significance was found (Figure 3B). These results indicated that MSC-ECFC-Matrigel–derived extramedullary bones provide a hematopoietic environment that is at least functionally equivalent to the native bones. Moreover, we fixed mouse femurs and extramedullary bones for sections; immunofluorescence staining results further confirmed the human cell engraftments at different time points after transplantation (Figure 3C-D). As reported for human HSC engraftment in mouse femur,36,37 we found ∼ 50% CD19+ and ∼ 30% CD33+ cells, indicative of lymphoid and myeloid engraftment in extramedullary bones.
HSC engraft onto extramedullary bones. (A) Human cord blood mononuclear cells (hMNC-CB) were injected into mice with established extramedullary bones. Mice were killed 4 weeks and 11 weeks after transplantation. Mononuclear cells were harvested from peripheral blood, femurs, and extramedullary bones, and then they were analyzed for human CD45 by flow cytometry. (B) Histograms showed the human CD45 percentage of individual mouse. (C) Histologic sections of femur (i) and extramedullary bones (ii) 4 weeks after transplantation were stained with anti–human CD45 antibody (green). (D) Femur (i) and extramedullary bone (ii) slides of 11-week transplantation mice were stained with anti–human CD19 and CD33. The figures show 1 representative result of 5 experiments.
HSC engraft onto extramedullary bones. (A) Human cord blood mononuclear cells (hMNC-CB) were injected into mice with established extramedullary bones. Mice were killed 4 weeks and 11 weeks after transplantation. Mononuclear cells were harvested from peripheral blood, femurs, and extramedullary bones, and then they were analyzed for human CD45 by flow cytometry. (B) Histograms showed the human CD45 percentage of individual mouse. (C) Histologic sections of femur (i) and extramedullary bones (ii) 4 weeks after transplantation were stained with anti–human CD45 antibody (green). (D) Femur (i) and extramedullary bone (ii) slides of 11-week transplantation mice were stained with anti–human CD19 and CD33. The figures show 1 representative result of 5 experiments.
Leukemic cells engraft into extramedullary bones
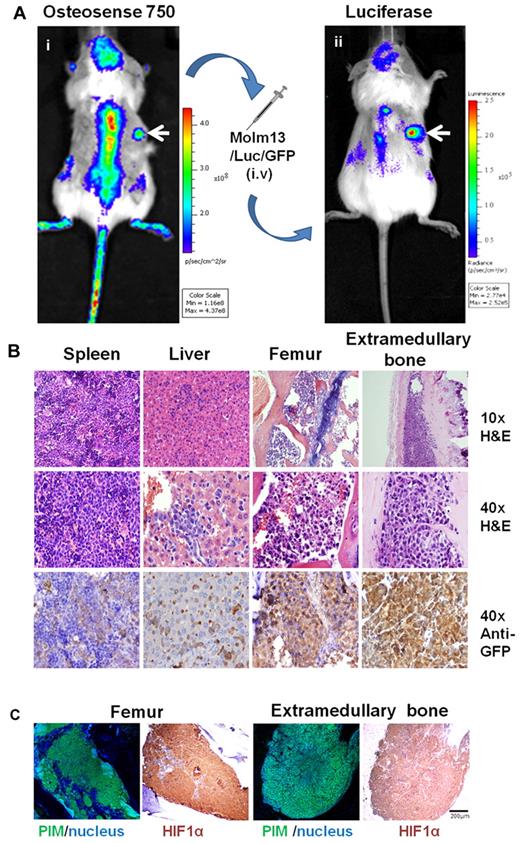
We sought to determine whether our model could be used to study human leukemia engraftment in a human microenvironment. Human MSCs and ECFCs mixed with Matrigel were first injected into mice to induce the formation of extramedullary bones (n > 5). Six weeks later, hydroxyl apatite–positive signals from the implants were captured by an IVIS-200 system (using ICR filters; Figure 4Ai). MOLM13/Luc/GFP acute myeloid leukemia cells were then intravenously transplanted into the mice. The tumor burden was monitored by bioluminescence imaging with the IVIS system. Two weeks after MOLM13/Luc/GFP-cell engraftment, luciferase signals were observed not only in the calvaria and vertebral column but also in the flanks of mice where the MSC-ECFC-Matrigel mixture had been implanted (Figure 4Aii), suggesting that the leukemic cells engrafted into both mouse skeletons and human cell–derived extramedullary bones.
Leukemic cells engraft onto extramedullary bones. (A) MSC-ECFC-Matrigel mixture was injected into the flanks of mice 6 weeks before the leukemic-cell transplantation. (i) Images captured by the IVIS system (using ICR filters) show the osteoblastic activity of the extramedullary bones (white arrow). Two weeks after MOLM13/Luc/GFP cell transplantation, the mice were imaged in the IVIS system after injection of the luciferase substrate. (ii) A positive luciferase signal was observed in both mouse skeletons and extramedullary bones (white arrows indicate the extramedullary bones). (B) Histologic sections of spleen, liver, femur, and extramedullary bones were stained with H&E or an anti-GFP antibody. (C) Staining with the hypoxia probe pimonidazole and HIF1α also was performed to determine the presence of hypoxic conditions after leukemia engraftment. Pimonidazole (PIM) staining was converted to fluorescent (green) signal using the CRi multispectral system. The results shown are representative of at least 5 separate experiments.
Leukemic cells engraft onto extramedullary bones. (A) MSC-ECFC-Matrigel mixture was injected into the flanks of mice 6 weeks before the leukemic-cell transplantation. (i) Images captured by the IVIS system (using ICR filters) show the osteoblastic activity of the extramedullary bones (white arrow). Two weeks after MOLM13/Luc/GFP cell transplantation, the mice were imaged in the IVIS system after injection of the luciferase substrate. (ii) A positive luciferase signal was observed in both mouse skeletons and extramedullary bones (white arrows indicate the extramedullary bones). (B) Histologic sections of spleen, liver, femur, and extramedullary bones were stained with H&E or an anti-GFP antibody. (C) Staining with the hypoxia probe pimonidazole and HIF1α also was performed to determine the presence of hypoxic conditions after leukemia engraftment. Pimonidazole (PIM) staining was converted to fluorescent (green) signal using the CRi multispectral system. The results shown are representative of at least 5 separate experiments.
To further analyze the leukemia burden, mice were humanely killed; the extramedullary bones and femurs were immediately isolated and imaged in the IVIS system. The luciferase signals confirmed that the leukemia cells engrafted into the extramedullary bones (supplemental Figure 2). The amount of leukemic infiltrates in different tissues was assessed by both H&E staining and antibody staining for GFP-positive leukemic cells. H&E staining revealed the dissemination of leukemic cells into the spleen and liver; all of the mouse femurs and extramedullary bones were abundantly populated with leukemic cells (Figure 4B). Moreover, as shown in Figure 4C, the leukemic cell engraftment resulted in widespread severe hypoxia in both femurs and extramedullary bones. These data suggest that the MSC-endothelial progenitor cell-Matrigel–derived ectopic extramedullary bones could be a useful model for investigating the interactions between leukemia cells and the BM microenvironment.
Leukemia cell engraftment can be altered in extramedullary bones by genetically modifying MSCs
To evaluate whether this extramedullary bone model can be used to investigate factors critical for leukemic hematopoiesis, we genetically manipulated the MSCs. MSCs from a large-scale expansion were infected with either control (NS) or HIF1α-specific shRNA lentiviral vectors. After puromycin selection, the NS-shRNA MSCs and HIF1α-shRNA MSCs exhibited similar morphology (Figure 5Ai-ii). We next measured the ability of these 2 MSCs to differentiate along multiple mesenchymal lineages. After ∼ 2- to 3-week induction in special media, both types of MSCs differentiated into adipocytes, chondrocytes, and osteoblasts (Figure 5Aiii). The degrees of differentiation were determined by quantitative RT-PCR analysis for the expression of lineage markers (Figure 5Aiv). No significant differences were found between NS-shRNA MSCs and HIF1α-shRNA MSCs. The 2 types of MSCs were exposed to hypoxic conditions for different times and then lysed. Western blot analysis of the cell lysates showed significant down-regulation of HIF1α in the HIF1α-shRNA MSCs (Figure 5Bi).
HIF1α-shRNA MSC–derived extramedullary bones show reduced leukemic cell engraftment. (A) After puromycin selection, the NS-shRNA MSCs (i) and HIF1α-shRNA (ii) MSCs exhibited similar morphology. Both of the MSCs differentiate along multiple mesenchymal lineages with induction of special media, including adipocytes (first row), chondrocytes (second row), and osteoblasts (last row; iii). The degrees of differentiation were determined by quantitative RT-PCR analysis for the expression of lineage markers, ABL1 was used as reference housekeeping gene (iv). No significant differences were found between NS-shRNA MSCs and HIF1α-shRNA MSCs. (B) NS-shRNA MSCs and HIF1α-shRNA MSCs were exposed to hypoxic conditions (1% oxygen) for different times. Western blot analysis showed significant down regulation of the HIF1α protein levels (i). SDF-1α transcription levels were diminished (∼ 30%; P < .01) in HIF1α-shRNA MSCs compared with control NS-shRNA MSCs (ii). Consistent with quantitative RT-PCR, HIF1α-shRNA MSCs secreted lower levels of SDF-1α than NS-shRNA MSCs (P < .05; n = 4; iii). (C) HIF1α accumulated in TRE HIF1α MSCs with the doxycycline administration in both hypoxia (1% pO2) and normoxia (21% pO2; i). This correlated with the significantly up-regulation of SDF-1α transcription and secretion (ii-iii). (D) Extramedullary bones with similar morphology and osteoblastic activity developed from NS-shRNA MSCs and HIF1α-shRNA MSCs. (E) Representative H&E (i-ii) and anti-GFP staining (iii-iv) shows different cell densities in the cavities of 2 types of extramedullary bones. (F) Slides stained with anti-GFP antibody were analyzed in a CRi system. Five images per slide were quantified and averaged at 3 different focal depths within the tissue section. The results showed a significant reduction of GFP+ leukemic cell density in HIF1α-shRNA MSC-derived extramedullary bones compared with the control MSC-derived bones (P = .0006). (1) NS-shRNA MSCs in panels A and B and extramedullary bones derived from NS-shRNA MSC-ECFC-Matrigel in panels D through F. (2) HIF1α-shRNA MSCs in panels A and B and extramedullary bones derived from HIF1α-shRNA MSC-ECFC-Matrigel in panels D through F.
HIF1α-shRNA MSC–derived extramedullary bones show reduced leukemic cell engraftment. (A) After puromycin selection, the NS-shRNA MSCs (i) and HIF1α-shRNA (ii) MSCs exhibited similar morphology. Both of the MSCs differentiate along multiple mesenchymal lineages with induction of special media, including adipocytes (first row), chondrocytes (second row), and osteoblasts (last row; iii). The degrees of differentiation were determined by quantitative RT-PCR analysis for the expression of lineage markers, ABL1 was used as reference housekeeping gene (iv). No significant differences were found between NS-shRNA MSCs and HIF1α-shRNA MSCs. (B) NS-shRNA MSCs and HIF1α-shRNA MSCs were exposed to hypoxic conditions (1% oxygen) for different times. Western blot analysis showed significant down regulation of the HIF1α protein levels (i). SDF-1α transcription levels were diminished (∼ 30%; P < .01) in HIF1α-shRNA MSCs compared with control NS-shRNA MSCs (ii). Consistent with quantitative RT-PCR, HIF1α-shRNA MSCs secreted lower levels of SDF-1α than NS-shRNA MSCs (P < .05; n = 4; iii). (C) HIF1α accumulated in TRE HIF1α MSCs with the doxycycline administration in both hypoxia (1% pO2) and normoxia (21% pO2; i). This correlated with the significantly up-regulation of SDF-1α transcription and secretion (ii-iii). (D) Extramedullary bones with similar morphology and osteoblastic activity developed from NS-shRNA MSCs and HIF1α-shRNA MSCs. (E) Representative H&E (i-ii) and anti-GFP staining (iii-iv) shows different cell densities in the cavities of 2 types of extramedullary bones. (F) Slides stained with anti-GFP antibody were analyzed in a CRi system. Five images per slide were quantified and averaged at 3 different focal depths within the tissue section. The results showed a significant reduction of GFP+ leukemic cell density in HIF1α-shRNA MSC-derived extramedullary bones compared with the control MSC-derived bones (P = .0006). (1) NS-shRNA MSCs in panels A and B and extramedullary bones derived from NS-shRNA MSC-ECFC-Matrigel in panels D through F. (2) HIF1α-shRNA MSCs in panels A and B and extramedullary bones derived from HIF1α-shRNA MSC-ECFC-Matrigel in panels D through F.
In our previous study, we found significantly reduced transwell migration of leukemic cells toward HIF1α-shRNA MSCs compared with NS-shRNA MSCs under hypoxic conditions.38 It has been reported that SDF-1α promoter contains 2 HIF1α binding sites.39 So, we measured the expression of SDF-1α in these MSCs. We found that SDF-1α transcription levels were diminished (∼ 30%; P < .01) in HIF1α-shRNA MSCs compared with control NS-shRNA MSCs (Figure 5Aii). Consistent with quantitative RT-PCR, HIF1α-shRNA MSCs secreted lower levels of SDF-1α than NS-shRNA MSCs (P < .05, n = 4; Figure 5Biii). In contrast, we introduced a doxycycline-inducible HIF1α P402A/P564A mutant into normal MSCs. In these HIF1α mutant cells (TRE HIF1α MSCs), the proline residues 402 and 564 within the oxygen-dependent degradation domain were mutated to alanine, and the mutant became insensitive to proteasomal degradation. As shown in Figure 5C, HIF1α accumulated in TRE HIF1α MSCs with the doxycycline administration in both hypoxia (1% pO2) and normoxia (21% pO2). This correlated with the significantly up-regulation of SDF-1α transcription and secretion.
We then mixed the NS-shRNA MSCs or HIF1α-shRNA MSCs with ECFCs and Matrigel and subcutaneously injected them into opposite flanks of the mice (n = 5). Six weeks later, we observed extramedullary bones with similar morphology were developing on both sides of the flanks (supplemental Figure 3), and then MOLM13/Luc/GFP cells were intravenously injected into these mice. Mice were killed 2 weeks after the leukemic cell transplantation, and extramedullary bones with high osteoblastic activity were observed on both the right and left flanks of the mice (Figure 5D). Interestingly, H&E staining revealed differences in leukemia cell engraftment between the extramedullary bones from the NS-shRNA MSC group and the HIF1α-shRNA MSC group (Figure 5E). More than 95% of the cells in extramedullary bone marrows were leukemic cells. To further confirm the results, slides were stained with an anti-GFP antibody and analyzed with the CRi spectral imaging system that identifies and enumerates GFP+ cell in the extramedullary bone cavity, representing the engraftment of MOLM13 cells. In brief, the system was trained to automatically count the number of GFP+ cells from different slides and fields, and the number of GFP+ cells was then divided by the calculated bone cavity area. The results showed a significant reduction (50% ± 6%; P = .0006) in the GFP+ cell density in HIF1α-shRNA MSC-ECFC–derived extramedullary bones (1449 ± 194 cells/mm2) compared with the NS-shRNA MSC-ECFC–derived bones (3037 ± 496 cells/mm2; Figure 5F). These results are consistent with our previous in vitro studies that showed that fewer leukemic cells migrated to HIF1α-shRNA MSCs under hypoxic conditions where the HIF1α was induced.38
Discussion
Numerous studies have demonstrated that bone marrow stroma cells do not engraft in human bone marrow, after bone marrow transplantation, as shown by the fact that stroma cells in bone marrow transplantation patients are always host-derived.14-16 Likewise, human stroma cells do not consistently engraft in murine BM. This is the first report of a genetically controlled human bone marrow microenvironment. We successfully developed a novel extramedullary bone marrow model using human MSCs and ECFCs with Matrigel in NOD/SCID/IL-2rγnull mice. Compared with normal bones, the extramedullary bones showed similar structures, with typical BM cavities that provide a robust hematopoietic environment.
After engraftment of normal human cord blood cells, the extramedullary bone marrow supported both murine and human cells. Similar frequencies of human CD45+ cells were seen in extramedullary and native long bones 4 weeks after injection. With long-term follow-up, we found the engrafted human hematopoietic cells became dominant population in the extramedullary bones after 11 weeks, and the human CD45+ percentages was higher than femurs in all 3 mice tested (1.14- ± 0.02-fold increase; n = 3). However, no statistical significance was found (Figure 3B). Both human lymphoid and myeloid lineage cells were observed by immunofluorescence staining with CD19 and CD33 antibodies. These data indicated that MSC-ECFC-Matrigel–derived extramedullary bones provide a hematopoietic environment that is at least functionally equivalent to the native bones. Using human MOLM13/Luc/GFP leukemia cells, we demonstrated that these acute myeloid leukemia cells could easily engraft into the extramedullary bones, suggesting that this leukemia model could be used to evaluate anti-leukemia drugs. Although rodent studies have provided important insight into both leukemia development and drug efficacy, there is much that needs to be learned about the effects of the human BM microenvironment on leukemia development and progression, human leukemia biology and the efficacy of new therapeutic modalities using this model. Some of the new anti-leukemia agents are modulating target genes in both hematopoietic and nonhematopoietic tissues. For example, we have recently shown that HDM2-mediated P53 activation suppresses SDF-1 production by MSCs.40 New HDM-2 inhibitors have selective activity against human HDM2 but not murine MDM2. For therapeutic purpose, human antibodies also have been developed that do not cross-react with murine targets. Hence, their effects on BM microenvironment cannot be examined in regular animal models. Our model would allow these types of investigation. Moreover, competing engraftment between leukemic and normal stem cell can be examined in this model. It is conceivably that specific genetically modified bone marrow environment can be created that favors engraftment and expansion of normal HSCs, whereas it creates an inhospitable microenvironment for leukemia. This seems increasingly likely because recent reports have demonstrated functional and genomic alterations in stroma cells derived from human myelodysplastic syndrome and acute myeloid leukemia bone marrow.17,21
The role of the microenvironment in tumor development was originally proposed by Paget in his “seed and soil” hypothesis.41 Increasing evidence supports the idea that leukemia development is strongly dependent on cellular interactions with BM niches and appropriate levels of signaling proteins.11,13,42,43 It has been suggested that disruption of the tumor microenvironment may serve as a critical therapeutic paradigm to kill leukemic cells. We and other groups have shown that leukemic cells undergo spontaneous apoptosis once they are removed from the in vivo BM microenvironment and placed in suspension cultures without supportive stroma, highlighting the importance of external signals from the microenvironment in maintaining these cells.10,44-47 Therefore, how leukemic cells are maintained in vivo and how to disrupt the interactions between leukemic cells and the microenvironment are important questions for both basic research and clinical drug development. Oxygen tension is an important component of the microenvironment, and local oxygen concentrations can directly influence stem cell self-renewal and differentiation.48,49 Recently, our group demonstrated a marked expansion of hypoxic niches in the BM of SCID mice engrafted with leukemic cells35 and in patients with acute myeloid leukemia (unpublished). To understand the mechanism by which the hypoxic niches in the BM expand, we investigated the role of HIF1α, the master regulator of hypoxia-induced responses, in the BM microenvironment and its effect on leukemia progression. In the current study, we used NS-shRNA MSCs and HIF1α-shRNA MSCs along with ECFCs and Matrigel to develop extramedullary bones. Similar extramedullary bones with high osteoblastic activity were developed from both types of cells. We observed a 50% reduction in leukemic cell engraftment in the HIF1α-shRNA MSC–derived extramedullary bones compared with the NS-shRNA MSC–derived bones. This finding establishes HIF1α as a critical component for the engraftment of leukemic cells by directly up-regulate SDF-1α expression in the physiologically hypoxic BM microenvironment. These results, for the first time, establish an in vivo bone and BM model with a genetically controlled human microenvironment. We propose that targeting HIF1α in the protective cells of the bone marrow niches may represent a new approach to improve therapeutic strategies for leukemia. Altogether, this set of experiments suggests that the extramedullary bone model could be a critical tool to investigate the gene on normal hematopoiesis and leukemic development by genetic manipulation of MSCs.
The evolving evidence from genomic studies suggests that MSCs in leukemia patients frequently carry genomic alterations that may contribute to the pathogenesis of the disease.50,51 The concepts can now be experimentally tested using this model. More studies are needed to focus on the BM microenvironment that will have important clinical implications in leukemia treatment. The genetically modifiable human cell–derived in vivo bone model presented in this study enables the systematic investigation of the impact of the BM microenvironment on the development and progression of leukemia, as well as the testing of new therapeutic targets and concepts.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate Dr E. J. Sphall's donation of cord blood stem cells and Dr Kate Newberry's help in editing the manuscript.
This work was supported by National Institutes of Health grant AML P01 CA55164 (M.A.), Cancer Center support grants CA016672, CA143805, CA049639, CA136411, and CA100632 (M.A.), a Paul and Mary Haas Chair in Genetics grant (M.A.), and National Cancer Institute grant 1R01CA155056-01 (M.K.).
National Institutes of Health
Authorship
Contribution: Y.C., D.S., and M.A. conceived the project, designed experiments, and interpreted data; Y.C., R.J., Y.-x.S., R.-y.W., V.L.B., D.S., N.A.H., and A.R. performed experiments or provided cells; S.K. provided help in the interpretation of bone and bone marrow morphology; and Y.C., R.J., M.K., and M.A prepared and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael Andreeff, Molecular Hematology and Therapy, Department of Leukemia, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 0448, Houston, TX 77030; e-mail: mandreef@mdanderson.org.