Abstract
T-cell acute lymphoblastic lymphomas commonly demonstrate activating Notch1 mutations as well as mutations or deletions in Fbxw7. However, because Fbxw7 targets Notch1 for degradation, genetic alterations in these genes are expected to be mutually exclusive events in lymphomagenesis. Previously, by using a radiation-induced Tp53-deficient mouse model for T-cell acute lymphoblastic lymphoma, we reported that loss of heterozygosity at the Fbxw7 locus occurs frequently in a Tp53-dependent manner. In the current study, we show that these thymic lymphomas also commonly exhibit activating Notch1 mutations in the proline-glutamic acid-serine-threonine (PEST) domain. Moreover, concurrent activating Notch1 PEST domain mutations and single-copy deletions at the Fbxw7 locus occur with high frequency in the same individual tumors, indicating that these changes are not mutually exclusive events. We further demonstrate that although Notch1 PEST domain mutations are independent of Tp53 status, they are completely abolished in mice with germline Fbxw7 haploinsufficiency. Therefore, Notch1 PEST domain mutations only occur when Fbxw7 expression levels are intact. These data suggest a temporal sequence of mutational events involving these important cancer-related genes, with Notch1 PEST domain mutations occurring first, followed by Fbxw7 deletion, and eventually by complete loss of Tp53.
Introduction
Activating Notch1 mutations occur in > 50% of human T-cell acute lymphoblastic lymphomas (T-ALLs).1 Comparable mutations arise in murine thymic lymphomas generated in genetically sensitized mouse models.2,3 These genetic alterations occur exclusively in the heterodimerization (HD) and proline-glutamic acid-serine-threonine (PEST) domains of Notch1, resulting in elevated levels of intracellular domain of Notch1 (ICN). HD domain mutations lead to unsolicited release of ICN,4 whereas PEST domain mutations prevent Fbxw7-mediated turnover of ICN.5-9
The mechanism by which elevated ICN levels lead to lymphomagenesis is not entirely clear. Some evidence suggests that elevated ICN may inhibit p19-Arf, leading to increased Mdm2-mediated degradation of Tp53.10 If indeed this pathway plays an important role in lymphomagenesis, activating Notch1 mutations and loss of Tp53 activity are expected to be mutually exclusive genetic events. Consistent with this hypothesis, a large-scale retroviral insertional mutagenesis screen demonstrated that Notch1 mutations occur more frequently in tumors from wild-type mice than from their Tp53-deficient counterparts.11
Genetic alterations in Fbxw7 are also commonly found in human T-ALL and murine thymic lymphomas.9,12 Fbxw7 is the F-box component of a Skp1-Cul1-F-box protein ubiquitin ligase complex and acts as a tumor suppressor in numerous types of human cancers.13 It targets several proto-oncogenes for ubiquitin-mediated degradation, including Notch1/4, Myc, Jun, Cyclin E, and mTOR.14-16 Nearly all ionizing radiation (IR)–induced thymic lymphomas from Tp53+/− mice exhibit single-copy deletion at the Fbxw7 locus, whereas those from Tp53−/− mice show no such aberration, indicating that Fbxw7 acts as a Tp53-dependent haploinsufficient tumor suppressor.12 Although FBXW7 mutations occasionally occur with concurrent NOTCH1 HD domain mutations in human T-ALL, the authors of several studies suggest that point mutations in FBXW7 and activating mutations in the NOTCH1 PEST domain may be mutually exclusive.8,17,18
We have previously demonstrated that IR-induced thymic lymphomas from Tp53+/− mice are genetically unstable, showing gene copy number gains in regions harboring known proto-oncogenes (ie, Aurka, Myc, and Notch1) and deletions or mutations in regions containing tumor suppressors (ie, Fbxw7).19,20 Although all of these tumors eventually lose the wild-type allele of Tp53, the order of the genetic events that takes place after radiation exposure has not been determined. In this study, we investigate the relationship of Notch1, Fbxw7, and Tp53 genetic alterations in thymic lymphomas to establish their sequential order of occurrence during lymphomagenesis. We show that activating Notch1 mutations in this model occur commonly in the PEST domain and are independent of Tp53 status.
In addition, Notch1 PEST domain mutations only occur in the presence of 2 functional copies of Fbxw7, whereas tumors from mice with only one germline copy of Fbxw7 fail to exhibit Notch1 PEST domain mutations. Interestingly, whereas point mutations in FBXW7 and the NOTCH1 PEST domain may be mutually exclusive changes as indicated in previous studies, we find that single-copy deletion of Fbxw7 and Notch1 PEST domain mutations occur at a high frequency in the same tumors. Because Notch1 PEST domain mutations are completely dependent on the presence of normal Fbxw7 expression levels, we conclude that in tumors harboring both of these genetic alterations, Notch1 PEST domain mutations are among the earliest genetic changes in IR-induced thymic lymphomas and are followed by Fbxw7 deletion and eventually by complete loss of Tp53.
Methods
Mice and tumor induction
Tp53+/−, Tp53−/−, Tp53+/−Fbxw7+/−, and Tp53−/−Fbxw7+/− mice were maintained on a 129/Sv background. F1 hybrid mice were produced by crossing female Tp53−/− 129/Sv mice with male Tp53+/+Mus spretus. Thymic lymphomas were generated as previously described.12 To summarize in brief, 5-week-old Tp53+/−, Tp53−/−, Tp53+/−Fbxw7+/−, Tp53−/−Fbxw7+/−, and F1 hybrid mice were exposed to a single dose of 4 Gy IR. Afterward, these mice were observed daily until they were moribund, then killed and autopsied. Thymic lymphomas were collected and used for subsequent analyses. Mice were bred and treated under the University of California San Francisco (UCSF) Institutional Animal Care and Use Committee regulations, and mouse experiments were approved by the UCSF Institutional Animal Care and Use Committee.
Mutation analysis of Notch1
The Notch1 gene was sequenced for the thymic lymphomas collected from 24 p53+/−, 23 Tp53−/−, 9 Tp53+/−Fbxw7+/−, 9 Tp53−/−Fbxw7+/−, and 35 F1 hybrid mice. First, thymic lymphoma samples were homogenized and total RNA was extracted via the use of TRIzol (Invitrogen). Subsequently, cDNA was synthesized from 0.25 μg of total RNA with the QuantiTect Reverse Transcription Kit (QIAGEN). PCR primers were designed on the basis of the Notch1 cDNA sequence (NM_008714). The HD and PEST domains for each thymic lymphoma were PCR amplified with the following primers: HD forward, 5′-AACAGTGCCGAATGTGAGTG-3′; HD reverse, 5′-CACAAAGAACAGGAGCACGA-3′; PEST1 forward, 5′-AGTCACCCCATGGCTACTTG-3′; PEST1 reverse, 5′- ACTGAGGTGTGGCTGTGATG-3′; PEST2 forward, 5′-ATAGCATGATGGGGCCACTA-3′; and PEST2 reverse, 5′-CCTGAAGCACTGGAAAGGAC-3′. The PCR products were purified, sequenced (Quintara), and analyzed for mutations.
Notch1 type 1 deletions were examined by PCR as described previously by Ashworth et al21 with minor modifications. The tumor genomic DNA was amplified with Taq DNA Polymerase (Allstar Scientific) with the forward primer 5′-ATGGTGGAATGCCTACTTTGTA-3′ and the reverse primer 5′-CGTTTGGGTAGAAGAGATGCTTTAC-3′. A positive control sample was kindly provided by Dr Jon Aster (Brigham and Women's Hospital).
Fbxw7 loss of heterozygosity and expression analysis
Thymic lymphoma DNA was extracted and examined for loss of heterozygosity (LOH) at the Fbxw7 locus as described previously.12 For Fbxw7 expression analysis, thymi were collected from 5-week-old wild-type, Tp53+/−, Tp53−/−, and Fbxw7+/− 129/Sv mice. Three animals for each genotype were used. Total RNA and cDNA synthesis were performed as described previously. Fbxw7 expression levels were determined by use of the TaqMan Gene Expression Assay for mouse Fbxw7 on a 7900 HT Fast Real-Time PCR System (Applied Biosystems). Beta-actin expression was used as an endogenous control.
Results
Notch1 PEST domain mutations are independent of Tp53 status
Because activating Notch1 mutations may be mutually exclusive toTp53 inactivation,10,11,22 we examined whether Notch1 mutations occur in thymic lymphomas from Tp53-deficient mice. IR-induced thymic lymphomas from Tp53−/− and Tp53+/− 129/Sv mice were interrogated for the most common Notch1 mutations, those in the HD and PEST domains. Of the thymic lymphomas from Tp53−/− mice, 9 of 23 (39%) contained Notch1 PEST domain mutations; similarly, 10 of 24 (42%) thymic lymphomas from Tp53+/− mice displayed Notch1 PEST domain mutations (Table 1). Interestingly, no mutations were observed in the HD domain. Instead, the mutations were exclusively within the PEST domain and result in the creation of a premature stop codon.
Type and frequency of Notch1 mutations in murine thymic lymphoma
| Genotype . | n* . | PEST domain mutations . | HD domain mutations . | Percent with Notch1 mutations . |
|---|---|---|---|---|
| Tp53+/− | 24 | 10 | 0 | 42 |
| Tp53−/− | 23 | 9 | 0 | 39 |
| Tp53+/− and Fbxw7+/− | 9 | 0 | 0 | 0† |
| Tp53−/− and Fbxw7+/− | 9 | 0 | 0 | 0† |
| F1 hybrid (Tp53+/−) | 35 | 16 | 0 | 46 |
| Genotype . | n* . | PEST domain mutations . | HD domain mutations . | Percent with Notch1 mutations . |
|---|---|---|---|---|
| Tp53+/− | 24 | 10 | 0 | 42 |
| Tp53−/− | 23 | 9 | 0 | 39 |
| Tp53+/− and Fbxw7+/− | 9 | 0 | 0 | 0† |
| Tp53−/− and Fbxw7+/− | 9 | 0 | 0 | 0† |
| F1 hybrid (Tp53+/−) | 35 | 16 | 0 | 46 |
HD indicates heterodimerization; and PEST, proline-glutamic acid-serine-threonine.
Total number of thymic lymphomas examined.
P < .03.
In all cases, the Fbxw7 recognition sequence within the Notch1 PEST domain was lost. The majority of these genetic alterations demonstrated complex insertions and deletions at a few mutational hotspots (Table 2). The type and frequency of mutations did not differ between thymic lymphomas from Tp53−/− and Tp53+/− mice. Taken together, these findings indicate that IR-induced thymic lymphomas from a Tp53-sensitized mouse model exhibit a high frequency of PEST domain mutations within Notch1. Furthermore, in contrast to the conclusions of earlier studies,10,11 Notch1 PEST domain mutations were independent of the original Tp53 status of the mice that harbored these tumors, being present in approximately the same frequency and showing similar changes in both Tp53−/− and Tp53+/− mice.
Notch1 mutations in Tp53+/− and Tp53−/− mouse thymic lymphomas
| Tp53+/− . | Tp53−/− . | ||
|---|---|---|---|
| Tumor ID . | Mutation status* . | Tumor ID . | Mutation status* . |
| RH-1 | Wild-type | RN-1 | 7015_7016insG |
| RH-2 | 7250_7253ATGT > GGAGTGGG | RN-2† | – |
| RH-3 | 7258_7259insA | RN-3 | Wild-type |
| RH-4 | Wild-type | RN-4 | 7082G > CC |
| RH-5 | 7260_7264CCGCT > ACCCCG | RN-5 | Wild-type |
| RH-6 | Wild-type | RN-6 | 7167_7168insA |
| RH-7 | Wild-type | RN-7 | Wild-type |
| RH-8 | 7194_7195insCGACGCAGAA | RN-8 | Wild-type |
| RH-9 | Wild-type | RN-9 | 6848_6849insG |
| RH-10 | Wild-type | RN-10 | Wild-type |
| RH-11 | Wild- type | RN-11 | 7132_7136AACTT > CCG |
| RH-12 | 7194_7195insAGGAGAA | RN-12 | 7315_7379del |
| RH-13 | Wild-type | RN-13 | Wild-type |
| RH-14 | 7299C > GGGGGT | RN-14 | Wild-type |
| RH-15 | Wild-type | RN-15 | 7194_7195insAA |
| RH-16 | Wild-type | RN-16 | Wild-type |
| RH-17 | Wild-type | RN-17 | Wild-type |
| RH-18 | Wild-type | RN-18 | Wild-type |
| RH-19 | Wild-type | RN-19 | 7325C > A |
| RH-20 | 7082G > CCCCAC | RN-20 | 7217_7218insC |
| RH-21 | 7082G > TC | RN-21 | Wild-type |
| RH-22 | 7226T > A | RN-22 | Wild-type |
| RH-23 | 7194_7195insTGGG | RN-23 | Wild-type |
| RH-24 | Wild-type | RN-24 | Wild-type |
| Tp53+/− . | Tp53−/− . | ||
|---|---|---|---|
| Tumor ID . | Mutation status* . | Tumor ID . | Mutation status* . |
| RH-1 | Wild-type | RN-1 | 7015_7016insG |
| RH-2 | 7250_7253ATGT > GGAGTGGG | RN-2† | – |
| RH-3 | 7258_7259insA | RN-3 | Wild-type |
| RH-4 | Wild-type | RN-4 | 7082G > CC |
| RH-5 | 7260_7264CCGCT > ACCCCG | RN-5 | Wild-type |
| RH-6 | Wild-type | RN-6 | 7167_7168insA |
| RH-7 | Wild-type | RN-7 | Wild-type |
| RH-8 | 7194_7195insCGACGCAGAA | RN-8 | Wild-type |
| RH-9 | Wild-type | RN-9 | 6848_6849insG |
| RH-10 | Wild-type | RN-10 | Wild-type |
| RH-11 | Wild- type | RN-11 | 7132_7136AACTT > CCG |
| RH-12 | 7194_7195insAGGAGAA | RN-12 | 7315_7379del |
| RH-13 | Wild-type | RN-13 | Wild-type |
| RH-14 | 7299C > GGGGGT | RN-14 | Wild-type |
| RH-15 | Wild-type | RN-15 | 7194_7195insAA |
| RH-16 | Wild-type | RN-16 | Wild-type |
| RH-17 | Wild-type | RN-17 | Wild-type |
| RH-18 | Wild-type | RN-18 | Wild-type |
| RH-19 | Wild-type | RN-19 | 7325C > A |
| RH-20 | 7082G > CCCCAC | RN-20 | 7217_7218insC |
| RH-21 | 7082G > TC | RN-21 | Wild-type |
| RH-22 | 7226T > A | RN-22 | Wild-type |
| RH-23 | 7194_7195insTGGG | RN-23 | Wild-type |
| RH-24 | Wild-type | RN-24 | Wild-type |
Number denotes position from Notch1 cDNA start site.
Sample was lost to degradation.
Tumors from Fbxw7+/− mice lack Notch1 PEST domain mutations
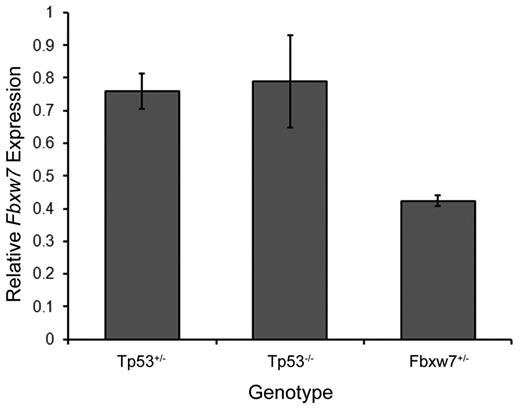
Because PEST domain mutations eliminate the Fbxw7 recognition sequence within the C-terminal region of Notch1, we proceeded to determine the frequency of Notch1 PEST domain mutations in thymic lymphomas from Tp53−/− and Tp53+/− mice with only one functional copy of Fbxw7. In contrast to mice with 2 functional copies of Fbxw7, no Notch1 mutations in either the HD or PEST domains were seen in thymic lymphomas from Tp53+/−Fbxw7+/− or Tp53−/−Fbxw7+/− mice (Table 1). As expected, further analysis by quantitative RT-PCR confirmed that Fbxw7+/− mice showed significantly lower Fbxw7 expression levels than Tp53−/− and Tp53+/− mice carrying 2 functional Fbxw7 alleles (Figure 1). Therefore, the acquisition of Notch1 PEST domain mutations is dependent on intact Fbxw7 levels and appears to proceed only when Fbxw7 expression levels exceed a specific threshold.
Fbxw7 mRNA levels in thymus.Fbxw7 expression levels are calculated relative to wild-type mice. Each genotype represents data taken from 3 animals.
Fbxw7 mRNA levels in thymus.Fbxw7 expression levels are calculated relative to wild-type mice. Each genotype represents data taken from 3 animals.
Conversely, Fbxw7 haploinsufficiency precludes the selection for Notch1 PEST domain mutations during thymic lymphoma development. These observations support the Tp53-independent nature of Notch1 mutations as presented in the previous section because both Tp53−/− and Tp53+/− mice showed approximately normal Fbxw7 expression levels. Of note, Ashworth et al recently reported new types of Notch1 mutations in murine T-ALLs in the form of deletions that lead to ligand-independent Notch1 activation.21 We analyzed a panel of 43 tumors for the presence of type 1 deletions in the Notch1 gene and found that only very rare mutations of this type were seen in tumors from Fbxw7+/− mice (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article; Figure 1). Only 1 of 19 tumors from Fbxw7+/− mice exhibited evidence for a deletion, whereas 9 of 24 tumors from Fbxw7+/+ mice contained similar deletions (P < .03 by Fisher exact test). These findings suggest that the frequency of this alternative method for Notch1 activation is also reduced in animals carrying only one copy of the Fbxw7 gene. However, additional routes to activation of Notch1 by larger deletions, gene amplification, or increased transcription have not been excluded. Further studies would be required to test these possibilities, as well as to investigate the potential roles of additional Notch gene family members in early stages of lymphomagenesis.
Concurrent Fbxw7 deletion and Notch1 PEST domain mutations occur frequently
We previously demonstrated that Fbxw7 LOH is a Tp53-dependent event that occurs in > 90% of murine thymic lymphomas from Tp53+/− F1 hybrids generated from a Mus spretus and Mus musculus cross.12 Because Notch1 PEST domain mutations also occur at a high frequency in approximately 40% of murine thymic lymphomas, it would be expected that many tumors will exhibit both Notch1 PEST domain mutations and LOH/deletion at the Fbxw7 locus. Therefore, we interrogated tumors from Tp53+/− F1 hybrids for Notch1 PEST domain mutations, and by using microsatellite markers, determined the status of their Fbxw7 loci. Sixteen of 35 (46%) tumors contained Notch1 PEST domain mutations, and 30 of 35 (86%) tumors exhibited Fbxw7 LOH (Table 3). The Notch1 PEST domain mutations were similar in proportion to those seen in the pure 129/Sv mice. Likewise, the types of mutations were comparable and occurred at similar mutational hotspots, indicating that the genetic background did not affect the frequency or spectrum of the Notch1 mutations. Of the 16 tumors with Notch1 PEST domain mutations, 13 (81%) demonstrated concurrent LOH at the Fbxw7 locus. Therefore, single-copy deletions of Fbxw7 and Notch1 PEST domain mutations occur simultaneously in thymic lymphomas at a high frequency and are not mutually exclusive events. This finding, combined with the fact that loss of a single Fbxw7 allele (as seen in Fbxw7+/− mice) abrogates completely the mutational activation of Notch1 through the PEST domain, suggests that these Notch1 PEST domain mutations must occur before Fbxw7 deletions in thymic lymphomas.
Notch1 mutations and Fbxw7 LOH in F1 hybrid Tp53+/− mouse thymic lymphomas
| Tumor ID . | Notch1 mutation status* . | Fbxw7 locus . |
|---|---|---|
| F1-1 | 7342_7343insTG | LOH |
| F1-2 | Wild-type | LOH |
| F1-3 | Wild-type | LOH |
| F1-4 | Wild-type | LOH |
| F1-5 | 7114_7115insGGCAACACAGCCTCACCTGGTGCAGACCC | LOH |
| F1-6 | Wild-type | ROH |
| F1-7 | Wild-type | LOH |
| F1-8 | Wild-type | LOH |
| F1-9 | 7265_7275GCCCCAGCAGT > CAGC | LOH |
| F1-10 | Wild-type | LOH |
| F1-11 | Wild-type | LOH |
| F1-12 | 7260_7261insGG | LOH |
| F1-13 | 7140_7141insCA | LOH |
| F1-14 | 7083_7084insT | LOH |
| F1-15 | 7039_7042AATA > GCCCT | LOH |
| F1-16 | Wild-type | LOH |
| F1-17 | Wild-type | LOH |
| F1-18 | Wild-type | LOH |
| F1-19 | Wild-type | LOH |
| F1-20 | Wild-type | LOH |
| F1-21 | 7216_7217insG | LOH |
| F1-22 | 7216_7217insACCTGGGC; 7229_7260del | LOH |
| F1-23 | Wild-type | LOH |
| F1-24 | Wild-type | LOH |
| F1-25 | 7082G > CCC | ROH |
| F1-26 | Wild-type | ROH |
| F1-27† | 7271_7272insGCCG | LOH |
| F1-28 | 7252G > CCC | ROH |
| F1-29 | Wild-type | LOH |
| F1-30 | Wild-type | LOH |
| F1-31 | 7391_7392insA | ROH |
| F1-32 | Wild-type | LOH |
| F1-33 | 7244_7344del | LOH |
| F1-34 | 7237_7485del | LOH |
| F1-35 | 7292_7596del | LOH |
| Tumor ID . | Notch1 mutation status* . | Fbxw7 locus . |
|---|---|---|
| F1-1 | 7342_7343insTG | LOH |
| F1-2 | Wild-type | LOH |
| F1-3 | Wild-type | LOH |
| F1-4 | Wild-type | LOH |
| F1-5 | 7114_7115insGGCAACACAGCCTCACCTGGTGCAGACCC | LOH |
| F1-6 | Wild-type | ROH |
| F1-7 | Wild-type | LOH |
| F1-8 | Wild-type | LOH |
| F1-9 | 7265_7275GCCCCAGCAGT > CAGC | LOH |
| F1-10 | Wild-type | LOH |
| F1-11 | Wild-type | LOH |
| F1-12 | 7260_7261insGG | LOH |
| F1-13 | 7140_7141insCA | LOH |
| F1-14 | 7083_7084insT | LOH |
| F1-15 | 7039_7042AATA > GCCCT | LOH |
| F1-16 | Wild-type | LOH |
| F1-17 | Wild-type | LOH |
| F1-18 | Wild-type | LOH |
| F1-19 | Wild-type | LOH |
| F1-20 | Wild-type | LOH |
| F1-21 | 7216_7217insG | LOH |
| F1-22 | 7216_7217insACCTGGGC; 7229_7260del | LOH |
| F1-23 | Wild-type | LOH |
| F1-24 | Wild-type | LOH |
| F1-25 | 7082G > CCC | ROH |
| F1-26 | Wild-type | ROH |
| F1-27† | 7271_7272insGCCG | LOH |
| F1-28 | 7252G > CCC | ROH |
| F1-29 | Wild-type | LOH |
| F1-30 | Wild-type | LOH |
| F1-31 | 7391_7392insA | ROH |
| F1-32 | Wild-type | LOH |
| F1-33 | 7244_7344del | LOH |
| F1-34 | 7237_7485del | LOH |
| F1-35 | 7292_7596del | LOH |
LOH indicates loss of heterozygosity; and ROH, retention of heterozygosity.
Number denotes position from Notch1 cDNA start site.
Loss of wild-type allele.
Discussion
Activating Notch1 mutations, single-copy Fbxw7 deletion, and complete loss of Tp53 are common mutational events in IR-induced thymic lymphomas from Tp53-deficent mice. In this study, we assessed the mutational frequency of the Notch1 PEST domain in Tp53+/−, Tp53−/−, Tp53+/−Fbxw7+/−, and Tp53−/−Fbxw7+/− mice to determine the relationship of these genetic alterations in lymphomagenesis. First, we found that Notch1 PEST domain mutations are independent of Tp53 status because similar mutations with approximate equal frequency occur in thymic lymphomas from Tp53−/− and Tp53+/− mice. Second, tumors from Tp53−/− and Tp53+/− mice with only one germline copy of Fbxw7 fail to develop Notch1 PEST domain mutations. Superficially, this finding may suggest that Notch1 PEST domain mutations and Fbxw7 haploinsufficiency are mutually exclusive genetic alterations. However, we demonstrate that a high frequency of concurrent single-copy Fbxw7 deletions and activating Notch1 PEST domain mutations occur in thymic lymphomas from Tp53+/− F1 hybrid mice. Therefore, although in previous reports authors indicate that point mutations in Fbxw7 may be mutually exclusive to Notch1 PEST domain mutations, our findings show that this is not the case for single-copy Fbxw7 deletions and that these genetic changes commonly occur together with Notch1 PEST domain mutations.
As a whole, these data point to a model of sequential mutation acquisition in thymic lymphomagenesis. Because Notch1 PEST domain mutations are absent in tumors from mice heterozygous for Fbxw7 yet simultaneous Fbxw7 deletion and Notch1 PEST domain mutations occur with high frequency in mice with 2 germline copies of Fbxw7, then it follows that Notch1 PEST domain mutations are dependent on intact Fbxw7 levels. Therefore, Notch1 PEST domain mutations must occur before Fbxw7 deletion. Furthermore, we have previously shown that Tp53 activation leads to increased Fbxw7 levels and that Fbxw7 LOH is a Tp53-dependent event.12 Thus, genetic changes in Fbxw7 must precede complete loss of the wild-type Tp53 allele. These conclusions suggest a model of thymic lymphoma development involving early-stage activation of Notch1 through mutations to the PEST domain, followed by Fbxw7 deletion/mutation, and late loss of the remaining Tp53 allele (Figure 2A). In contrast, Fbxw7 heterozygous mice can bypass the requirement for Notch1 PEST domain mutations because Fbxw7 expression levels are compromised and proceed directly to eliminate Tp53 activity to yield malignant transformation (Figure 2B).
Model of sequential genetic alterations in T-ALL. After ionizing radiation exposure, Tp53 levels increased as a result of DNA damage, which elicited an increase in Fbxw7 levels and a subsequent reduction in overall Notch1 activity. Genetic alterations opposing these changes arose sequentially in Notch1, Fbxw7, and Tp53 to yield T-ALL formation (A). In contrast, thymocytes with only 1 copy of Fbxw7 bypassed Notch1 mutations during malignant transformation (B). Paradoxically, thymocytes entirely lacking Tp53 underwent malignant transformation without Fbxw7 alteration but still displayed Notch1 mutations (C).
Model of sequential genetic alterations in T-ALL. After ionizing radiation exposure, Tp53 levels increased as a result of DNA damage, which elicited an increase in Fbxw7 levels and a subsequent reduction in overall Notch1 activity. Genetic alterations opposing these changes arose sequentially in Notch1, Fbxw7, and Tp53 to yield T-ALL formation (A). In contrast, thymocytes with only 1 copy of Fbxw7 bypassed Notch1 mutations during malignant transformation (B). Paradoxically, thymocytes entirely lacking Tp53 underwent malignant transformation without Fbxw7 alteration but still displayed Notch1 mutations (C).
The Tp53-independent nature of Notch1 PEST domain mutations was unexpected given that the thymic target cells have no functional Tp53 and should not be able to transcriptionally activate Fbxw7, a well-characterized downstream transcriptional target of Tp53. However, we show that thymus from both Tp53−/− and Tp53+/− mice contain approximately normal Fbxw7 expression levels, whereas Fbxw7+/− mice demonstrate significantly lower Fbxw7 expression levels. Consequently, reduced Fbxw7 expression levels in Fbxw7+/− mice abrogate the requirement for Notch1 PEST domain mutations whereas in Tp53−/− and Tp53+/− mice, Fbxw7 expression levels are maintained at normal levels, leading to continued selection pressure for Notch1 PEST domain mutations. These findings suggest that in both Tp53−/− and Tp53+/− mice, the intact Fbxw7 levels, which lead to sufficient Fbxw7-mediated ICN turnover, necessitate activating Notch1 PEST domain mutations to overcome this barrier to malignant transformation. Another possibility is that Notch1 signaling may be rewired in Tp53−/− compared with Tp53+/− cells, and that there is a different stimulus for Notch1 PEST domain mutations in the Tp53−/− cells. Possibly, other targets for Fbxw7-mediated degradation, such as Myc, c-Jun, or mTor, may be important in this context and will require further investigation.
Currently, although many common genetic alterations have been reported in numerous tumor types, the sequence in which these mutational events occur during tumorigenesis remains largely unknown. In this study, we are able to establish the sequential order of a few key mutational events during lymphomagenesis by examining the frequency of these genetic alterations in tumors from various genetically engineered mouse models. Additional studies using this strategy may help further establish a chronologic description of other genetic mutations that contribute to tumor development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Jon Aster (Brigham and Women's Hospital) for providing a DNA sample from a tumor carrying a Notch1 type1 deletion.
This work is supported by the National Institutes of Health (NIH) National Research Service Award 5T32 CA108462 (K.-Y.J.), the NIH, National Cancer Institute (NCI) grant R01 CA116481 (J.H.M.), the Department of Energy Low Dose Radiation Research Program DESC0003679 (A.B.), and the NIH, NCI grant U01 CA141455 (A.B.).
National Institutes of Health
Authorship
Contribution: K.-Y.J. designed and performed the research and analyzed and interpreted the data; J.-H.M. designed and performed the early stages of the research and interpreted the data; I.Y.S. analyzed patterns of deletions of Notch1 in tumors, and K.L.B. and D.W. provided excellent technical assistance; and A.B. obtained the research funding, designed the research, and interpreted the data. The manuscript was prepared primarily by K.Y.J and A.B., with contributions from the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Allan Balmain, PhD, FRSE, Helen Diller Family Comprehensive Cancer Center, University of California–San Francisco, 1450 3rd St, San Francisco, CA 94158; e-mail: abalmain@cc.ucsf.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal