Abstract
Gene-targeting studies in mice have identified the essential roles of most prosurvival Bcl-2 family members in normal physiology and under conditions of stress. The function of one member, Bcl2a1/Bfl-1/A1, is only poorly understood because of quadruplication of its gene locus in mice, hindering conventional knockout studies. To overcome this problem, we generated mouse models allowing traceable constitutive or reversible ablation of A1 in the hematopoietic system by RNA interference. Knockdown of A1 impaired early stages of T-cell differentiation, B-cell homeostasis, and sensitized transitional as well as follicular B cells to apoptosis induced by ligation of the B-cell receptor. As a consequence, B-cell proliferation in response to mitogens was severely impaired, whereas that of T cells appeared unaffected. Furthermore, depending on the extent of A1 knockdown, granulocytes showed increased spontaneous death in culture or failed to accumulate in significant numbers in vivo. These models highlight the critical role of A1 in leukocyte development and homeostasis, constituting valuable tools for investigating presumed roles of this Bcl-2 family member in immunity, tumorigenesis, and drug resistance.
Introduction
The prosurvival Bcl-2 family member A1 is mainly expressed in the hematopoietic system and signaling via T- and B-cell receptors (TCR/BCR), members of the TNF receptor family (eg, CD40, TNF-R1) or certain cytokines, such as G-CSF or the TLR4 agonist LPS lead to NF-κB dependent up-regulation of A1 mRNA in white blood cells.1 Furthermore, it was shown that the A1 protein is posttranslationally regulated by ubiquitin-dependent proteasomal degradation2,3 and stabilization of A1 half-life–facilitated oncogene-driven tumorigenesis in mice.4 Overexpression of A1 in lymphoma cell lines afforded significant protection from apoptosis induced by BCR ligation,5 IL-3 deprivation, staurosporine,3 or etoposide2 treatment, whereas its knockdown sensitized B lymphoma cells to the apoptotic effects of CD20 cross-linking and DNA-damaging agents.6 Transgenic expression of A1 in the lymphoid compartment of mice increased survival of immature B- and T-cell precursors and protected thymocytes from cell death triggered by ionomycin or γ-irradiaton as well as mature splenic T cells from TCR ligation-induced apoptosis.7,8 Similar phenotypes were, however, also observed in mice overexpressing related Bcl-2 prosurvival proteins (eg, Bcl-2 or Mcl-1), leaving the question of the physiologic relevance of A1 in lymphocyte homeostasis largely unanswered.9,10 A1 gene knockout studies have been hampered by the fact that its gene locus in mice underwent quadruplication, leading to the presence of 3 functional A1 genes encoding A1-a, A1-b, and A1-d isoforms and one pseudogene, A1-c. Selective deletion of A1-a in mice led to increased spontaneous apoptosis of neutrophils and augmented apoptosis susceptibility of allergen sensitized and activated mast cells in vitro.11,12 No other obvious defects were reported, but this may be explained by the fact that A1 isoforms, which are more than 95% homologous on mRNA as well as on protein level are redundant in function,13 or the fact that in contrast to the other A1 isoforms, A1-a is only poorly expressed in T and B cells.14,15 In contrast to mice, the human genome only contains one A1/Bfl-1 gene, whose overexpression has been implicated in the pathology of B16 and T-cell lymphomagenesis14 as well as drug resistance phenotypes.17 Furthermore, increased levels of A1 mRNA expression have also been reported in solid cancers, such as those of breast, stomach, or colon and in some tumor types (ie, melanoma or hepatocellular carcinoma), it correlated with metastatic disease.18 In addition, autoimmune disorders, such as systemic lupus erythematosus19 or rheumatoid arthritis,20 also associate with increased levels of A1, making it overall an attractive target for therapeutic intervention in multiple human pathologies. However, model systems allowing the preclinical validation of these observations in vivo are lacking. An initial attempt to knockdown all A1 isoforms by RNA-interference (RNAi), expressing short hairpin (sh)RNAs from the DNA-pol-III–responsive U6 promoter in mice, failed to reveal significant insights, possibly because of the strong cell type dependent variation in knockdown efficiency and/or choice of promoter.15
Here we report on the development of 2 novel mouse models, in which the constitutive or reversible (doxycycline-regulated) ablation of all functional A1 isoforms was achieved by expression of a micro-RNA30 (miR30)–based precursor, harboring a shRNA sequence (mi-shRNA) that is processed into a small interfering (si)RNA that targets all A1 isoforms. Expression of mi-shRNAs was restricted to hematopoietic cells in vivo by the use of the DNA-pol-II–dependent Vav-gene promoter. This strategy allowed the identification of critical checkpoints in leukocyte development that depend on the expression of A1.
Methods
Generation of transgenic mice
Animal experiments were performed in accordance with Austrian legislation (BGBl no. 501/1988, idF BGBl I no. 162/2005) and were granted by the Ministry of Education, Science and Research (code: 66-011/0058-II/3b/2011). Vav-Venus-miR30-A1 or Vav-Venus miR30-FF transgenic mice, referred to as lines VVA1.1, VVA1.2, or VVFF, expressing mi-shRNAs targeting A1 or firefly luciferase, respectively, were generated by microinjection of approximately 2 to 3 μg/μL AseI-PvuI fragment, gel purified from the Vav-Venus-miR30 plasmid, into fertilized oocytes derived from C57BL/6xCBA F1 mice and maintained on this mixed genetic background. Vav-Venus miR30-FF transgenic mice were generated directly on C57BL/6 background. Mice harboring a miR30-based cassette to express mi-shRNAs21 targeting A1 under control of the Tet-CMVmin promoter (TREA1) were generated in accordance with German legislation (proposal 621-2531.01-54/02) granted by the government of Unterfranken, by injecting concentrated lentiviral particles into the perivitelline space of fertilized mouse oocytes obtained from superovulated C57BL/6 female mice. The injected zygotes were cultured overnight and subsequently transferred into the oviduct of pseudopregnant CD1 mice. The generation and genotyping of Vav-Venus22 and Vav-tTA transgenic mice have been described.23 Transgenic mice were selected on the basis of PCR genotyping on tail DNA using Vav-tTA primers: 5′-3′ forward, CTCTCTTGCCTGCCTGTG; reverse, GTAAACTTCTGACCCACTGGAAT; and TREA1 primers: 5′-3′ forward, CCCACCAAGGCAAAGAGAAGAG; and reverse, ACTCCAACTAGCATTCCAAGGC. Experiments using double-transgenic (DT) and relevant single or nontransgenic control mice were all performed using F1 offspring, representing a mixed (FVBxB6) genetic background. To repress Vav-tTA–mediated mi-shRNA expression, 5 mg/mL doxycycline hyclate (Sigma-Aldrich) was added to the drinking water containing 1% sucrose.
Inducible lentiviral shRNA and miR30-containing constructs
shRNA sequences targeting all isoforms of A1 were cloned into the miR30 containing LMP vector.21 For the expression of the miR30-A1 under the control of a Tet-responsive element promoter (TRE), the following oligonucleotide shA1-4 was subcloned via XhoI and EcoR1 restriction sites into the TMP vector: 5′-ctcgagaaggtatattgctgttgacagtgagcgcCCATAGATACCGCC AGAATAATAgtgaagccacagatgTATTATTCTGGCGGTATCTATGGatgcctactgcctcggaattc-3 (sequence targeting A1 mRNA in capital letters). To generate an inducible lentiviral construct containing an eGFP marker, the previously reported FUGW vector21 was modified by inserting the PCR-amplified TRE-miR-A1 cassette using the aforementioned TMP miR30-A1 vector upstream of the human ubiquitin promoter (Ubi-P)–driven eGFP cassette by Pac1.
In a parallel approach, additional alternative shRNA sequences targeting all A1 isoforms were embedded in a pHR-THT-eGFP vector24 and tested for efficacy in transient transfection assays in 293T-FLAG-A1 cells, generated via stable transfection of 293T cells with pEF-FLAG-A1-puro, sorted according to the levels of eGFP expression: sh-A1.1 5′-tttccaaaaaGAGTTGCTTTCTCCGTTCatctcttgaaTGAACGGAGAAAGCAACTCggggatc-3′; sh-A1.2 5′-tttccaaaaaGGATGACTTTCACGTGGAAtctcttgaaTTCCACGTGAAAGTCATCCggggatc-3′. Subsequently, the shA1.1 core sequence or a sequence targeting firefly luciferase was embedded into the context of the miR30 backbone, subcloned into pENTR207, and recombined into Gateway-compatible versions of the pEGFP-C1 plasmid (Clontech) or the VavP HS21/45 transgenic vector.22 The following oligonucleotides were amplified by PCR, using primers containing XhoI and EcoRI overhangs, digested, and subcloned into pENTR-207: miR30-A1 5′-tgctgttgacagtgagcgaaaGAGTTGCTT TCTCCGTTCATagtgaagccacagatgtatGAACGGAGAAAGCAACTCTTctgcctactgcctcgga-3′; miR30-FF 5′-tgctgttgacagtgagcgaaaCTTACGCTGAGTACTTCGATagtgaagccacagatgtaTCGAAGTACTCAGCGTAAGttctgcctactgcctcgga-3′. Recombination into pEGFP-dest and pVav HS21/45-Venus-dest was performed using LR-clonase (Invitrogen).
Cell culture, viral transduction, and transfection assays
HEK293T cells were transfected with 10 μg of FTmiR-A1-UGW DNA together with 5 μg of pMDL-RRE, 2.5 μg of pRSV-REV, and 3 μg of pVSV-g using CaPO4 precipitation to generate lentiviral particles. Forty-eight hours after transfection virus-containing supernatants were collected, passed through a 0.45-μm filter, and concentrated by ultracentrifugation. Virus containing pellet was resuspended in 30 μL of PBS/0.1% BSA. (For additional information, see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article.)
Immunoblotting
Cell extracts were prepared in CHAPS lysis buffer and analyzed by immunoblotting. Antibodies used include M2 mouse anti-FLAG (Sigma-Aldrich) and 137F5 rabbit anti-ERK1/2 and DM1A mouse anti-alpha tubulin mAb (Abcam). HRP-conjugated goat anti–mouse Ig antibodies (Jackson ImmunoResearch Laboratories) or goat anti–rabbit antibodies (Dako Denmark) served as secondary reagents and the enhanced chemiluminescence (GE Healthcare) system was used for detection.
Immunofluorescence analysis, cell sorting, and antibodies used
The monoclonal antibodies used were purchased from either eBioscience or BioLegend. Their specificities are: RA3-6B2, anti-B220; GK1.5, anti-CD4; H129.19.6.8, anti-CD4; 53.6.72, anti-CD8; YTS 169, anti-CD8; RB6-8C5, anti–Gr-1; S7, anti-CD43; 5.1, anti-IgM; 11/26C, anti-IgD; MI/70, anti–Mac-1; Ter119, anti-erythroid cell surface marker; T24.31.2, anti–Thy-1; IM7, anti-CD44; 3C7, anti-CD25; H57-59, anti-TCRβ; anti-CD3, 2C11; 6D5, anti-CD19; 2B8, anti–c-Kit; D7, anti-Sca1. Flow cytometric analysis was performed using a FACSCalibur or LSR-Fortessa cell analyzer. Isolation of cells was performed using a FACSVantage cell sorter (all BD Biosciences). (For additional information, see supplemental Methods.)
RNA isolation and Northern blot analysis
Total RNA was isolated using TRIzol (Invitrogen); 10 μg of total RNA was denatured for 3 minutes at 95°C in 1 times RNA loading dye (47.5% formamide, 0.0125% SDS, 0.0125% bromophenol blue) and separated on a denaturing 8% polyacrylamide gel (7M urea, 1 × TBE). Oligonucleotides used include the following: VVA1 probe 5′-GAGTTGCTTTCT-3′; TREA1 probe 5′-CATAGATACCGCCAGAATAATAGTGAAGCCACAGATGTATTATTCTGGCGGTATCTATG-3′. Oligonucleotides were 5′-labeled with [γ-32P] ATP using T4 polynucleotide kinase (Promega). Hybridization was carried out at 42°C in 1M sodium phosphate buffer (pH 6.2), 7% (weight/volume) SDS for 18 hours. Subsequently, membranes were washed for 5 minutes at room temperature in washing solution I (2 × saline sodium citrate, 0.1% weight/volume SDS) followed by a second washing step at room temperature for 5 minutes with washing solution II (0.2 × saline sodium citrate, 0.1% weight/volume SDS). Membranes were exposed in a phospho-imager for up to 4 days.
Quantitative RT-PCR analysis of mRNA or miRNA expression levels
Quantitative RT-PCRs on mRNA were performed with Omniscript RT kit (QIAGEN) using 1 μg total RNA pretreated with 1 U/μL RQ1 DNase (Promega). PCR conditions are as follows: 95°C for 7 minutes, 40 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. Quantitative RT-PCR was performed using Platinum SYBR Green qPCR SuperMix-UDG reagent (Invitrogen). Primers used include: A1 forward, 5′-AGAGCAGATTGCCCTGGATGTA-3; A1 reverse, 5′-GATAACCATTCTCGTGGGAG-3′; HPRT: forward, 5′-GCTGGTGAAAAGGACCTC-3′; and HPRT reverse, 5′-CACAGGACTAGAACACCTGC-3′. Reactions were performed in triplicates. Relative quantification was performed using the ΔΔCT method. Relative expression levels of each A1 isoform were monitored as described in Hatakeyama et al 1998.13 For miRNA detection, cDNA was synthesized using the NCode miRNA First-Strand cDNA Synthesis Kit (Invitrogen) using 2.5 μg of total RNA. PCR conditions include: 50°C for 2 minutes, 95°C for 2 minutes, and 40 cycles of 95°C for 15 seconds and 57°C for 30 seconds. Primers used to detect include: miR30-A1 in VVA1 mice, 5′-GAGTTGCTTTCTCCGTTCA-3′; miR30-A1 in DT mice, 5′-TATTCTGGCGGTATCTATG-3′; miR30-FF in VVFF mice, 5′-CTTACGCTGAGTACTTCGA-3′; miR16, 5′-TAGCAGCACGTAAATATTGGCG-3′; miR142b, 5′-TGTAGTGTTTCCTACTTTATGGA-3′; and U6, 5′-CGATACAGAGAAGATTTAGCATGG-3′. Expression of U6 was evaluated as internal control.
Luciferase assays
Lipofectamine was used to transiently transfect 293T cells with 2 μg pGL2-Bim-0.8 kb reporter plasmid plus 0.5 or 1 or 2 or 4 μg/mL pEYFP-C1-miR30FF for 18 hours, followed by a change of medium. After an additional 24 hours, cells were lysed and Firefly-luciferase activity was quantified using the Luciferase Reporter Assay kit (Promega). Protein content of lysates was quantified by Bradford analysis.
Generation of B- and T-cell blasts
Splenocytes were stimulated with 100 U/mL of mIL-2, 10 ng/mL mIL-4, 10 ng/mL mIL-5 (all PeproTech) and 2 μg/mL F(ab′)2 fragments goat anti–mouse IgM (Jackson ImmunoResearch Laboratories), 2 μg/mL hamster anti–mouse CD40 mAb (3 of 23, BD Biosciences), or 100nM ODN74 5′-AAAAAAAAAAAAAACGTTAAAAAAAAAAA-3′ (Microsynth). To induce T-cell proliferation, splenocytes were stimulated with 2 μg/mL concanavalin A (Sigma-Aldrich) or 1 μg/mL anti-CD3 (2C11).
Colony-forming cell assay
Mouse colony-forming cell assay was performed with 2 × 104 bone marrow cells that were added to 1 mL MethoCult 3234 (1% methylcellulose, 15% FBS, 1% BSA, 10 μg/mL bovine pancreatic insulin, 200 μg/mL human transferrin, in IMDM) according to the user manual. The MethoCult was supplemented immediately or with a delay of 24-hour with 50 ng/mL mG-CSF, 10 ng/mL mIL-3, and 10 ng/mL mIL-6 (PeproTech) in the presence or absence of 1 μg/mL doxycycline (Sigma-Aldrich). After 7 days in culture, colonies were scored as clusters containing more than 50 cells.
Statistical analysis
Statistical analysis was performed using ANOVA followed by Fisher PLSD posthoc test, applying the StatView Version 4.1 software program. P values less than .05 were considered to indicate statistically significant differences.
Results
Development of miR30-based vectors targeting all functional A1 isoforms
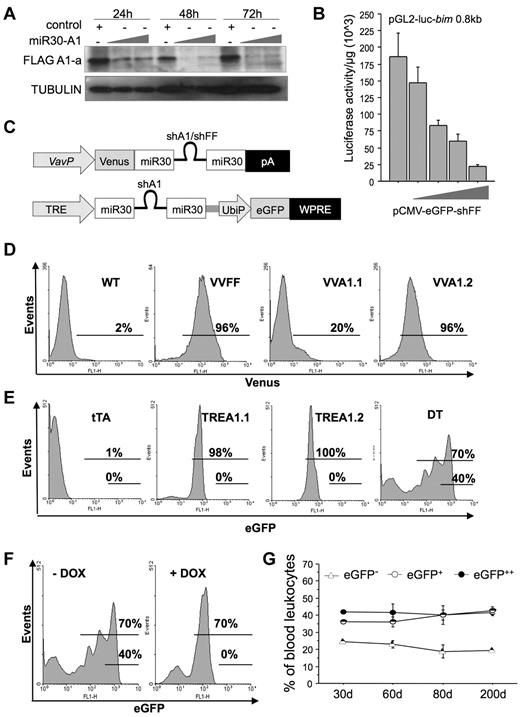
To overcome the obstacle that mRNA encoding A1 can be expressed from 3 of 4 gene loci present in mice, we designed different siRNA sequences complementary to regions homologous to A1-a, A1-b, and A1-d. Two different sequences were tested in the context of shRNA-expressing lentiviral vectors used to transiently transduce 293T cells stably expressing FLAG-tagged mouse A1-a (supplemental Figure 1A). Both tested sequences effectively reduced A1 protein levels below detection limit. One such sequence (#1) was embedded in the context of miR30 in the 3′-untranslated region of eGFP under control of the DNA-pol-II–dependent CMV promoter and tested for its efficacy to produce mature siRNA, assessed by Northern blotting (supplemental Figure 1B), and to knockdown A1 protein levels when transiently expressed in 293T FLAG-A1 cells (Figure 1A). For control purposes, a sequence targeting firefly luciferase (FF) was subcloned into the same vector and tested for its efficacy to reduce luciferase promoter reporter activity (Figure 1B). Finally, the miR30-A1 or miR30-FF backbones were subcloned into a construct driving expression of Venus, a brighter version of YFP, under the control of the hematopoiesis-specific Vav-gene promoter,25 positioning miR30-A1 or miR30-FF sequences downstream of the open reading frame encoding for Venus but upstream of the poly-A site (Figure 1C). To produce transgenic A1-RNAi mice, the constructs were linearized, trimmed of bacterial sequences, and used for pronuclear injection of zygotes. Offspring were screened by PCR genotyping on tail DNA and Venus expression in peripheral blood leukocytes. Two founders, showing expression of the transgene in approximately 20% or more than 90% of all cells in the peripheral blood (Figure 1D), were selected to establish transgenic lines referred to as Vav-Venus-miR30-A1 strain 1 or strain 2 (VVA1.1; VVA1.2). As a control, a transgenic line expressing miR30-FF was also established, referred to herein as Vav-Venus-miR30-FF (VVFF). Neither VVA1 nor VVFF mice showed any gross phenotypic abnormalities or breeding problems over an observation period of up to 12 months.
Generation of miR30-A1 mice. (A) The 293T cells expressing FLAG-tagged mouse A1-a were transiently transfected with graded doses of the pEGFP-miR30-A1 vector that targets all A1 isoforms. The GFP+ cells were FACS-sorted, lysed, and extracts subjected to Western blot analysis using a FLAG-specific antibody and anti-GAPDH, to compare loading. (B) The 293T cells were transiently transfected with 2 μg of pGL2-Bim 0.8 kbp reporter plasmid, driving expression of the luciferase gene under the control of the first 800 bp of the Bim gene promoter plus graded doses of pEGFP-miR30-FF (0.5, 1.0, 2.0, or 4.0 μg), targeting firefly luciferase. At 24 hours after transfection, cells were lysed and a luciferase reporter assay performed. Bars represent mean of triplicates ± SD (n = 2). (C) Schematic representation of plasmids used for mouse transgenesis. (D) Transgenic expression of Venus in peripheral blood leukocytes of F1 offspring of PCR+ founders was monitored by flow cytometric analysis. Percentages given refer to Venus+ cells. (E) Transgenic eGFP expression was quantified in peripheral blood leukocytes of single-transgenic tTA, TREA1.1, TREA1.2, and DT mice. Percentages given refer to eGFP+ cells. (F) Transgene expression in peripheral blood of double-transgenic mice was monitored before and during doxycycline application in the drinking water by flow cytometric analysis. Percentages given refer to eGFP+(low) and eGFP++(high) cells at the beginning of doxycycline treatment, day 0, or after 19 days. (G) eGFP expression in DT mice was followed in the peripheral blood over an observation period of 200 days and analyzed by flow cytometry. Data are mean of n = 4 animals ± SEM.
Generation of miR30-A1 mice. (A) The 293T cells expressing FLAG-tagged mouse A1-a were transiently transfected with graded doses of the pEGFP-miR30-A1 vector that targets all A1 isoforms. The GFP+ cells were FACS-sorted, lysed, and extracts subjected to Western blot analysis using a FLAG-specific antibody and anti-GAPDH, to compare loading. (B) The 293T cells were transiently transfected with 2 μg of pGL2-Bim 0.8 kbp reporter plasmid, driving expression of the luciferase gene under the control of the first 800 bp of the Bim gene promoter plus graded doses of pEGFP-miR30-FF (0.5, 1.0, 2.0, or 4.0 μg), targeting firefly luciferase. At 24 hours after transfection, cells were lysed and a luciferase reporter assay performed. Bars represent mean of triplicates ± SD (n = 2). (C) Schematic representation of plasmids used for mouse transgenesis. (D) Transgenic expression of Venus in peripheral blood leukocytes of F1 offspring of PCR+ founders was monitored by flow cytometric analysis. Percentages given refer to Venus+ cells. (E) Transgenic eGFP expression was quantified in peripheral blood leukocytes of single-transgenic tTA, TREA1.1, TREA1.2, and DT mice. Percentages given refer to eGFP+ cells. (F) Transgene expression in peripheral blood of double-transgenic mice was monitored before and during doxycycline application in the drinking water by flow cytometric analysis. Percentages given refer to eGFP+(low) and eGFP++(high) cells at the beginning of doxycycline treatment, day 0, or after 19 days. (G) eGFP expression in DT mice was followed in the peripheral blood over an observation period of 200 days and analyzed by flow cytometry. Data are mean of n = 4 animals ± SEM.
In a second approach, 2 additional pan-A1–selective miR30-A1 sequences were tested in the context of the miR30 backbone in a retroviral vector. One of these sequences successfully reduced A1 protein expression in WEHI-231 immature mouse B cells stably expressing FLAG-tagged A1-a (supplemental Figure 1C). Generation of mature siRNA was again confirmed by Northern analysis in transiently transfected 293T cells (supplemental Figure 1D). Sequence 4 in the miR30 backbone was then inserted into a lentiviral vector downstream of the tetracycline responsive CMVmin promoter (TRE), followed by a woodchuck hepatitis virus posttranscriptional regulatory element and eGFP, driven from the human ubiquitin promoter, UbiP (Figure 1C). This lentivirus was injected into the perivitelline space of fertilized oocytes before transfer into pseudo-pregnant females. Two strains were successfully established from transgene-positive founders showing eGFP expression in all hematopoietic cells, but also other cell types, such as mouse embryonic fibroblasts (not shown); these strains are referred to as TRE-miR30-A1 (TREA1.1 or TREA1.2; Figure 1E). TREA1 mice were subsequently intercrossed with mice expressing the Tet transactivator (tTA) under control of the Vav-gene promoter23 to selectively direct mi-shA1 expression to hematopoietic cells in bitransgenic mice already early in embryogenesis, referred to here as DT mice. Data from both TREA1 strains were comparable and therefore pooled unless indicated otherwise. DT mice also showed increased eGFP expression because of binding of tTA to the upstream CMV-TRE promoter (Figure 1E). Of note, we also detected a fraction of cells that no longer expressed eGFP, suggesting silencing of the transgene in some cells (Figure 1F). This effect was not progressive, and percentages of eGFP−, eGFP+, and eGFP++ cells remained largely constant over an observation period of approximately 200 days (Figure 1G). Importantly, the addition of doxycycline into the drinking water of animals to inhibit tTA function led to the disappearance of GFP++ cells with a concomitant increase in GFP+ cells, whereas the percentage of GFP− cells remained unchanged in peripheral blood (Figure 1F; supplemental Figure 2A), bone marrow, thymus, spleen, or lymph node (supplemental Figure 2B). This finding is indicative of stable transgene silencing in a subset of leukocytes during early development. Nonetheless, the system allowed reversible mi-shRNA expression in a significant percentage of hematopoietic cells.
Quantification of A1/Bfl-1 knockdown in hematopoietic cells in vivo
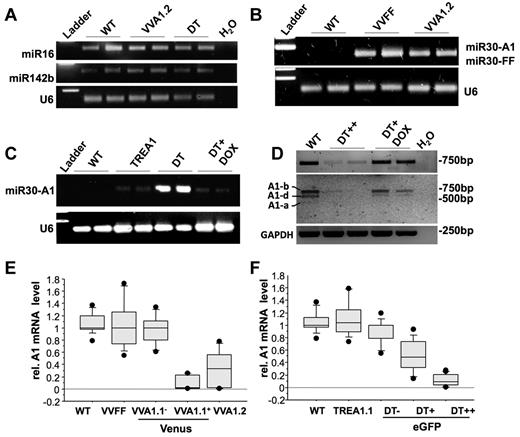
To rule out a negative side effect because of RNA-induced silencing complex overloading by transgenic expression of mi-shRNAs, we first tested for the presence of 2 miRNAs widely transcribed and processed in the hematopoietic compartment, miR142b and miR16,26 by PCR on total RNA from spleen from WT, VVA1.2, and DT mice. U6 rRNA was amplified for reference (Figure 2A). Levels of both miRNAs were comparable with those found in nontransgenic littermates, excluding gross interference of mi-shRNA expression with the biogenesis of endogenous miRNAs. We documented expression and maturation of the A1- or FF-selective siRNAs in VVFF, VVA1.2, or DT-derived spleen RNA by RT-PCR using a universal poly-T primer to synthesize cDNA from the polyA-tailed miRNAs (Figure 2B-C). These experiments confirmed the presence of mature siRNA against A1 or FF in the relevant transgenic lines, as well as their absence in TREA1 single-transgenic mice or nontransgenic controls. Furthermore, addition of doxycycline into the drinking water shut down the expression of A1-selective siRNAs (Figure 2C; supplemental Figure 1F).
Normal miRNA biogenesis and effective knockdown of A1 mRNA in transgenic mice. (A) To assess the impact of transgene expression on endogenous miRNA levels of miR16 and miR142b in spleens from WT, VVA1.2, and DT mice were assessed by N-Code PCR analysis on reverse-transcribed miRNAs. U6 ribosomal RNA was amplified for reference. (B-C) siRNAs targeting firefly luciferase or A1 were assessed by N-Code PCR analysis on reverse-transcribed miRNA samples extracted from the spleen of mice of the indicated genotypes and in spleen cells derived from DT mice after 32 days of doxycycline application in the drinking water. (D) cDNA was generated from splenocytes of WT mice, sorted eGFP++ cells from untreated DT mice or DT mice kept on doxycycline for 32 days. Knockdown efficiency was evaluated by quantitative RT-PCR on cDNA using primers amplifying all A1 isoforms yielding a 743-bp product (top panel). PCR products were purified and digested by restriction enzymes BglII and NsiI to confirm knockdown of all major A1 isoforms present in spleen (see also supplemental Figure 1 for further details). Images have been gray-scale inverted for easier visualization of cleaved isoforms. (E) cDNA was generated from total spleen of WT or VVA1.2 mice as well as from FACS-sorted Venus+ or Venus− spleen cells from VVA1.1 mice. Knockdown efficiency was evaluated by quantitative RT-PCR on cDNA using primers amplifying all A1 isoforms or HPRT for normalization. (F) cDNA was generated from splenocytes of WT, TREA1 single-transgenic, or DT mice, expressing different levels of eGFP. Knockdown efficiency was evaluated as in panel D. Results of 3 independent experiments and cDNA samples from 5 animals/genotype are represented as box plots. Box length equals interquartile range. Circles represent minimal and maximal values.
Normal miRNA biogenesis and effective knockdown of A1 mRNA in transgenic mice. (A) To assess the impact of transgene expression on endogenous miRNA levels of miR16 and miR142b in spleens from WT, VVA1.2, and DT mice were assessed by N-Code PCR analysis on reverse-transcribed miRNAs. U6 ribosomal RNA was amplified for reference. (B-C) siRNAs targeting firefly luciferase or A1 were assessed by N-Code PCR analysis on reverse-transcribed miRNA samples extracted from the spleen of mice of the indicated genotypes and in spleen cells derived from DT mice after 32 days of doxycycline application in the drinking water. (D) cDNA was generated from splenocytes of WT mice, sorted eGFP++ cells from untreated DT mice or DT mice kept on doxycycline for 32 days. Knockdown efficiency was evaluated by quantitative RT-PCR on cDNA using primers amplifying all A1 isoforms yielding a 743-bp product (top panel). PCR products were purified and digested by restriction enzymes BglII and NsiI to confirm knockdown of all major A1 isoforms present in spleen (see also supplemental Figure 1 for further details). Images have been gray-scale inverted for easier visualization of cleaved isoforms. (E) cDNA was generated from total spleen of WT or VVA1.2 mice as well as from FACS-sorted Venus+ or Venus− spleen cells from VVA1.1 mice. Knockdown efficiency was evaluated by quantitative RT-PCR on cDNA using primers amplifying all A1 isoforms or HPRT for normalization. (F) cDNA was generated from splenocytes of WT, TREA1 single-transgenic, or DT mice, expressing different levels of eGFP. Knockdown efficiency was evaluated as in panel D. Results of 3 independent experiments and cDNA samples from 5 animals/genotype are represented as box plots. Box length equals interquartile range. Circles represent minimal and maximal values.
RT-PCR analysis followed by restriction enzyme digest of PCR products confirmed efficient and reversible knockdown of the dominant A1 isoforms expressed in spleen, ie A1-b and A1-d (Figure 2D; supplemental Figure 1E-F). Finally, we examined knockdown efficiency by measuring A1 mRNA levels by quantitative RT-PCR. RNA was isolated by cell sorting from total splenocytes from WT, VVFF, or VVA1.2 mice (> 90% Venus+ cells), as well as Venus+ or Venus− thymocytes (not shown) or splenocytes isolated by cell sorting from VVA1.1 mice. Similarly, thymocytes (not shown) or splenocytes expressing no, low, or high levels of eGFP were sorted from DT mice or control littermates. Consistent with the presence of processed siRNAs (Figure 2B-C), we confirmed knockdown of all A1 mRNA isoforms in spleen cells derived from VVA1 mice (Figure 2E; supplemental Figure 1E); albeit the knockdown efficacy varied between the different transgenic lines. Of note, the extent of A1 knockdown appeared to be lower in the VVA1.2 mice, although they contained higher percentages of Venus-expressing cells than VVA1.1 mice (Figure 1D). In contrast, A1 mRNA levels in cells derived from VVFF mice did not differ significantly from levels found in Venus− cells or cells from nontransgenic animals (Figure 2E). In cells isolated from DT mice, relative A1 mRNA expression levels were found to be comparable with that in nontransgenic controls in eGFP− cells, significantly reduced in eGFP+ cells from DT mice, suggesting low-level expression of the siRNA in that pool of cells but nearly undetectable expression of all A1 isoforms in eGFP++ cells (Figure 2F; supplemental Figure 1F). In addition, doxycycline treatment of DT animals for 32 days led to reaccumulation of A1 mRNA, indicating reversibility of the system (Figure 2D, supplemental Figure 1F).
Loss of A1/Bfl-1 expression impairs early T lymphopoiesis
First, we determined the overall composition of the hematopoietic system in the Venus-positive versus Venus-negative fraction of VVA1 mice as well as in the eGFP++, eGFP+, and eGFP− fractions of DT mice. Although the overall cellularity of all primary and secondary lymphoid organs was comparable with those found in nontransgenic or single-transgenic control mice (not shown), a more detailed analysis of the cellular subsets expressing A1 specific siRNAs revealed significant differences. Although the overall distribution of double-negative (DN), CD4+CD8+ double-positive (DP), and CD4+ or CD8+ single-positive (SP) cells in the thymus was comparable between the different controls, knockdown of A1 led to a significantly reduced percentage of DP cells with a relative increase in DN and CD8+ SP cells in the reporter gene-positive fraction of the thymus of the VVA1.1 and DT strain (Figure 3A-C). This effect was not immediately obvious in the VVA1.2 strain (Figure 3A-B), which showed weaker A1 knockdown (Figure 2E). However, a more detailed analysis of the DN subset using surface marker specific antibodies for CD25 and CD44 revealed a clear defect in the progression of DN thymocytes from the DN3 to the DN4 stage of development in the reporter gene-positive fraction of all A1 knockdown strains. This block in development was accompanied by the accumulation of DN2 and DN3 cells with a corresponding reduction of DN4 cells (Figure 3D-F). Again, the severity of the defect correlated with the extent of A1 knockdown with VVA1.2 mice showing the mildest phenotype. Of note, although doxycycline treatment of DT mice led to a loss of cells expressing high levels of eGFP in the peripheral blood already after 15 days (supplemental Figure 2A), the skewed DN3 to DN4 profile still persisted in mice that were kept on doxycycline for 32 days (supplemental Figure 2C). However, the abnormalities previously noted in the relative percentages of DP and CD8+ SP thymocytes (Figure 3C) were corrected at that stage (supplemental Figure 2C). Surprisingly, isolation and in vitro culture of DN3, DN4, as well as DP thymocytes failed to reveal any differences in spontaneous as well as drug-induced cell death between A1 knockdown and control cells (supplemental Figure 3A-B).
A1 knockdown impairs early T-cell development. Flow cytometric analysis of thymocytes was performed by staining single-cell suspensions derived from mice of the indicated genotypes with cell surface-marker specific antibodies recognizing CD4 or CD8, either alone or in combination with antibodies specific for CD25 and CD44 in subfractions, positive or negative for Venus or eGFP. (A) Representative dot blots of flow cytometric analysis of mice of the indicated genotypes using antibodies against CD4 or CD8. Numbers refer to percentages of quadrant analysis. (B-C) The percentages of CD4−CD8− DN, CD4+CD8+ DP, as well as CD4+CD8− and CD4−CD8+ SP thymocytes found in VVA1, DT, and relevant control mice. (D) Representative dot blots of flow cytometric analysis using antibodies against CD24 and CD44, gated on CD4−CD8− DN thymocytes. Numbers refer to percentages of quadrant analysis. (E-F) DN stages 1 to 4 (CD25−CD44+ DN1, CD25+CD44+ DN2, CD25+CD44− DN3, CD25−CD44− DN4) are quantified in VVA1, DT, and relevant control mice of the indicated genotypes. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic mice; **P ≤ .01, compared with WT or single-transgenic mice.
A1 knockdown impairs early T-cell development. Flow cytometric analysis of thymocytes was performed by staining single-cell suspensions derived from mice of the indicated genotypes with cell surface-marker specific antibodies recognizing CD4 or CD8, either alone or in combination with antibodies specific for CD25 and CD44 in subfractions, positive or negative for Venus or eGFP. (A) Representative dot blots of flow cytometric analysis of mice of the indicated genotypes using antibodies against CD4 or CD8. Numbers refer to percentages of quadrant analysis. (B-C) The percentages of CD4−CD8− DN, CD4+CD8+ DP, as well as CD4+CD8− and CD4−CD8+ SP thymocytes found in VVA1, DT, and relevant control mice. (D) Representative dot blots of flow cytometric analysis using antibodies against CD24 and CD44, gated on CD4−CD8− DN thymocytes. Numbers refer to percentages of quadrant analysis. (E-F) DN stages 1 to 4 (CD25−CD44+ DN1, CD25+CD44+ DN2, CD25+CD44− DN3, CD25−CD44− DN4) are quantified in VVA1, DT, and relevant control mice of the indicated genotypes. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic mice; **P ≤ .01, compared with WT or single-transgenic mice.
A1/Bfl-1 knockdown impairs B-cell homeostasis and proliferation
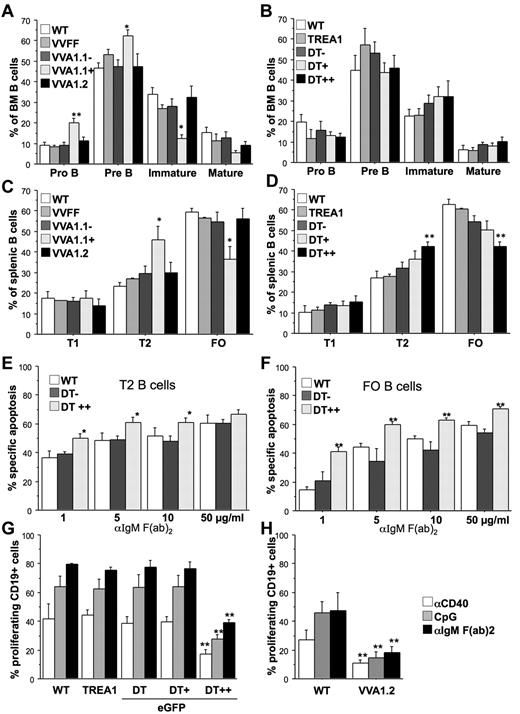
Analysis of the corresponding developmental stage in the B-cell lineage revealed largely normal development of CD19+c-kit+ pro-B and CD19+CD25+ pre-B cells in the Venus-positive fraction of the VVA1.2 and eGFP++ fraction of DT mice, whereas the Venus-positive fraction of VVA1.1 mice showed a mild but significant increase in these precursors and a concomitant reduction in IgM+IgD− immature B cells in the bone marrow (Figure 4A-B; supplemental Figure 4A). Similarly, the percentage of IgM+IgD+ transitional T2 B cells in spleen was found increased, which coincided with a relative drop in IgMlowIgD+ follicular B cells in the Venus+ fraction of VVA1.1 as well as the eGFP++ fraction of DT mice (Figure 4C-D; supplemental Figure 4B). This phenotype was reversed in DT mice that were kept on doxycycline for 32 days (supplemental Figure 2D). Of note, spontaneous death in culture of isolated pre-B, T2, or FO B cells with reduced A1 expression was comparable with that of control cells (supplemental Figure 3C-D). As a previous report correlated impaired survival of BCR-stimulated PLCγ-mutant mature B cells with lack of A1 induction,27 we explored the consequences of A1 knockdown on BCR ligation in sorted T1, T2, and mature follicular B cells. FACS-sorted eGFP++, eGFP+, and eGFP− cells from spleens of DT mice were cultured in the absence or presence of graded doses of plate-bound anti-IgM F(ab)2 fragments, and cell survival was monitored by flow cytometric analysis. T1 B cells were most sensitive to BCR crosslinking, but A1 knockdown had no impact on their death (not shown). In contrast, in T2 B cells as well as FO B cells, expression of mi-shRNA against A1 significantly enhanced BCR stimulation-induced apoptosis (Figure 4E-F).
A1 knockdown impairs B-cell responsiveness. Flow cytometric analysis of splenocytes was performed by staining single-cell suspensions derived from mice of the indicated genotypes with cell surface-marker–specific antibodies in the different subfractions, positive or negative for Venus or eGFP. (A) Early B-cell development was analyzed in the bone marrow of VVA1 mice by flow cytometric analysis, using cell surface-marker–specific antibodies to identify CD19+c-Kit+ pro-B cells, CD19+CD25+ pre-B cells, IgM+IgD− immature B cells, and IgMlowIgD+ recirculating mature B cells. (B) Analysis of early B-cell development in DT and relevant control mice performed as in panel A. (C) B-cell distribution in spleens of VVA1 and relevant control mice. Transitional type 1 (T1) B cells (IgM+IgD−), transitional type 2 (T2) B cells (IgM+IgD+), and follicular (FO) B cells (IgMlowIgD+). (D) B-cell distribution in spleens of DT and relevant control mice performed as in panel C. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls. (E) T2 or (F) FO B cells from DT mice were sorted based on eGFP as well as levels of surface IgM and IgD expression (T2, IgM+IgD+; FO, IgMlowIgD+) and cultured in the presence of graded concentrations of plate-bound IgM F(ab′)2 fragments. Cell viability was assessed over time by 7-amino-actinomycin D exclusion and flow cytometric analysis. The extent of apoptosis induced specifically by BCR ligation was calculated by the following equation: (induced apoptosis/spontaneous cell death) × 100. Data are mean ± SEM of 5 independent experiments performed in duplicates. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with DT− or WT controls; **P ≤ .01, compared with DT− or WT controls. Total spleen cells from WT, TREA1, DT (G) or VVA1.2 mice (H), labeled with CDP proliferation dye were incubated with the indicated mitogens (anti-CD40 mAb, 1 μg/mL; CpG oligonucleotide, 100nM; anti-IgM F(ab′)2 fragments, 2 μg/mL). The percentages of CD19+ cells that underwent cell division were assessed in cells expressing different eGFP levels after 72 hours by flow cytometric analysis. Data are mean ± SEM of 3 independent experiments performed in duplicates. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic mice; **P ≤ .01 compared with WT or single-transgenic mice.
A1 knockdown impairs B-cell responsiveness. Flow cytometric analysis of splenocytes was performed by staining single-cell suspensions derived from mice of the indicated genotypes with cell surface-marker–specific antibodies in the different subfractions, positive or negative for Venus or eGFP. (A) Early B-cell development was analyzed in the bone marrow of VVA1 mice by flow cytometric analysis, using cell surface-marker–specific antibodies to identify CD19+c-Kit+ pro-B cells, CD19+CD25+ pre-B cells, IgM+IgD− immature B cells, and IgMlowIgD+ recirculating mature B cells. (B) Analysis of early B-cell development in DT and relevant control mice performed as in panel A. (C) B-cell distribution in spleens of VVA1 and relevant control mice. Transitional type 1 (T1) B cells (IgM+IgD−), transitional type 2 (T2) B cells (IgM+IgD+), and follicular (FO) B cells (IgMlowIgD+). (D) B-cell distribution in spleens of DT and relevant control mice performed as in panel C. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls. (E) T2 or (F) FO B cells from DT mice were sorted based on eGFP as well as levels of surface IgM and IgD expression (T2, IgM+IgD+; FO, IgMlowIgD+) and cultured in the presence of graded concentrations of plate-bound IgM F(ab′)2 fragments. Cell viability was assessed over time by 7-amino-actinomycin D exclusion and flow cytometric analysis. The extent of apoptosis induced specifically by BCR ligation was calculated by the following equation: (induced apoptosis/spontaneous cell death) × 100. Data are mean ± SEM of 5 independent experiments performed in duplicates. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with DT− or WT controls; **P ≤ .01, compared with DT− or WT controls. Total spleen cells from WT, TREA1, DT (G) or VVA1.2 mice (H), labeled with CDP proliferation dye were incubated with the indicated mitogens (anti-CD40 mAb, 1 μg/mL; CpG oligonucleotide, 100nM; anti-IgM F(ab′)2 fragments, 2 μg/mL). The percentages of CD19+ cells that underwent cell division were assessed in cells expressing different eGFP levels after 72 hours by flow cytometric analysis. Data are mean ± SEM of 3 independent experiments performed in duplicates. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic mice; **P ≤ .01 compared with WT or single-transgenic mice.
Assessing proliferation by CPD-labeling and flow cytometric analysis revealed that A1 knockdown impaired the growth of eGFP++ splenic B cells from DT or VV1.2 mice in response to mitogenic stimulation using either anti-IgM F(ab)2 fragments, a CD40 crosslinking antibody or a CpG-motive containing oligonucleotide (Figure 4G-H; supplemental Figure 5A). Of note, the proliferative response of mature splenic CD4+ or CD8+ T cells, stimulated with anti-CD3 mAb or the lectin ConA, were unaffected by knockdown of A1 (supplemental Figure 5B).
A1 is critical for the survival of mature granulocytes and their progenitors
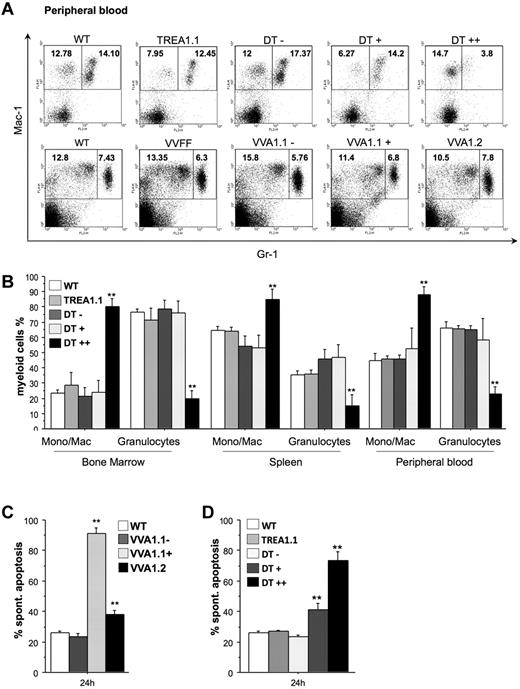
Flow cytometric analysis using antibodies specific for the cell surface markers Mac-1 and Gr-1 to identify monocytes/macrophages (Mac-1+Gr-1−) and granulocytes (Mac-1+Gr-1+) revealed comparable percentages of granulocytes in the VVFF and both VVA1 strains, but a severe reduction of these cells was observed in the eGFP++ fraction in bone marrow, spleen, and peripheral blood of DT mice (Figure 5A-B; supplemental Figure 6). Accordingly, there was a relative increase in the percentages of monocytes/macrophages in this population. Importantly, this phenotype was largely reversed in DT mice kept on doxycycline (supplemental Figure 2E). Furthermore, when we isolated granulocytes from VVA1.1 mice by cell sorting, the Venus-positive cells lacking A1 expression showed most rapid cell death in culture, and granulocytes derived from VVA1.2 bone marrow showed higher cell death rates than Venus-negative cells or granulocytes from nontransgenic mice (Figure 5C). In addition, granulocytes from the eGFP+ and eGFP++ fraction of DT bone marrow died more rapidly in culture than those from TREA1 single-transgenic or nontransgenic control mice (Figure 5D). This indicates that the low level expression of the siRNA in eGFP+ cells derived from DT mice and the minor reduction of A1 mRNA achieved in this pool of cells (Figure 2E) was already sufficient to compromise granulocyte survival (Figure 5D). When Gr-1+ granulocytes were sorted from the bone marrow of DT mice that were kept on doxycycline, their spontaneous death in culture was no longer different from that of granulocytes from TREA1 mice (supplemental Figure 2F).
Impaired myelopoiesis and survival of granulocytes on A1 knockdown. (A) Representative dot blots of flow cytometric analysis of peripheral blood of mice of the indicated genotypes using antibodies to identify Mac-1+Gr-1− monocytes/macrophages and Mac-1+Gr-1+ granulocytes, respectively, are shown. Numbers refer to percentages of region analysis. (B) The composition of myeloid cells in the bone marrow, spleen, and blood of WT, TREA1 single-transgenic, and the different eGFP fractions (−, +, ++) from DT mice is shown. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls. Gr1high granulocytes were sorted from the bone marrow of (C) WT, the Venus+, or Venus− pool of bone marrow cells from VVA1.1 or VVA1.2 mice. Alternatively, granulocytes were isolated from (D) WT, TREA1 single-transgenic mice, or the eGFP−, eGFP+, or eGFP++ fraction of DT mice. Cells were put in culture without further treatment. Viability was assessed by 7-amino-actinomycin D exclusion in a flow cytometer. Bars represent mean ± SD of 4 mice per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls.
Impaired myelopoiesis and survival of granulocytes on A1 knockdown. (A) Representative dot blots of flow cytometric analysis of peripheral blood of mice of the indicated genotypes using antibodies to identify Mac-1+Gr-1− monocytes/macrophages and Mac-1+Gr-1+ granulocytes, respectively, are shown. Numbers refer to percentages of region analysis. (B) The composition of myeloid cells in the bone marrow, spleen, and blood of WT, TREA1 single-transgenic, and the different eGFP fractions (−, +, ++) from DT mice is shown. Data are mean ± SEM of n = 4 to 8 animals per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls. Gr1high granulocytes were sorted from the bone marrow of (C) WT, the Venus+, or Venus− pool of bone marrow cells from VVA1.1 or VVA1.2 mice. Alternatively, granulocytes were isolated from (D) WT, TREA1 single-transgenic mice, or the eGFP−, eGFP+, or eGFP++ fraction of DT mice. Cells were put in culture without further treatment. Viability was assessed by 7-amino-actinomycin D exclusion in a flow cytometer. Bars represent mean ± SD of 4 mice per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; **P ≤ .01, compared with WT or single-transgenic controls.
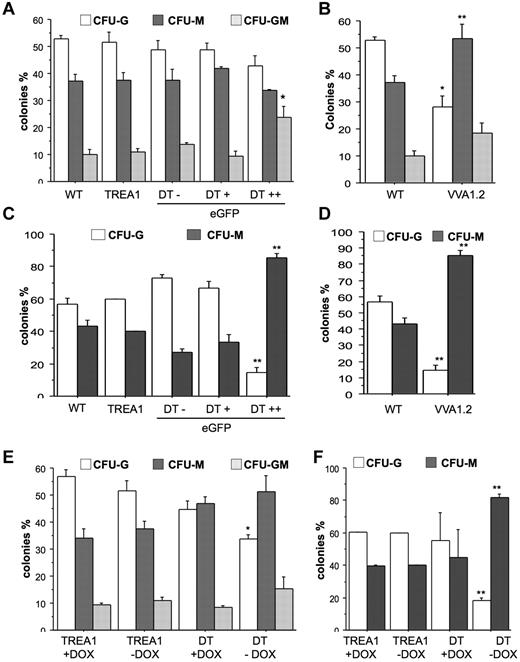
To investigate whether A1 knockdown mainly affects spontaneous death of mature granulocytes or may also impact on the clonogenic capacity of myeloid progenitors, we performed colony formation assays in soft agar with bone marrow cells from VVA1.2 mice or sorted, based on fluorescence marker-gene expression, from DT mice and assessed their colony-forming potential when growth- and survival-promoting cytokines were added either immediately or with a 24-hour delay. When cytokines were present from the start, we found that A1 knockdown caused a bias toward the development of mixed granulocyte/macrophage (GM) colonies in the eGFP++ fraction derived from DT mice (Figure 6A). This effect was not noted in bone marrow cells derived from VV1.2 mice that showed preferential outgrowth into CFU-M colonies (Figure 6B). Regardless, when cytokines were added with a 24-hour delay, A1 knockdown caused a significant reduction in the numbers and percentage of granulocytic colonies and most colonies that formed showed macrophage morphology whereas mixed GM colonies were no longer detected. This phenotype was consistent between the different transgenic lines (Figure 6C-D). Importantly, when we sorted bone marrow from DT mice that were kept on doxycycline for 32 days to shut off miR30-A1 expression and plated these cells in the presence of doxycycline in methylcellulose, we maintained their potential to form CFU-G colonies. The CFU-G–forming potential was comparable with that of TREA1-derived controls, insenstive to doxycycline (Figure 6C-D). When cultured in the absence of doxycycline, the consequent reinduction of the A1 siRNA caused a reduction of CFU-G colonies (Figure 6E). Again, this effect was more pronounced after delayed cytokine addition (Figure 6F). Collectively, these studies demonstrate that A1 expression is critical for granulopoiesis.
Reduced colony formation of myeloid progenitors on A1 knockdown. (A) A total of 2 × 104 cells derived from bone marrow from WT, TREA1, or DT mice, sorted into eGFP−, eGFP+, and eGFP++ fractions, or (B) WT and VVA1.2 mice were plated in Methocult medium in the presence of G-CSF, IL-6, and IL-3. (C-D) Alternatively, cytokines were added after a 24-hour delay. Colonies were counted blinded after 7 days of incubation. Data are mean ± SE of 3 independent experiments performed in duplicates. (E) Bone marrow cells were isolated from single-transgenic TREA1 or DT mice that had been treated for 32 days with doxycycline in the drinking water. Cells were isolated and seeded into cytokine-complemented Methocult medium in the absence or presence of doxycycline. (F) Alternatively, cytokines were added after 24-hour delay. Colonies were counted in duplicate cultures blinded after 7 days of incubation. Data are mean ± SD of 2 or 3 mice per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; and **P ≤ .01, compared with WT or single-transgenic controls.
Reduced colony formation of myeloid progenitors on A1 knockdown. (A) A total of 2 × 104 cells derived from bone marrow from WT, TREA1, or DT mice, sorted into eGFP−, eGFP+, and eGFP++ fractions, or (B) WT and VVA1.2 mice were plated in Methocult medium in the presence of G-CSF, IL-6, and IL-3. (C-D) Alternatively, cytokines were added after a 24-hour delay. Colonies were counted blinded after 7 days of incubation. Data are mean ± SE of 3 independent experiments performed in duplicates. (E) Bone marrow cells were isolated from single-transgenic TREA1 or DT mice that had been treated for 32 days with doxycycline in the drinking water. Cells were isolated and seeded into cytokine-complemented Methocult medium in the absence or presence of doxycycline. (F) Alternatively, cytokines were added after 24-hour delay. Colonies were counted in duplicate cultures blinded after 7 days of incubation. Data are mean ± SD of 2 or 3 mice per genotype. ANOVA followed by Fisher PLSD posthoc test: *P ≤ .05, compared with WT or single-transgenic controls; and **P ≤ .01, compared with WT or single-transgenic controls.
Discussion
Despite the high interest in Bcl-2–regulated cell death mechanisms, little information is available on the Bcl-2 prosurvival homolog A1/Bfl-1. Using constitutive as well as conditional expression of mi-shRNAs targeting all A1 isoforms expressed in the mouse (Figures 1 and 2; supplemental Figure 1), we have delineated biologic roles for A1 in early T-cell development, B-cell homeostasis, as well as granulopoiesis.
The observed accumulation of immature thymocytes at the DN3 stage of development and subsequent loss of DP thymocytes on reduced expression of A1 (Figure 3) is in line with the notion that A1 mRNA is induced on expression of a functional pre-T-cell receptor and T cell–restricted A1 overexpression is enhancing DN → DP maturation and positive selection of thymocytes in RAG1-deficient as well as TCR-transgenic mice.14,28 Interestingly, we observed a proportional increase of CD8 SP thymocytes, which might be the result of increased loss or impaired maturation of CD4 SP cells. Recently, Singer et al demonstrated that CD8 lineage choice is dependent on IL-7 signaling, whereas CD4 specification is determined by prolonged TCR signaling.29 Because A1 expression in the thymus is TCR-dependent, CD4 SP thymocytes probably may have a survival disadvantage compared with CD8 SP cells, leading to the observed bias. Consistently, CD8 SP thymocytes do express significantly less A1 mRNA compared with their CD4 SP thymocytes28 and therefore may be less affected by A1 knockdown allowing their accumulation.
Surprisingly, abrogation of the A1 mi-shRNA on doxycycline treatment of mice did not reverse the block at the DN3 stage of thymocyte development, whereas that on DN2 cells was already relieved (supplemental Figure 2C). This may be explained by low-level mi-siRNA production, even in the presence of doxycycline in these cells that may suffice to compromise DN3 maturation, although this awaits experimental validation. Curiously, DN3 and DN4 cells showed extremely high rates of cell death in culture because of cytokine deprivation (supplemental Figure 3) irrespective of their genotype (> 70% death after 24 hours), suggesting that an effect of A1 knockdown under these conditions might be difficult to reveal. Similarly, reduced expression of A1 also failed to sensitize DP thymocytes to the apoptotic effects of ionomycin, dexamethasone, or TCR ligation (supplemental Figure 3). Although not explored experimentally, compensation appears plausible (eg, by Mcl-1 shown to be critical for normal thymocyte development and their stress resistance).30
Besides its effect on the developmental progression of thymocytes, A1 knockdown in our model system appears also beyond a critical threshold for unperturbed mature B-cell survival, leading to enhanced demand of progenitors, most prominently noted in the Venus-positive fraction of VV1.1 and eGFP++ fraction of DT mice (Figure 4A-D). Our observations are consistent with the perturbed B-cell development and pro-B cell accumulation noted in Eμ-A1 transgenic mice.8 The lack of an apparent cell death phenotype in culture, however, may be the result of the reported low-level mRNA expression in pro- and pre-B cells31 or the aforementioned compensatory action of Mcl-1.30 A1 knockdown enhanced BCR-induced apoptosis in T2 and FO, but not T1 B cells (Figure 4E-F; and data not shown). This is in accordance with the lack of A1 induction in BCR-stimulated T1 B cells but consistent with increased mRNA levels observed on mitogenic stimulation in mature FO B cells ex vivo.32 In addition, it was reported that overexpression of A1 protected against BCR ligation-mediated killing in WEHI2315 or in FO B cells.27
The growth disadvantage of splenic A1-deficient B cells in response to BCR, CD40, or TLR-9 signals indicates that induction of A1 is a general prerequisite for the survival of mature B cells during mitogen-induced stimulation (Figure 4G-H). This observation is in line with earlier investigations into A1 mRNA expression during B-cell maturation31,33 or the analysis of B-cell lines derived from mice lacking the NF-κB family member c-Rel.34 Consistently, loss of the BH3-only protein Noxa, a proposed A1 antagonist induced on B-cell stimulation, was recently reported to facilitate the outgrowth of low-affinity B cell clones from germinal centers.35
Interestingly, we observed different phenotypes with graded A1 knockdown in the myeloid lineage, ranging from a strong reduction of granulocytes in DT mice with the strongest A1 knockdown to a shorter life span of granulocytes ex vivo isolated from VVA1 mice with a lower knockdown efficiency (Figure 5). This is in line with observations made using granulocytes from A1-a deficient mice, lacking one of 3 A1-encoding genes,11 and suggests that a critical threshold of A1 expression is required to allow the successful maturation of granulocytes, dictating their overall life span. In addition, we observed reduced clonogenic capacity of myeloid precursors and a preference to differentiate into the monocytic/macrophage lineage (Figure 6). This phenotype is strikingly similar to the one observed on conditional LysM-cre mediated deletion of Mcl-1 in myeloid progenitors,36 suggesting that both Mcl-1 and A1 are nonredundant survival factors in granulopoiesis, whereas monocyte survival during development must be regulated by different Bcl-2 family members, possibly also by Bcl-2 itself.37
Our model systems allowed a detailed analysis of the relevance of A1 in the development of the hematopoietic system and will enable studies into its role in malignant transformation as well as allergy and autoimmunity in the near future.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank A. Strasser and T. Huenig for critical reading of the manuscript, D. Largeaspada for Vav-tTA mice, I. Gaggl for mouse genotyping, K. Rossi and C. Bauer for animal care, G. Böck for cell sorting, N. Yannoutsos for oocyte injections (MUI Transgenesis Unit), C. Ploner for help with lentiviral transduction, P. Bouillet for the pGL-luc-Bim reporter construct, and I. Berberich for A1-a cDNA.
This work was supported by Graduate School for Molecular Biology and Oncology, (E.O., F.G., A.V., and S.G.; and SFB021, A.V.), the Austrian Science Fund, the Tiroler Krebshilfe (E.O. and D.T.), and the German Research Council (Deutsche Forschungsgemeinschaft, Re 1631/7-1; H.M.R.). M.J.H. was supported by the Deutsche Forschungsgemeinschaft (postdoctoral fellowship He 5740/1-1).
Authorship
Contribution: E.O. performed experiments, analyzed data, wrote the paper, and prepared the figures; F.G., D.T., C.S., and A.P. performed experiments; S.G. and H.M.R. designed research and provided reagents; A.V. designed research, analyzed data, prepared figures, wrote the paper, and conceived the study; and M.J.H. provided reagents, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for F.G. is Medical Research Council, Toxicology Unit, Leicester University, Leicester, United Kingdom.
Correspondence: Andreas Villunger, Division of Developmental Immunology, Biocenter, Innsbruck Medical University, Innrain 80-82, A-6020 Innsbruck, Austria; e-mail: andreas.villunger@i-med.ac.at; and Marco J. Herold, Walter and Eliza Hall Institute, Parkville, 3050 Victoria, Australia; e-mail: herold@wehi.edu.au.
References
Author notes
A.V. and M.J.H. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal