Abstract
Traditionally, GRP78 is regarded as protective against hypoxia and nutrient starvation prevalent in the microenvironment of solid tumors; thus, its role in the development of hematologic malignancies remains to be determined. To directly elucidate the requirement of GRP78 in leukemogenesis, we created a biallelic conditional knockout mouse model of GRP78 and PTEN in the hematopoietic system. Strikingly, heterozygous knockdown of GRP78 in PTEN null mice is sufficient to restore the hematopoietic stem cell population back to the normal percentage and suppress leukemic blast cell expansion. AKT/mTOR activation in PTEN null BM cells is potently inhibited by Grp78 heterozygosity, corresponding with suppression of the PI3K/AKT pathway by GRP78 knockdown in leukemia cell lines. This is the first demonstration that GRP78 is a critical effector of leukemia progression, at least in part through regulation of oncogenic PI3K/AKT signaling. In agreement with PI3K/AKT as an effector for cytosine arabinoside resistance in acute myeloid leukemia, overexpression of GRP78 renders human leukemic cells more resistant to cytosine arabinoside-induced apoptosis, whereas knockdown of GRP78 sensitizes them. These, coupled with the emerging association of elevated GRP78 expression in leukemic blasts of adult patients and early relapse in childhood leukemia, suggest that GRP78 is a novel therapeutic target for leukemia.
Introduction
One of the most frequently mutated tumor suppressor genes in human cancer is PTEN (phosphatase and tension homolog deleted on chromosome 10), which encodes for a nonredundant, plasma-membrane lipid phosphatase that antagonizes the phosphatidylinositol-3-kinase (PI3K) signaling pathway.1,2 On loss of PTEN, the PI3K/AKT signaling pathway is activated, leading to promotion of cell survival, proliferation, and angiogenesis, as well as activation of the mTOR and S6 kinases, resulting in enhanced protein translation commonly observed in cancers.3 PTEN also has a role in the maintenance of the hematopoietic stem cells (HSCs), as shown by ablation of PTEN function in adult HSCs through crossing of polyinosine-polycytidine (pIpC)–inducible Mx-1–Cre transgenic mouse line4 with a Pten flox/flox (Ptenf/f) mouse line.5 Induced Cre expression in postnatal mice exhausted normal HSCs and promoted excessive proliferation of leukemogenic stem cells, resulting in the development of myeloproliferative disorders (MPDs) and eventually leukemia.6,7 These studies further showed that the mTOR inhibitor rapamycin effectively suppressed growth of the leukemogenic stem cells and prevented exhaustion of normal HSCs. Furthermore, recent studies have shown that PTEN is down-regulated by BCR-ABL in leukemic stem cells, and PI3K and AKT play critical roles in BCR-ABL–induced leukemia in mice.8,9 Thus, inhibition of the PTEN/AKT/mTOR pathway not only can suppress solid tumor growth but could also represent a novel therapeutic strategy against stem cell disorders that are leukemogenic in nature.
GRP78, also referred to as BiP or HSPA5, is a member of the HSP70 protein family.10 Traditionally, it is regarded as a major endoplasmic reticulum (ER) chaperone facilitating protein folding, protein quality control, calcium binding, and regulation of transmembrane ER inducers.11 However, evidence is emerging that a subfraction of GRP78 can localize to the cell surface under pathophysiologic conditions, such as in cancer cells, and serves as a coreceptor for growth and pro–survival signaling mediated by PI3K/AKT.12-14 Strikingly, Pten null prostate tumorigenesis and AKT activation are potently blocked by targeted knockout of GRP78 in the prostate epithelium.15 Furthermore, in a variety of cell culture systems, specific knockdown of GRP78 results in inhibition of AKT activation.11,14 Although GRP78 is established to protect cancer cells against the adverse hypoxic and nutrient-deprived microenvironment of solid tumors,16,17 its role in initiation and progression of hematologic cancers is not known. Through creation of a biallelic conditional knockout mouse model of GRP78 and PTEN in the hematopoietic system, we demonstrate here that GRP78 haploinsufficiency potently suppresses leukemogenesis and AKT/mTOR signaling in PTEN null BM cells. In agreement with PI3K/AKT as an effector for cytosine arabinoside (AraC) resistance in leukemia,18 we observed that manipulation of GRP78 expression level alters the sensitivity of human leukemic cells to AraC-induced apoptosis. These, coupled with the emerging association of elevated GRP78 expression in leukemic blasts of adult patients and early relapse in childhood leukemia reported here and by others,19-21 suggest that GRP78 is a novel therapeutic target for leukemia.
Methods
Mice
The construction of the Ptenf/fGrp78f/f mouse was previously described.15 The transgenic Mx1-Cre mouse in C57BL/6 background was purchased from The Jackson Laboratory. The breeding of the Ptenf/fGrp78f/f mice with the Mx1-Cre mice to generate the mice cohorts was described in supplemental Figure 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
All protocols for animal use were reviewed and approved by the University of Southern California Institutional Animal Care and Use Committee.
Flow cytometry
BM cells were flushed from long bones (tibias and femurs) with Dulbecco PBS without calcium and magnesium, and then filtered through nylon screen (70 μm; BD Biosciences) to obtain single-cell suspension. BM cells were resuspended in Dulbecco PBS without calcium and magnesium with 0.5% BSA and 0.1% sodium azide. A total of 3 × 106 cells were used to stain for HSC percentage, using LSK markers (Lin−Sca-1+c-Kit+) and 3.5 × 106 cells used for immunophenotyping for leukemic blast cells. Cells were incubated with fluorescence conjugated antibodies for Lin markers [B220 (RA3-6B2/FITC), CD4 (RM4-5/FITC), CD8 (53-6.7/FITC), Gr-1 (RB6-8C5/FITC), Mac-1 (WT.5/FITC), TER-119 (FITC)], c-Kit (2B8/APC-H7), and Sca-1 (D7/PE-cy7), which were all purchased from BD Biosciences PharMingen. To identify leukemic blast cells, anti-CD45 (30-F11/APC) from eBioscience was used. After washing, cells were resuspended in PBS with 0.1% sodium azide, and 2 μL 7-amino-actinomycin D (7-AAD) was added to exclude dead cells. All FACS analyses were performed on LSR II flow cytometer.
Cell cycle analysis
A total of 1 × 107 whole BM cells were incubated with 10 μg/mL Hoechst 33342 (Sigma-Aldrich) at 37°C for 45 minutes and then stained with primitive hematopoietic cell antibodies (Lin, Sca-1, and c-Kit). The stained cells were resuspended in 10% buffered formalin and incubated at 4°C overnight. To stain for RNA content, pyronin Y (Polysciences) was added to the cells at a final concentration of 0.75 μg/mL and incubated at 4°C for 30 minutes. Cell cycle status was examined using a LSR II flow cytometer.
Cell culture and transfection
The human leukemia NB4 and HL60 cells were maintained in RPMI medium supplemented with 10% FBS and 1% penicillin/streptomycin. For knockdown of GRP78, the siRNA against Grp78 is 5′-ggagcgcauugauacuagatt-3′; for knockdown of GRP94, the siRNA against Grp94 is 5′-aucugggacaagcgaguuuuu-3′; and the control siRNA is 5′-aaggagacguauagcaacggu-3′. Transfection of siRNA was described.15 Forty-eight hours after siRNA transfection, the cells were treated with 300nM thapsigargin (Tg) or AraC (24 hours) before harvesting. For serum stimulation experiments, 24 hours after siRNA transfection, HL60 cells were serum starved for 16 hours, followed by 10% serum stimulation before harvesting. For overexpression of GRP78, NB4 cells at 60% confluency were transfected with either pcDNA or Flag-tagged GRP78 expression vector (pcDNA-F-GRP78) as previously described.13 Transfection was performed using BioT (Bioland Scientific) following the manufacturer's instructions. Forty-eight hours after transfection, the cells were treated with AraC as indicated for 24 hours before harvesting.
Western blot analysis
Whole cell lysates were prepared from Ficoll-Paque isolated peripheral blood mononuclear cells of normal control and leukemia patients, leukemia cell lines, single-cell suspensions of BM cells, or splenocytes from the indicated mice. Cells were lysed in RIPA buffer supplemented with competent protease-inhibitor mixture (Roche Diagnostics) and phosphatase-inhibitor mixture (Roche Diagnostics), and 25 μg cell lysate was subjected to SDS-PAGE. The immunoblot membranes were incubated with primary antibodies at 4°C overnight, and the protein signals were detected with ECL reagent (Roche Diagnostics) or Supersignal chemiluminescence reagent (Pierce Chemical) after reacting with HRP-conjugated secondary antibody. The primary antibodies used are as follows. Monoclonal mouse anti-GRP78 (1:2000) and monoclonal mouse anti–caspase-7 (1:4000) are from BD Biosciences PharMingen. Mouse anti–PTEN (26H9, 1:1000), rabbit anti-AKT (1:1000), rabbit anti–p-AKT (Ser473, 1:1000), rabbit anti–p-AKT (Thr308, 1:1000), rabbit anti-S6K (1:1000), rabbit anti–p-S6K (Thr389, 1:1000), rabbit anti–p-PI3K p85 (Tyr458, 1:1000), mouse anti-p44/42 MAPK (ERK1/2, 1:1000), mouse anti–p-p44/42 MAPK (ERK1/2, Thr202/Tyr204, 1:1000), rabbit anti-GSK3β (1:1000), rabbit anti–p-GSK3β (Ser9, 1:1000), rabbit anti-p38 MAPK (1:1000), and rabbit anti–p-p38 MAPK (Thr180/Tyr182, 1:1000) are from Cell Signaling. Mouse anti-PARP (1:500) is from Santa Cruz Biotechnology. Mouse anti–β-actin (1:5000) is from Sigma-Aldrich.
Microscopy and immunofluorescence analysis
Transfected HL60 cells were serum starved for 12 hours before serum stimulation for 3 minutes or remained untreated. The cells were then fixed and processed for immunofluorescence staining for phosphatidylinositol 3,4,5-triphosphate (PI(3,4,5)P3) according to the manufacturer's protocol (Echelon Biosciences). Briefly, after TBS wash, the cells were rinsed with distilled H2O and applied to lysine-coated slides (Snowcoat X-tra; Surgipath Medical Instruments) and allowed to dry before saponin permeabilization. The cells were blocked with 10% goat serum and then incubated with mouse anti–PI(3,4,5)P3 IgG antibody (Z-P345b, Echelon Biosciences) at 1 μg/mL for 60 minutes at 37°C, followed by incubation with AlexaFluor-488 goat anti–mouse antibody (Invitrogen) for 30 minutes at 37°C. After washing, the cells were mounted with Vectashield anti–fade medium with DAPI (H-1200, Vector Laboratories). Cells were analyzed by a Zeiss LSM510 confocal microscope equipped with a Hamamatsu R6357 photomultiplier and LSM 510 Version 4.2 SP1 acquisition software (Carl Zeiss). Representative images (Figure 5F left) were taken with an EC Plan-Neofluar 40×/1.30 oil objective without zoom and cropped in Photoshop CS2 (Adobe). Images for quantification (Figure 5F right) were taken with a Plan-Apochromat 100×/1.4 oil DIC objective at 4× zoom, with thickness of 500 nm. Fluorescence quantification was performed as described with modifications.22 Briefly, color images were digitally processed into monochrome with LSM Image Browser R4.2. Digital images (n ≥ 30 for each condition) were analyzed by McMaster Biophontonics Facility ImageJ Version 1.43 software. Pixel intensities at the cell perimeter around the leading edge were collected using a 4-μm tangent line and averaged by the software. The edge intensities from the serum-stimulated cells were averaged and normalized to the edge intensities of nonstimulated cells.
Apoptosis assays
For LSK apoptotic measurements, 1 × 106 whole BM cells were stained with primitive hematopoietic cell antibodies (Lin, c-Kit, and Sca-1) and were resuspended in 150 μL 1× annexin V binding buffer (BD Biosciences PharMingen). For HL60 apoptosis detection, cells were resuspended at the concentration of 1 to 1.5 × 106 cells/mL in the annexin V binding buffer. Cells in the binding buffer were incubated with annexin V and 7-AAD (both from BD Biosciences PharMingen) at room temperature for 15 minutes and then analyzed on BD LSR II flow cytometer within an hour.
Clinical specimens
Leukemic blasts from BM and peripheral blood samples were provided by Dr Allen Yang (University of Southern California Keck School of Medicine) and the University of Southern California Translational Pathology Core. They were obtained from diagnosed leukemia patients in University of Southern California Norris Comprehensive Cancer Center according to institutional guidelines. The normal peripheral blood mononuclear cells were purchased from ALLCELLS (catalog #PB003F). The ethical use of the human tissues for research was approved by the University of Southern California Institutional Review Board.
RT-PCR
Total RNA from peripheral blood or BM samples of normal persons or leukemia patients were extracted by Trizol reagent (Invitrogen) followed by DNase I treatment. First-strand cDNA was synthesized with the Superscript II reverse transcriptase (Invitrogen) with oligo d(T) primer according to the manufacturer's instructions.
Microarray analysis for clinical specimens
For the comparison of Grp78 expression between normal BM and acute myeloid leukemia (AML) patient samples, expression values for Grp78 mRNA were acquired from previously published microarray data, series accession number GSE1159.23 For the association of Grp78 expression level in relapsed acute lymphoblastic leukemia (ALL) patients, Grp78 mRNA expression values were acquired from a previously published genome-wide study of 60 patients with relapsed childhood ALL on the Affymetrix U133A microarray platform.24 Gene expression data are publically available through Gene Expression Omnibus (http://www.ncbi.nlm.gov/geo); series accession number GSE3912. BM blasts were obtained from patients enrolled on the Children's Oncology Group protocol AALL01P2 for first medullary relapse.25 Thirty-seven patients relapsed early (< 36 months from initial diagnosis) and 23 patients relapsed late (≥ 36 months). The Affymetrix probe set 211936_at represents the gene HSPA5 (also known as GRP78). Normalization was performed by the method of Robust Multiarray Analysis,26 and the normalized probe set intensity values were used for subsequent analyses.
Statistical analyses
Kaplan-Maier survival curves were constructed using GraphPad (Prism Version 5 software), and log-rank analysis was used to analyze the results. For bar graphs, the unpaired 2-tailed Student t test was used to compute P values; the error bars represent SE.
Results
Creation of mouse models with conditional biallelic deletion of Pten and Grp78 in the hematopoietic system
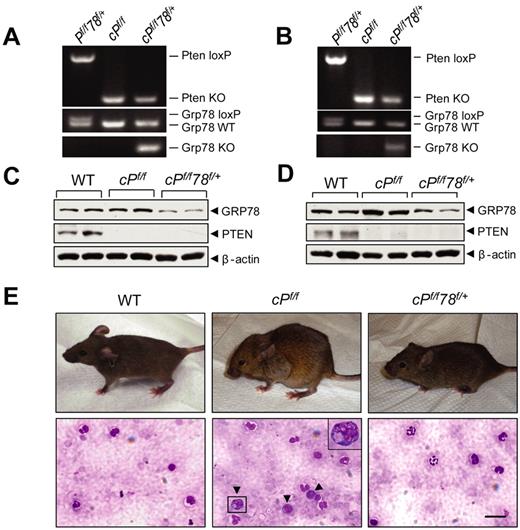
Pten is commonly deleted or inactivated in cancers, including hematopoietic malignancies.6 To test the requirement of GRP78 for leukemogenesis, we created a new mouse model containing the Pten floxed allele, the Grp78 floxed allele, and the Mx-1-Cre transgene. Pten and Grp78 were deleted in the hematopoietic system from 6- to 8-week-old Pten, Grp78 floxed mice carrying the Mx-1-Cre transgene by administrating pIpC every other day for a total of 7 doses to induce Cre expression.4 The mice were analyzed 6 days after pIpC injection. Littermates without the Cre transgene (Ptenf/fGrp78+/+ and Ptenf/fGrp78f/+) are phenotypically equivalent to animals with a wild-type (WT) Pten allele; they served as WT normal controls and were also injected with pIpC. Because mice with homozygous deletion of both Pten and Grp78 (cPtenf/fGrp78f/f) died during the pIpC administration interval (data not shown), this study focused on Pten null, Grp78 heterozygous (cPtenf/fGrp78f/+) mice. Partial reduction of GRP78 in heterozygotes is a valuable model mimicking anti–GRP78 therapeutic agents that achieve partial suppression of GRP78 in human diseases. The status of Pten and Grp78 deletion was validated by PCR in isolated BM cells (Figure 1A), splenocytes (Figure 1B), and peripheral blood (supplemental Figure 2A). Western blot analysis of BM cells (Figure 1C) and splenocytes (Figure 1D) confirmed that PTEN expression was completely ablated in the cPtenf/f and cPtenf/fGrp78f/+ mice, and GPR78 expression was reduced in the cPtenf/fGrp78f/+ mice. As reported previously,6,7 cPtenf/f mice developed MPD, which includes hunched posture, enlarged lymph nodes in the axillary areas, and an increased number of immature progenitor cells in peripheral blood (Figure 1E). Strikingly, despite loss of PTEN, cPtenf/fGrp78f/+ mice resembled WT normal controls with normal posture and no apparent increase in immature progenitor cells in the peripheral blood (Figure 1E).
Grp78 heterozygosity impedes PTEN null-induced leukemogenesis. (A-B) Representative PCR genotyping results from WT (Pf/f78f/+), Pten null (cPf/f) and Pten null, Grp78 heterozygous mice (cPf/f78f/+) BM (A) and splenocytes (B) 6 days after completion of pIpC treatment. (C-D) Western blot results for detection of GRP78 and PTEN protein level in the BM (C) and splenocytes (D) performed in duplicates. (E) Top panel: Hunched posture of PTEN null mice compared with normal posture of WT and cPf/f78f/+ mice. Bottom panel: Detection of immature progenitor cells (dark arrows) in the blood smear of PTEN null mice. A magnified image of one of these cells is shown in the upper right corner. Blood smears were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635). Images were acquired with a Leica Type DMLB2 microscope (63×/1.4 oil objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). Scale bar represents 30 μm.
Grp78 heterozygosity impedes PTEN null-induced leukemogenesis. (A-B) Representative PCR genotyping results from WT (Pf/f78f/+), Pten null (cPf/f) and Pten null, Grp78 heterozygous mice (cPf/f78f/+) BM (A) and splenocytes (B) 6 days after completion of pIpC treatment. (C-D) Western blot results for detection of GRP78 and PTEN protein level in the BM (C) and splenocytes (D) performed in duplicates. (E) Top panel: Hunched posture of PTEN null mice compared with normal posture of WT and cPf/f78f/+ mice. Bottom panel: Detection of immature progenitor cells (dark arrows) in the blood smear of PTEN null mice. A magnified image of one of these cells is shown in the upper right corner. Blood smears were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635). Images were acquired with a Leica Type DMLB2 microscope (63×/1.4 oil objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). Scale bar represents 30 μm.
Grp78 heterozygosity suppresses Pten null-mediated MPD characteristics and blast cell expansion
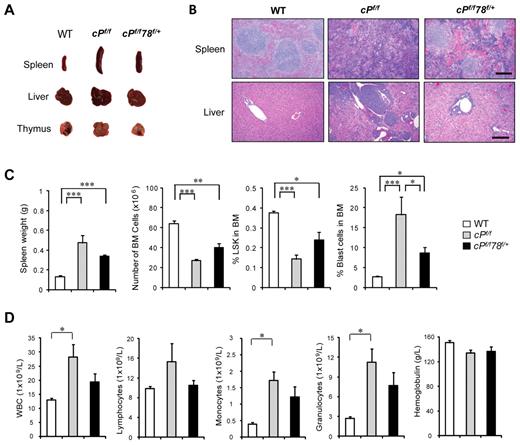
Compared with WT controls, the cPtenf/f mice showed substantially enlarged spleen, liver, and thymus (Figure 2A), and hematoxylin and eosin staining of the paraffin tissue sections showed effaced splenic architecture and myeloid cell infiltration in the liver (Figure 2B). The cPtenf/fGrp78f/+ mice exhibited intermediate enlargement of spleen and liver size but not the thymus, close to normal splenic architecture, and absence of the myeloid cell infiltration in the liver (Figure 2A-C). In agreement with previous reports, PTEN null mice showed a significant decrease in BM cells and HSCs as measured by the percentage of LSK cells, which include both long-term and short-term HSCs.27 Grp78 heterozygosity reversed these trends such that, in the cPtenf/fGrp78f/+ mice, BM cell numbers were partially restored and the percentage of HSCs in the BM was close to normal level (Figure 2C). By examining the cell cycle profiles of the HSCs (LSK cells), we further determined that the increase in proliferation in cPtenf/f HSC was suppressed in cPtenf/fGrp78f/+ HSC (Figure 2D), whereas apoptosis was not affected (Figure 2E). For comparison, we also analyzed cGrp78f/+ mice, which did not exhibit any obvious abnormality compared with littermate WT mice, exhibiting normal organ size and morphology, and no loss in total BM cell number or HSC population (Figure 2A-C). Collectively, these results provide proof of principle that anti–GRP78 agents that partially suppress GRP78 can potentially suppress development of PTEN null-mediated myeloproliferative disease without affecting normal organ function.
Grp78 heterozygosity suppresses PTEN null-mediated MPD characteristics. All analyses were performed 6 days after completion of pIpC treatment. (A) Organ size and morphology from mice of the indicated genotypes. (B) H&E staining of paraffin sections of spleen (top) and liver (bottom) of the indicated genotypes. The scale bar represents 200 μm. Sections of spleen and liver were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635) and stained with H&E. Images were acquired with a Leica Type DMLB2 microscope (10×/0.3 objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). (C) Quantitation of the spleen weight (n = 8), total BM (BM) cell number (n = 8), and percentage of LSK cells in BM (n = 5) for each indicated genotype. (D) Quantitation of cell cycle distribution of LSK cells from WT (n = 3), cPf/f (n = 5), and cPf/f78f/+ (n = 5). (E) Flow cytometric analysis of apoptotic LSK cells using annexin V and 7-AAD. Data are as mean ± SE. *P < .05 (Student t test). **P < .01 (Student t test). ***P < .001 (Student t test).
Grp78 heterozygosity suppresses PTEN null-mediated MPD characteristics. All analyses were performed 6 days after completion of pIpC treatment. (A) Organ size and morphology from mice of the indicated genotypes. (B) H&E staining of paraffin sections of spleen (top) and liver (bottom) of the indicated genotypes. The scale bar represents 200 μm. Sections of spleen and liver were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635) and stained with H&E. Images were acquired with a Leica Type DMLB2 microscope (10×/0.3 objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). (C) Quantitation of the spleen weight (n = 8), total BM (BM) cell number (n = 8), and percentage of LSK cells in BM (n = 5) for each indicated genotype. (D) Quantitation of cell cycle distribution of LSK cells from WT (n = 3), cPf/f (n = 5), and cPf/f78f/+ (n = 5). (E) Flow cytometric analysis of apoptotic LSK cells using annexin V and 7-AAD. Data are as mean ± SE. *P < .05 (Student t test). **P < .01 (Student t test). ***P < .001 (Student t test).
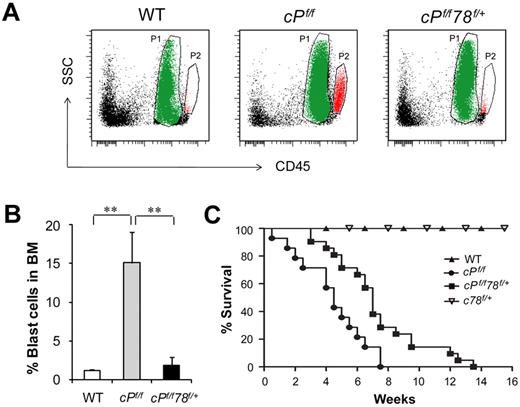
To further assess whether Grp78 heterozygosity affects leukemic blast cell expansion, we analyzed for leukemic blast cell presence using side scatter pattern and CD45 staining in BM cell suspensions.28 Dramatic increase of blast cell frequency was observed in PTEN null mice as previously described.6 In contrast, despite Pten deletion, the percentage of blast cells in the BM in cPtenf/fGrp78f/+ mice was similar to normal WT level (Figure 3A-B). This implies that partial suppression of one cellular moiety, GRP78, is sufficient to suppress PTEN null-induced leukemic blast expansion. In agreement, cPtenf/fGrp78f/+ mice showed a statistically significant prolonged life span than the cPtenf/f mice (P < .05; Figure 3C). In analyzing these 2 groups of mice at a later time point (21 days) after injection, we observed that, compared with 6 days after injection, the spleen and the thymus of the cPtenf/fGrp78f/+ mice were further enlarged with evident signs of effacement of the splenic architecture and myeloid cell infiltration in the liver (Figure 4A-B). There was a further decrease in BM cells and the HSC (LSK cells) population (Figure 4C). The percentage of blast cells in the BM was increased by 4-fold, corresponding with the trend of increase in white blood cells, specifically monocytes and granulocytes in blood count analysis, whereas the other organs remained phenotypically normal (Figure 4D). These data strongly suggest that the cPtenf/fGrp78f/+ mutant mice showed delayed development of leukemia, and this could be a contributing cause for their eventual death between 3 and 14 weeks.
Grp78 heterozygosity suppresses PTEN null-mediated blast cell expansion and prolongs survival. (A) Representative results of leukemic blast cell identification. P1 population in green represents the neutrophil region. P2 population in red represents leukemic blast cell region. (B) Quantitation of the leukemic blast cell percentages of WT (n = 7), cPf/f (n = 7), and cPf/f78f/+ (n = 10) shown in panel A. Data are mean ± SE. **P < .01 (Student t test). (C) Kaplan-Meier survival curve of WT (n = 10), Pten null (cPf/f, n = 14), Pten null, Grp78 heterozygous mice (cPf/f78f/+, n = 21), and Grp78 heterozygous mice (c78f/+, n = 5).
Grp78 heterozygosity suppresses PTEN null-mediated blast cell expansion and prolongs survival. (A) Representative results of leukemic blast cell identification. P1 population in green represents the neutrophil region. P2 population in red represents leukemic blast cell region. (B) Quantitation of the leukemic blast cell percentages of WT (n = 7), cPf/f (n = 7), and cPf/f78f/+ (n = 10) shown in panel A. Data are mean ± SE. **P < .01 (Student t test). (C) Kaplan-Meier survival curve of WT (n = 10), Pten null (cPf/f, n = 14), Pten null, Grp78 heterozygous mice (cPf/f78f/+, n = 21), and Grp78 heterozygous mice (c78f/+, n = 5).
Pten null Grp78 heterozygous mice show MPD characteristics after longer latency period. All analyses were performed 21 days after completion of pIpC treatment. (A) Organ size and morphology from mice of the indicated genotypes. (B) H&E staining of paraffin sections of spleen (top) and liver (bottom) of the indicated genotypes. Scale bar represents 200 μm. Sections of spleen and liver were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635) and stained with H&E. Images were acquired with a Leica Type DMLB2 microscope (10×/0.3 objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). (C) Quantitation of the spleen weight (n = 6), total BM cell number (n = 6), and percentage of LSK cells in BM (n = 5) and percentage blast cells in BM (n = 6) for each indicated genotype. (D) Complete blood count with tail peripheral blood from WT (n = 7), cPf/f (n = 7), and cPf/f78f/+ (n = 7) mice. Peripheral blood was collected via tail bleeding and analyzed using an auto hematology analyzer BC-2800 vet (Mindray) according to the manufacturer's instructions. Data are mean ± SE. *P < .05 (Student t test). **P < .01 (Student t test). ***P < .001 (Student t test).
Pten null Grp78 heterozygous mice show MPD characteristics after longer latency period. All analyses were performed 21 days after completion of pIpC treatment. (A) Organ size and morphology from mice of the indicated genotypes. (B) H&E staining of paraffin sections of spleen (top) and liver (bottom) of the indicated genotypes. Scale bar represents 200 μm. Sections of spleen and liver were mounted with Pro-Texx mounting medium (Lerner Laboratories, #137635) and stained with H&E. Images were acquired with a Leica Type DMLB2 microscope (10×/0.3 objective) fitted with SPOT RT KE SE Model 7.3 Three Shot Color camera and SPOT 4.0.5 computer software (Diagnostic Instruments Inc). (C) Quantitation of the spleen weight (n = 6), total BM cell number (n = 6), and percentage of LSK cells in BM (n = 5) and percentage blast cells in BM (n = 6) for each indicated genotype. (D) Complete blood count with tail peripheral blood from WT (n = 7), cPf/f (n = 7), and cPf/f78f/+ (n = 7) mice. Peripheral blood was collected via tail bleeding and analyzed using an auto hematology analyzer BC-2800 vet (Mindray) according to the manufacturer's instructions. Data are mean ± SE. *P < .05 (Student t test). **P < .01 (Student t test). ***P < .001 (Student t test).
Knockdown of GRP78 inhibits AKT/S6K activation in BM and leukemia cells
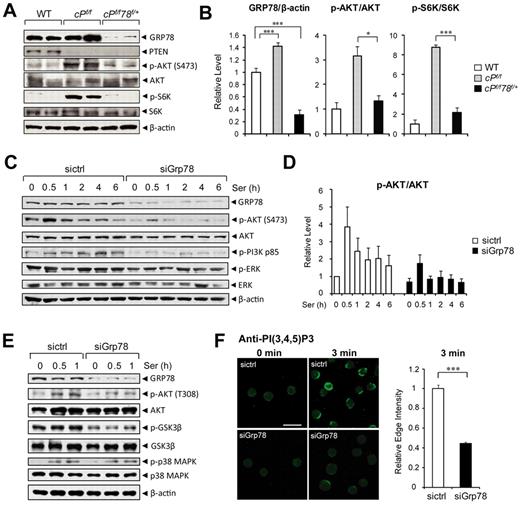
To understand the potential mechanisms underlying the relieved leukemic phenotype of cPtenf/fGrp78f/+ mice compared with PTEN null mice, we examined the PI3K/AKT signaling pathway, in which PTEN is the major negative regulator. Cellular lysates were prepared from the BM cells of WT, PTEN null, and the cPtenf/fGrp78f/+ mice and subjected to Western blot analysis. Consistent with up-regulation of GRP78 in diverse cancers,16 GRP78 level was up-regulated by 1.4-fold (P < .001) in the BM cells of the PTEN null mice compared with WT (Figure 5A-B). In the BM of the PTEN null mice, strong activation of the AKT/mTOR pathway was detected with high levels of phosphorylation of AKTS473 (p-AKT) and the p70 S6 kinase (p-S6K), whereas p-AKT level was considerably lower and p-S6K was barely detectable in the cPtenf/fGrp78f/+ mice (Figure 5A). The levels of total AKT and S6K were similar in either genotype. The relative levels of p-AKTS473 and p-S6K after normalization with the total AKT and p-S6K were summarized in Figure 5B.
Knockdown of GRP78 suppresses AKT signaling. (A) Representative Western blot results using BM cell lysates (n = 2 for each genotype) for detection of the indicated protein levels. (B) Quantitation of Western blot results of relative GRP78 level (n = 9 for each genotype) normalized against β-actin and the ratio of p-AKT to total AKT and p-S6K to total S6K in panel A. The ratio of one of the WT levels of GRP78, p-AKT, and p-S6K was set as 1. (C) Western blot results of lysates from HL60 cells transfected with siRNA against Grp78 (siGrp78) or control siRNA (sictrl), followed by serum starvation for 16 hours, and then stimulated with 10% serum for the indicated time (hours). (D) Quantitation of the ratio of p-AKT to total AKT in panel C. The ratio at the 0-hour time point in cells transfected with sictrl was set as 1. (E) Western blot results of lysates from HL60 cells transfected with siRNA against Grp78 (siGrp78) and control siRNA (sictrl), followed by 16-hour serum starvation, and then stimulated with 10% serum for the indicated time (hours). (F) Left: Representative immunofluorescent images of untreated (0 minutes) or serum-stimulated (3 minutes) HL60 cells transfected with either sictrl or siGrp78 and stained with anti–PI(3,4,5)P3 antibody. Scale bar represents 20 μm. Right: Quantification of relative edge fluorescence intensity of PI(3,4,5)P3 staining in serum-stimulated siRNA-transfected cells normalized to nonstimulated siRNA-transfected cells (n ≥ 30 cells per condition). The normalized relative intensity of the 3-minute stimulated sictrl cells was set as 1. Data are mean ± SE. *P < .05 (Student t test). ***P < .001 (Student t test).
Knockdown of GRP78 suppresses AKT signaling. (A) Representative Western blot results using BM cell lysates (n = 2 for each genotype) for detection of the indicated protein levels. (B) Quantitation of Western blot results of relative GRP78 level (n = 9 for each genotype) normalized against β-actin and the ratio of p-AKT to total AKT and p-S6K to total S6K in panel A. The ratio of one of the WT levels of GRP78, p-AKT, and p-S6K was set as 1. (C) Western blot results of lysates from HL60 cells transfected with siRNA against Grp78 (siGrp78) or control siRNA (sictrl), followed by serum starvation for 16 hours, and then stimulated with 10% serum for the indicated time (hours). (D) Quantitation of the ratio of p-AKT to total AKT in panel C. The ratio at the 0-hour time point in cells transfected with sictrl was set as 1. (E) Western blot results of lysates from HL60 cells transfected with siRNA against Grp78 (siGrp78) and control siRNA (sictrl), followed by 16-hour serum starvation, and then stimulated with 10% serum for the indicated time (hours). (F) Left: Representative immunofluorescent images of untreated (0 minutes) or serum-stimulated (3 minutes) HL60 cells transfected with either sictrl or siGrp78 and stained with anti–PI(3,4,5)P3 antibody. Scale bar represents 20 μm. Right: Quantification of relative edge fluorescence intensity of PI(3,4,5)P3 staining in serum-stimulated siRNA-transfected cells normalized to nonstimulated siRNA-transfected cells (n ≥ 30 cells per condition). The normalized relative intensity of the 3-minute stimulated sictrl cells was set as 1. Data are mean ± SE. *P < .05 (Student t test). ***P < .001 (Student t test).
To determine whether the decrease in AKT activation in the BM of the cPtenf/fGrp78f/+ mice is a consequence of suppressed leukemogenesis or because of the requirement of GRP78 in AKT signaling, we performed GRP78 knockdown experiments in 2 human leukemia cell lines (HL60 and NB4) using 2 different stimuli (serum and ER stress inducer) for AKT activation. The human acute promyelocytic leukemia cell line HL60 was transfected with siRNA specific for Grp78 or with random siRNA serving as negative control. Knockdown of GRP78 expression by siRNA in HL60 cells suppressed serum induced AKTS473 phosphorylation throughout the entire course of treatment with no effect on total AKT level (Figure 5C-D). Serum stimulation led to tyrosine phosphorylation of the p85 regulatory subunit of PI3K, relieving its inhibitory activity on PI3K.29 We observed that knockdown of GRP78 suppressed serum-stimulated PI3K p85 phosphorylation, consistent with lower AKTS473 activation in the same cells (Figure 5C). Furthermore, knockdown of GRP78 not only suppressed serum-induced AKTS473 but also AKTT308 phosphorylation (Figure 5E). Phosphorylation of GSK3β, which is a downstream substrate of AKT, was also suppressed because of GRP78 knockdown (Figure 5E). In contrast, GRP78 knockdown minimally affected the level of phosphorylated ERK, which showed high constitutive level and remained elevated throughout serum treatment (Figure 5C) and did not affect serum induced p38 MAPK phosphorylation (Figure 5E). Similarly, knockdown of GRP78 expression in human acute promyelocytic leukemic cell line NB4 suppressed ER stress induced AKTS473 phosphorylation. Previously, it has been demonstrated that the ER stress inducer Tg was able to induce AKT activation.15,30 In NB4 cells treated with Tg, Western blot analysis showed that p-AKT levels were reduced at all treatment time points in siGrp78-treated cells expressing a much lower level of GRP78, whereas the total levels of AKT were unaffected (supplemental Figure 3A). The ratio of p-AKT level to total AKT level was quantitated and summarized in supplemental Figure 3B. We further determined that knockdown of GRP78 has no effect on the level of endogenous PTEN expressed in NB4 cells (supplemental Figure 3A); thus, knockdown of GRP78 suppresses AKT activation in leukemic cells independent of PTEN status. To determine whether the suppression of AKT activation is specific for GRP78 knockdown or a consequence of ER chaperone disruption, another essential multifunctional ER chaperone, GRP94,31,32 was examined. Our results showed that knockdown of GRP94 expression in HL60 cells did not suppress serum-induced AKT activation as observed for GRP78 knockdown (supplemental Figure 4).
To assess at which step GRP78 is regulating AKT activation, we measured the effect of GRP78 knockdown on serum-stimulated production of PI(3,4,5)P3, indicative of PI3K activity. PI(3,4,5)P3 production at the leading edge of serum-stimulated HL60 cells was measured using a monoclonal anti–PI(3,4,5)P3 antibody previously established for sensitive detection of PI(3,4,5)P3 levels in cells.33,34 To examine the requirement of GRP78 in PI(3,4,5)P3 production, HL60 cells were transfected with siRNA against Grp78 (siGrp78) or control siRNA (sictrl) followed by 12-hour serum starvation, and then stimulated with serum to activate leading edge PI(3,4,5)P3 production. At 3 minutes of serum stimulation, anti–PI(3,4,5)P3 staining showed clear strong structure in a narrow band at the cell edge in HL60 cells treated with sictrl (Figure 5F). In contrast, GRP78 knockdown resulted in a marked inhibition of PI(3,4,5)P3 production, evidenced by a significant decrease of anti–PI(3,4,5)P3 staining at the cell edge of siGrp78-treated cells compared with sictrl-treated cells after serum stimulation (Figure 5F). Thus, this suggests that GRP78 knockdown suppresses AKT signaling at the level of PI3K activation.
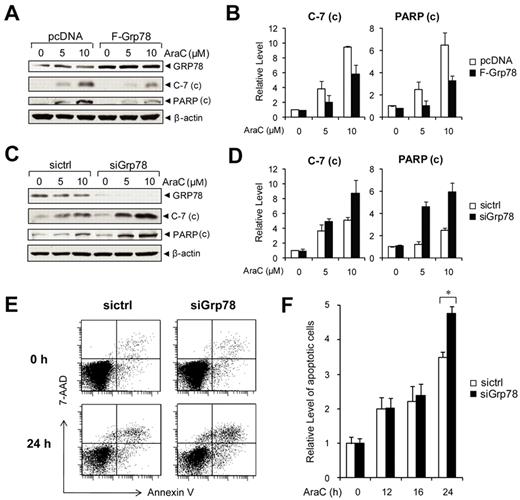
Knockdown of GRP78 in human leukemic cells enhances AraC-induced apoptosis
The PI3K/AKT pathway is constitutively active in primary AML cells, and PI3K inhibitor potentiates the response to AraC, a frontline chemotherapy drug against leukemia, including AML.18 As a regulator of PI3K/AKT signaling, GRP78 could contribute to chemoresistance. To test this, NB4 cells were transiently transfected with an expression vector for GRP78, or empty vector as a negative control. The cells were then treated with AraC, a DNA-damaging agent with IC50 value between 5 and 10μM in leukemic cells.35 To monitor for AraC-induced onset of apoptosis, we measured the cleavage of caspase-7 (C-7) and poly(ADP-ribose) polymerase (PARP) as indicators for their activation and onset of apoptosis. In NB4 cells transfected with empty vector, thus only expressing endogenous level of GRP78, we observed an increase in the level of cleaved C-7 and PARP in an AraC–dosage-dependent manner (Figure 6A). GRP78 overexpression suppressed C-7 and PARP activation in AraC-treated cells (Figure 6A-B). Using siRNA specifically targeting human Grp78,36,37 we obtained efficient knockdown of GRP78 in NB4 cells. When treated with AraC, cells with reduced level of GRP78 showed enhanced C-7 and PARP activation (Figure 6C-D). In the trypan blue exclusion assays, live cells possess intact cell membranes that exclude the trypan blue dye, whereas dead cells take up the dye. In cells treated with control siRNA, the percentage of trypan blue-positive dead cells at 5 and 10μM AraC were 18% and 25%, respectively. GRP78 siRNA knockdown in combination with 5 and 10μM AraC treatment resulted in an increase to 30% and 40% trypan blue-positive cells, respectively (supplemental Figure 5). To confirm the protective role of GRP78 in chemoresistance in other leukemia cell lines, HL60 cells were either transfected with control siRNA or siRNA against Grp78 and then subjected to treatment with 10μM AraC. Apoptosis was monitored by annexin V/7-AAD FACS analysis (Figure 6E). We observed that knockdown of GRP78 sensitized the HL60 cells to AraC treatment in a time-dependent manner where the enhanced sensitization was most evident at 24-hour treatment (Figure 6F). Collectively, these results showed that suppression of GRP78 expression in human leukemic cells enhances AraC-induced apoptosis and lowers viability.
Knockdown of GRP78 sensitizes human leukemia cells to AraC. (A) Western blot analysis of NB4 cells transfected with pcDNA or Flag-tagged GRP78 expression vector and then treated with the indicated AraC concentration for 24 hours for detection of cleaved caspase-7 and PARP with β-actin as loading control. (B) Quantitation of cleaved caspase-7 and PARP normalized to β-actin in panel A. The ratio at 0μM AraC in pcDNA cells was set as 1. (C) Western blot analysis of lysates from NB4 cells transfected with siGrp78 or sictrl and treated with the indicated AraC concentration for 24 hours for detection of cleaved caspase-7 and cleaved PARP with β-actin as loading control. (D) Quantitation of cleaved caspase-7 and PARP normalized against β-actin in panel C. The ratio at 0μM AraC in sictrl cells was set as 1. (E) Representative annexin V/7-AAD flow cytometric apoptosis analysis of HL60 cells transfected with control siRNA (sictrl) or siRNA against Grp78 (siGrp78). The cells were either nontreated (0 hours) or treated with 10μM AraC for 24 hours. (F) Time course analysis of apoptotic HL60 cells treated with AraC. HL60 cells were transfected with sictrl or siGrp78, followed by treatment with 10μM AraC for the indicated time (hours). The percentage of apoptotic cells was measured by annexin V/7-AAD flow cytometry. The level of apoptosis at 0 hours in cells treated with sictrl was set as 1. Data are mean ± SE. *P < .05 (Student t test).
Knockdown of GRP78 sensitizes human leukemia cells to AraC. (A) Western blot analysis of NB4 cells transfected with pcDNA or Flag-tagged GRP78 expression vector and then treated with the indicated AraC concentration for 24 hours for detection of cleaved caspase-7 and PARP with β-actin as loading control. (B) Quantitation of cleaved caspase-7 and PARP normalized to β-actin in panel A. The ratio at 0μM AraC in pcDNA cells was set as 1. (C) Western blot analysis of lysates from NB4 cells transfected with siGrp78 or sictrl and treated with the indicated AraC concentration for 24 hours for detection of cleaved caspase-7 and cleaved PARP with β-actin as loading control. (D) Quantitation of cleaved caspase-7 and PARP normalized against β-actin in panel C. The ratio at 0μM AraC in sictrl cells was set as 1. (E) Representative annexin V/7-AAD flow cytometric apoptosis analysis of HL60 cells transfected with control siRNA (sictrl) or siRNA against Grp78 (siGrp78). The cells were either nontreated (0 hours) or treated with 10μM AraC for 24 hours. (F) Time course analysis of apoptotic HL60 cells treated with AraC. HL60 cells were transfected with sictrl or siGrp78, followed by treatment with 10μM AraC for the indicated time (hours). The percentage of apoptotic cells was measured by annexin V/7-AAD flow cytometry. The level of apoptosis at 0 hours in cells treated with sictrl was set as 1. Data are mean ± SE. *P < .05 (Student t test).
Expression of GRP78 in patient leukemic blasts and association with relapse
Recent studies reveal that GRP78 expression is up-regulated in various forms of human leukemia, including Ph-positive primary acute leukemia,19 B-chronic lymphocytic leukemia (B-CLL),20 and high-grade B-lineage lymphoid malignancies and relapsed childhood B-acute lymphoblastic leukemia (B-ALL).21 Extending these findings, we analyzed Grp78 mRNA and protein levels from mononuclear cells purified from the BM or peripheral of patients diagnosed with AML, chronic myeloid leukemia (CML), ALL, and chronic lymphocytic leukemia (CLL). We observed that compared with normal controls, Grp78 mRNA and protein levels are generally elevated at various levels in multiple types of adult patient leukemic blasts, notably AML (Figure 7A-B). Analysis of the Valk leukemia microarray database of 285 AML patients23 further revealed significant elevation of Grp78 mRNA compared with normal controls (P = .0488; Figure 7C).
Grp78 expression in leukemia and relapse in childhood ALL. (A) Grp78 mRNA levels measured by RT-PCR from peripheral blood or BM (*) samples of the indicated types of leukemia patients, with β-actin levels as control. Below is a summary of Grp78 transcript level in an expanded study after normalization against β-actin. (B) GRP78 protein expression in Ficoll-Paque isolated peripheral blood mononuclear cells of normal control and leukemia patients (AML, CML, ALL, and CLL). Below is the quantitation of the relative GRP78 level normalized against β-actin. (C) Comparison of Grp78 expression between normal BM (n = 5) and AML patient samples (n = 285) using microarray database. (D) Grp78 expression level determined from microarray database of relapse ALL patients (n = 60) was plotted against time to relapse. The horizontal bar represents median Grp78 expression (456) of all ALL patients; and the vertical bar at the 36-month time point separates patients with early relapse (left) and late relapse (right). *P < .05 (Student t test). **P < .01 (Student t test).
Grp78 expression in leukemia and relapse in childhood ALL. (A) Grp78 mRNA levels measured by RT-PCR from peripheral blood or BM (*) samples of the indicated types of leukemia patients, with β-actin levels as control. Below is a summary of Grp78 transcript level in an expanded study after normalization against β-actin. (B) GRP78 protein expression in Ficoll-Paque isolated peripheral blood mononuclear cells of normal control and leukemia patients (AML, CML, ALL, and CLL). Below is the quantitation of the relative GRP78 level normalized against β-actin. (C) Comparison of Grp78 expression between normal BM (n = 5) and AML patient samples (n = 285) using microarray database. (D) Grp78 expression level determined from microarray database of relapse ALL patients (n = 60) was plotted against time to relapse. The horizontal bar represents median Grp78 expression (456) of all ALL patients; and the vertical bar at the 36-month time point separates patients with early relapse (left) and late relapse (right). *P < .05 (Student t test). **P < .01 (Student t test).
In addition to promoting tumorigenesis, GRP78 is known to confer resistance to anti–cancer agents.16 The outcome for relapse leukemia is extremely poor. Duration of first remission is an important prognostic factor after retrieval therapy, and early relapse portends a poor response. In a genome-wide microarray analysis of BM blasts obtained from 60 patients with relapsed childhood ALL,24 we observed an inverse correlation of Grp78 expression with time to relapse. Among the 37 patients who relapsed early (< 36 months from initial diagnosis) and 23 patients who relapsed late (≥ 36 months), Grp78 level was significantly higher in patients who relapsed early versus late (mean of 779 vs 450; P = .008, Figure 7D).
Discussion
In this study, through the creation of a biallelic conditional knockout mouse model of GRP78 and PTEN in the hematopoietic system, we demonstrated that partial reduction of a single molecular entity, GRP78, in the form of Grp78 heterozygosity, is sufficient to restore the HSC population back to the normal percentage and suppress AKT activation and leukemic blast cell expansion mediated by loss of PTEN. We observed that the increase in proliferation in cPf/f HSCs was suppressed in cPf/f78f/+ HSCs, whereas apoptosis was not affected. This raises the interesting question of how might Grp78 heterozygosity restore the HSC population to near normal levels? It is recently reported that depletion of PTEN null HSCs is caused by increases in expression of p53 and p16, which promote the HSCs to differentiate/mature and migrate out of the BM HSC pool and this is dependent on mTOR activation.38 Because GRP78 deficiency reduces mTOR activation, this could explain why HSC depletion is suppressed, resulting in higher number of HSCs in the cPf/f78f/+ mice versus cPf/f mice. Furthermore, we showed that Grp78 heterozygosity by itself has no apparent effect on development and survival39,40 and, as analyzed here, has no effect on total BM cell number or HSC population. Thus, partial GRP78 expression is sufficient to maintain normal organ homeostasis, whereas tumor progression requires optimal level of GRP78.
How might GRP78 contribute to leukemogenesis? Although GRP78 is able to confer multiple antiapoptotic effects on cancer cells,15,16 here we propose a key mechanistic explanation, by which a prominent effector AKT, which is activated by the loss of PTEN, is compromised by reduction of GRP78 in the hematopoietic system. This is based on our discovery that PTEN-null mediated AKT/mTOR signaling is potently suppressed in the BM of the Pten null Grp78 heterozygous mice. In cell culture experiments, knockdown of GRP78 by siRNA reduced AKT activation independent of PTEN status and with minimal effect on the ERK and the p38 MAPK pathways. The requirement of GRP78 for AKT activation may involve multiple mechanisms. For example, GRP78 as a major ER chaperone protein may be required for the processing and cell surface expression of growth factor receptors mediating AKT activation in the PTEN null model.41 In addition, although GRP78 is traditionally regarded as primarily an ER chaperone, its ability to regulate AKT signaling in the BM and in human leukemic cells is consistent with the recent exciting discovery that under pathophysiologic conditions such as cancer, a subfraction of GRP78 relocalizes to the cell surface and acts as coreceptor promoting growth signaling.11,14,42 It has been reported that ligation of cell surface GRP78 in human cancer cells with antibodies directed against its carboxyl domain suppresses PI3K/AKT/mTOR signaling.43 Our observation that knockdown of GRP78 suppresses serum-stimulated phosphorylation of the p85 regulatory subunit of PI3K and inhibits PI(3,4,5)P3 production suggests that GRP78 regulates AKT activation through functional regulation of PI3K, and this warrants further investigation. Importantly, ER stress or overexpression of GRP78 is sufficient to promote cell surface expression of GRP78.13 Because PTEN mutation and AKT/mTOR signaling represent a major oncogenic pathway in human cancer, promoting both proliferative and survival pathways, GRP78 suppression will have wide implications in blocking both solid and hematologic cancer progression.
With regard to drug resistance in leukemia, PI3K/AKT is constitutively active in primary AML cells from patients and blocking PI3K with inhibitor (LY294002) potentiates the response to AraC.18 In agreement with our earlier findings in prostate cancer cells,15 we demonstrated here that GRP78 is required for PI3K/AKT signaling in leukemic cells and modulation of GRP78 expression alters sensitivity to AraC. We recently established that siRNA knockdown of GRP78 down-regulates both intracellular and cell surface GRP78 expression,44 both forms of GRP78 could contribute to AraC resistance. Recently, it is reported that resistance of B-ALL cells to the anti–leukemic drug vincristine was suppressed by (−)–epigallocatechin gallate, which inhibits the antiapoptotic function of GRP78 by targeting to its ATP-binding domain.21,45 Furthermore, chemoresistant B-ALL cells underwent apoptosis when exposed to a doxorubicin-conjugated penetrating cyclic anti–GRP78 peptide targeting cell surface GRP78.21 Although the role of GRP78 in human leukemia and recurrence remains to be determined, emerging evidence suggests elevated Grp78 expression in patient leukemic blasts. For example, Grp78 mRNA level is significantly elevated in Ph-positive primary acute leukemic cells.19 It is reported that B-CLL patients expressed high constitutive level of GRP78 and siRNA knockdown of GRP78 increases B-CLL cell apoptosis.20 Another recent study showed that Grp78 is significantly up-regulated in high-grade B-lineage lymphoid malignancies and relapsed childhood B-ALL (particularly in early relapse) compared with initial diagnosis and that inhibitors targeting GRP78 induces apoptosis to chemoresistant B-ALL cells.21 Furthermore, proteomic analysis reveals Grp78 is differentially expressed in HSC-like fractions from the BM of leukemic patients.46 Of note is our finding linking Grp78 overexpression to early relapse in childhood B-ALL compared with late relapse, complementing the finding of Grp78 overexpression in relapsed B-ALL compared with initial diagnosis.21 Collectively, these discoveries identify GRP78 as a novel therapeutic target against chemoresistance in leukemic cells.
How might GRP78 protect leukemic cells against chemotoxic treatment? One explanation is that this is mediated by the role of GRP78 as a master regulator of ER homeostasis and apoptosis.11 Furthermore, ER stress induces alternative splicing of the Grp78 transcript, leading to the production of a cytosolic isoform of GRP78 (GRP78va), which also protects HL60 cells from ER stress-induced cell death.47 Therapeutic drugs in clinical treatment or preclinical test for leukemia, such as arsenic trioxide and imatinib mesylate, have been shown to up-regulate ER stress markers and induce leukemic cell death through ER stress-mediated apoptosis.47 AraC is a nucleoside analog, and its incorporation into DNA causes localized alterations in the DNA duplex, resulting in inhibition of DNA polymerase as well as stabilization of covalent topoisomerase I-DNA complexes contributing to cytotoxicity.48 Previously, we established that GRP78 protects cells from apoptosis induced by other DNA-damaging agents, such as topoisomerase inhibitors through inhibition of caspase-7, which associates with the ER and can be activated by ER stress.49,50 Here we showed that treatment of leukemic cells with AraC leads to activation of caspase-7. Because GRP78 is known to complex with caspase-7 and suppress its activation,45,49,50 GRP78 level can play an important role in responsiveness to AraC treatment; however, future investigations are required to address other potential mechanisms.
In conclusion, here we provide proof of principle that partial reduction of GRP78 can arrest leukemogenesis and sensitize leukemic cells to chemotherapeutic treatment while having no harmful effect on the hematopoietic system. Therapeutics targeting GRP78 have recently emerged and have shown anti–tumor activity in solid tumors.16,51-54 Whether these and other agents capable of suppressing GRP78 expression or activity can be applied for suppression of leukemogenesis warrants vigorous investigation.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Hung Wu (University of California, Los Angeles, CA) for providing the floxed Pten mice Dr Allen Yang for providing clinical samples and cell line; Drs Cheng Ji and Parkash Gill for the gift of antibodies; Drs Michael Kahn, Darryl Shibata, Parkash Gill, Preet Chaudhary, and Jonathan Backer (Albert Einstein College of Medicine, Bronx, NY) for helpful discussions; the University of Southern California Norris Comprehensive Cancer Center Flow Cytometry Core Facility for assistance with the flow cytometry analysis; and the Translational Pathology Core Facility for providing clinical samples and preparation of histologic sections.
This work was supported by the National Institutes of Health (grant CA027607, A.S.L.). The University of Southern California core facilities described were supported by the National Institutes of Health, National Cancer Institute (P30 CA014089). Microscopy was performed at the Cell and Tissue Imaging Core of the University of Southern California Research Center for Liver Diseases, supported by the National Institutes of Health (P30 DK048522).
National Institutes of Health
Authorship
Contribution: S.W. designed and performed research, contributed vital new reagents or analytical tools, analyzed data, and wrote the paper; B.L. and C.-C.T. performed research and analyzed data; M.N. designed and performed research and analyzed data; H.Z. performed research; Y.F. contributed vital new reagents or analytical tools; D.B. designed and performed research, analyzed data, and wrote the paper; W.L.C. designed research and analyzed data; and A.S.L. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for M.N. is Dana-Farber Cancer Institute, Boston, MA. The current affiliation for Y.F. is UCLA Medical School, Los Angeles, CA.
Correspondence: Amy S. Lee, University of Southern California Norris Comprehensive Cancer Center, 1441 Eastlake Ave, Los Angeles, CA 90089-9176; e-mail: amylee@ccnt.usc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal