Abstract
Considering the important advances in treating specific types of systemic amyloidoses, unequivocal typing of amyloid deposits is now essential. Subcutaneous abdominal fat aspiration is the easiest, most common diagnostic procedure. We developed a novel, automated approach, based on Multidimensional Protein Identification Technology, for typing amyloidosis. Fat aspirates were obtained from patients with the most common systemic amyloidoses (ALλ, ALκ, transthyretin, and reactive amyloidosis), with Congo red score more than or equal to 3+, and nonaffected controls. Peptides from extracted and digested proteins were analyzed by Multidimensional Protein Identification Technology. On semiquantitative differential analysis (patients vs controls) of mass spectrometry data, specific proteins up-represented in patients were identified and used as deposit biomarkers. An algorithm was developed to classify patients according to type and abundance of amyloidogenic proteins in samples; in all cases, proteomic characterization was concordant with fibril identification by immunoelectron microscopy and consistent with clinical presentation. Our approach allows reliable amyloid classification using readily available fat aspirates.
Introduction
Multiple unrelated autologous proteins can cause systemic amyloidoses.1,2 The various forms differ for pathogenesis, prognosis, and treatment but present overlapping clinical manifestations, making their differentiation on a clinical basis very difficult. The most common type in Western countries is light chain (AL) amyloidosis, caused by misfolded monoclonal immunoglobulin light chains (LCs). However, several hereditary forms also exist, with high prevalence in selected geographic regions. Precise typing is key for adequate treatment because the various forms require different approaches, which can range from hematopoietic stem cell transplantation in AL amyloidosis to liver transplantation in transthyretin (TTR) amyloidosis (ATTR).2 Diagnosis and classification are based on histologic demonstration of amyloid deposits and identification of which proteins originate the fibrils. Abdominal subcutaneous fat is the tissue of choice for diagnostic examination when a systemic form is suspected.3 Because of the frequent unreliability of traditional, histochemistry-based typing techniques,4-6 novel proteomic strategies, based on mass spectrometry (MS) identification of the protein constituents of the deposits, have been proposed.7,8 Multidimensional Protein Identification Technology (MudPIT)9,10 is an automated, high-throughput proteomic approach that allows identifying hundreds of proteins in complex samples.
We used MudPIT profiling for typing amyloid deposits in whole, nonfixed subcutaneous fat aspirates from patients affected by the most common forms of systemic amyloidoses: ALλ, ALκ, ATTR, and reactive (AA) amyloidosis. A simple diagnostic algorithm (α-value) was developed to use proteomic data for precise amyloid type assignment.
Methods
Subcutaneous abdominal fat was obtained by fine needle aspiration from 26 systemic amyloidosis patients (12 ALλ, 4 ALκ, 5 ATTR, and 5 AA) and 11 nonaffected controls (Table 1; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Sample weight ranged between 10 and 20 mg both in patients (median, 12 mg) and controls (median, 15 mg). The use of the tissue for research purposes was approved by the Ethical Committee of Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo, Pavia, Italy. All persons gave written informed consent in accordance with the Declaration of Helsinki for storing and using their biologic samples for research purposes, according to the Institutional Review Board guidelines. Specific amyloidosis type was confirmed by immunoelectron microscopy (IEM), a reference method developed at our center.13 All patients' samples had Congo red positivity score more than or equal to 3+.11 Proteins were extracted from tissue as described,7 dialyzed against 50mM ammonium bicarbonate (18 hours, 4°C), and digested with trypsin. Resulting peptide mixtures were analyzed in 2 replicates by MudPIT, based on 2-dimensional chromatography coupled to tandem MS.14 The software MAProMa14,15 was used to identify up-represented proteins in patients and select those forming the amyloid deposits in each amyloidosis type, by comparing each patient's fat sample protein profile against the profile of the same tissue from the nonaffected population. For estimating which specific amyloid protein was prevalent in each patient, a parameter (α-value) was calculated; this is obtained by normalizing the patient over control ratio of each biomarker's spectral count16 (further details in supplemental Methods).
Clinical features, at time of fat tissue acquisition, of the 26 systemic amyloidosis patients included in the study
| Sample . | Amyloid type* . | Age (y), sex . | CR score† . | Amyloid organ involvement‡ . | Serum MC (HR-IFE) . | κ/λ ratio§ . |
|---|---|---|---|---|---|---|
| P1 | AL-λ | 61, F | 3+ | Kidney | IgGλ + λ-FLC | 0.28 |
| P2 | AL-λ | 53, M | 3+ | Heart, kidney | λ-FLC | 0.05 |
| P3 | AL-λ | 67, M | 3+ | Heart | IgGλ + λ-FLC | 0.005 |
| P4 | AL-λ | 59, F | 3+ | Heart, kidney, PNS, ANS | λ-FLC | 0.06 |
| P5 | AL-λ | 64, M | 3+ | Heart, kidney | IgAλ | 0.009 |
| P6 | AL-λ | 64, M | 3+ | Heart | λ-FLC | 0.06 |
| P7 | ATTR (Val30Met) | 68, M | 3+ | PNS | 0.69 | |
| P8 | AL-λ | 68, F | 3+ | Heart, kidney, liver | λ-FLC | 0.06 |
| P9 | AL-λ | 58, M | 3+ | Heart, soft tissues | λ-FLC | 0.03 |
| P10 | AL-λ | 67, F | 3+ | Heart, kidney | IgAλ + λ-FLC | 0.05 |
| P11 | AL-λ | 73, F | 3+ | Heart, kidney, PNS | λ-FLC | 0.05 |
| P12 | AL-λ | 59, M | 4+ | Heart, kidney, PNS, ANS | λ-FLC | 0.02 |
| P13 | AL-λ | 70, F | 4+ | Kidney | IgMλ + λ-FLC | 0.08 |
| P14 | AL-κ | 75, M | 4+ | Heart, kidney, liver | κ- FLC + faint λ-FLC | 7.7 |
| P15 | AL-κ | 60, M | 3+ | Heart, soft tissues | κ-FLC + faint IgMλ | 158 |
| P16 | ATTR (Tyr78Phe) | 66, M | 3+ | Heart | 0.64 | |
| P17 | AA | 70, F | 3+ | Kidney | 0.48 | |
| P18 | ATTR (Leu68Ile) | 76, M | 3+ | Heart, PNS | 1.31 | |
| P19 | ATTR (Ser50Arg) | 37, M | 3+ | Heart, PNS, ANS | IgAκ | 2 |
| P20 | ATTR (Ser50Arg) | 55, F | 3+ | Heart, PNS, ANS | 0.94 | |
| P21 | AA | 70, M | 4+ | Kidney | 1.08 | |
| P22 | AA | 75, F | 4+ | Kidney | 1.4 | |
| P23 | AA | 66, F | 4+ | Kidney | 1.33 | |
| P24 | AA | 79, M | 3+ | Kidney | 1.62 | |
| P25 | AL-κ | 80, M | 4+ | Heart, soft tissues, PNS | IgGκ + κ-FLC | 22.3 |
| P26 | AL-κ | 63, M | 4+ | Kidney, PNS | IgGκ | 21.8 |
| Sample . | Amyloid type* . | Age (y), sex . | CR score† . | Amyloid organ involvement‡ . | Serum MC (HR-IFE) . | κ/λ ratio§ . |
|---|---|---|---|---|---|---|
| P1 | AL-λ | 61, F | 3+ | Kidney | IgGλ + λ-FLC | 0.28 |
| P2 | AL-λ | 53, M | 3+ | Heart, kidney | λ-FLC | 0.05 |
| P3 | AL-λ | 67, M | 3+ | Heart | IgGλ + λ-FLC | 0.005 |
| P4 | AL-λ | 59, F | 3+ | Heart, kidney, PNS, ANS | λ-FLC | 0.06 |
| P5 | AL-λ | 64, M | 3+ | Heart, kidney | IgAλ | 0.009 |
| P6 | AL-λ | 64, M | 3+ | Heart | λ-FLC | 0.06 |
| P7 | ATTR (Val30Met) | 68, M | 3+ | PNS | 0.69 | |
| P8 | AL-λ | 68, F | 3+ | Heart, kidney, liver | λ-FLC | 0.06 |
| P9 | AL-λ | 58, M | 3+ | Heart, soft tissues | λ-FLC | 0.03 |
| P10 | AL-λ | 67, F | 3+ | Heart, kidney | IgAλ + λ-FLC | 0.05 |
| P11 | AL-λ | 73, F | 3+ | Heart, kidney, PNS | λ-FLC | 0.05 |
| P12 | AL-λ | 59, M | 4+ | Heart, kidney, PNS, ANS | λ-FLC | 0.02 |
| P13 | AL-λ | 70, F | 4+ | Kidney | IgMλ + λ-FLC | 0.08 |
| P14 | AL-κ | 75, M | 4+ | Heart, kidney, liver | κ- FLC + faint λ-FLC | 7.7 |
| P15 | AL-κ | 60, M | 3+ | Heart, soft tissues | κ-FLC + faint IgMλ | 158 |
| P16 | ATTR (Tyr78Phe) | 66, M | 3+ | Heart | 0.64 | |
| P17 | AA | 70, F | 3+ | Kidney | 0.48 | |
| P18 | ATTR (Leu68Ile) | 76, M | 3+ | Heart, PNS | 1.31 | |
| P19 | ATTR (Ser50Arg) | 37, M | 3+ | Heart, PNS, ANS | IgAκ | 2 |
| P20 | ATTR (Ser50Arg) | 55, F | 3+ | Heart, PNS, ANS | 0.94 | |
| P21 | AA | 70, M | 4+ | Kidney | 1.08 | |
| P22 | AA | 75, F | 4+ | Kidney | 1.4 | |
| P23 | AA | 66, F | 4+ | Kidney | 1.33 | |
| P24 | AA | 79, M | 3+ | Kidney | 1.62 | |
| P25 | AL-κ | 80, M | 4+ | Heart, soft tissues, PNS | IgGκ + κ-FLC | 22.3 |
| P26 | AL-κ | 63, M | 4+ | Kidney, PNS | IgGκ | 21.8 |
CR indicates Congo red; MC, monoclonal component; HR-IFE, high-resolution electrophoresis and immunofixation; FLC, free light chains; PNS, peripheral nervous system; ANS, autonomic nervous system; LC-κ, immunoglobulin light chain-κ; and LC-λ, immunoglobulin light chain-λ.
Confirmed by IEM.
Graded on subcutaneous fat tissue.11
Defined according to the International Consensus Panel criteria.12
Reference interval: 0.26-1.65.
Results and discussion
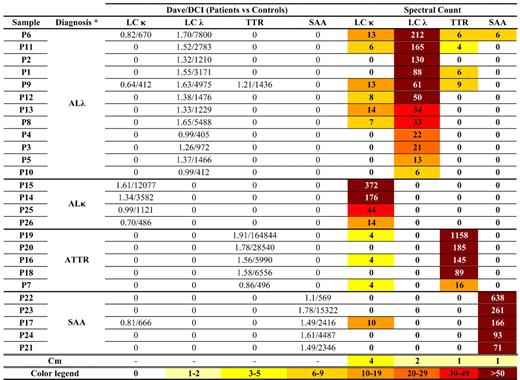
MudPIT allowed identifying hundreds of proteins in each fat sample, among which a few carried over serum proteins (supplemental Table 2). Protein profiles from amyloidosis patients were compared against that of the control counterpart; this allowed minimizing the contribution of contaminating blood proteins and selecting up-represented proteins in patients. Within each of the 4 considered groups of patients (clustered by amyloidosis type), only a single amyloidogenic protein was up-represented in 100% of cases. These 4 proteins (LC λ, LC κ, TTR, and serum amyloid A [SAA]), identified by proteomic analysis, matched those expected to constitute the fibrils according to IEM (Figure 1; supplemental Table 3). In addition, other proteins (supplemental Table 3) were found to be over-represented across the various amyloidoses, including clusterin,17 apolipoprotein E,18,19 apolipoprotein A-IV,20 vitronectin,18 basement membrane-specific heparan sulfate proteoglycan core protein,21 sushi-repeat-containing protein, and serum amyloid P.22 Most of these had previously been described as associated with amyloid fibrils and involved in the mechanisms of amyloid formation; this confirms that MudPIT can fully characterize the deposits in whole adipose tissue and constitutes an internal validation parameter. In addition, sushi-repeat–containing protein may play a role in amyloid disease; indeed, it is secreted by adipocytes23 and may intervene in adipogenic differentiation,24 regulation of body weight, and metabolism.24,25 Moreover, evidence of its involvement in tumor suppression, possibly through apoptosis induction,26 emphasizes its potential interest in amyloid cell toxicity. However, its potential role may be limited to subcutaneous fat because it has not been reported in other studies on amyloid.
Amyloid proteins detected by proteomics analysis in patients' abdominal subcutaneous fat tissue. Results of semiquantitative differential analysis (patients vs controls) and absolute spectral count of the amyloid proteins are shown. Specific amyloid proteins were identified by comparing the protein lists of each patient against the average protein profile of nonaffected adipose tissue, by means of MAProMa software (through calculation of Dave/DCI values; Dave ≥ +0.4 and DCI ≥ +400 correspond to proteins significantly up-represented in patients).14 For LC-κ and LC-λ, the contributions of both constant and variable regions were considered. SAA indicates serum amyloid A; Cm, the average value of each amyloid protein in nonaffected tissue, acquired from 11 controls and deposited in our database; and —, not applicable. *Confirmed by IEM.
Amyloid proteins detected by proteomics analysis in patients' abdominal subcutaneous fat tissue. Results of semiquantitative differential analysis (patients vs controls) and absolute spectral count of the amyloid proteins are shown. Specific amyloid proteins were identified by comparing the protein lists of each patient against the average protein profile of nonaffected adipose tissue, by means of MAProMa software (through calculation of Dave/DCI values; Dave ≥ +0.4 and DCI ≥ +400 correspond to proteins significantly up-represented in patients).14 For LC-κ and LC-λ, the contributions of both constant and variable regions were considered. SAA indicates serum amyloid A; Cm, the average value of each amyloid protein in nonaffected tissue, acquired from 11 controls and deposited in our database; and —, not applicable. *Confirmed by IEM.
The 4 amyloid proteins identified by MS in the previous step (LC λ, LC κ, TTR, and SAA), specific for each amyloidosis type, were used to design a diagnostic algorithm, based on calculation of the parameter α-value (Figure 2; supplemental Methods). α-value allows to estimate the abundance of each amyloid protein relative to the remaining ones, eliminating the confounding effect of carried-over LC, SAA, and TTR from blood (Figure 1). The diagnostic capability of the algorithm was tested by assigning in blind the amyloid type to the aforedescribed patients. The α-values of each of the 4 amyloid proteins were calculated for all patients; amyloid was assigned to the type whose amyloid protein α-value was predominant. In all cases, amyloid-type definition according to α-value was in agreement with IEM and could be visualized in a user-friendly graphical representation (Figure 2). Notably, disease classification was correct also in the 2 AL amyloidosis patients (P14 and P15) who had biclonal gammapathies (P14, both κ- and λ-free LC bands; P15, κ-free LC and IgMλ bands), and in the ATTR patient (P19) with a coincidental serum IgAκ monoclonal component. The observation that, in cases such as P9 and P13, minor amounts of other amyloid proteins (TTR and κ LC, respectively) were identified in conjunction with λ LC indicates the possibility that “normal” proteins can adsorb on amyloid fibrils. Combining MudPIT with complementary approaches, such as laser capture microdissection or IEM, would determine whether these proteins colocalize with fibrils.
Typing of systemic amyloidosis through MS-based diagnostic algorithm in 26 patients. Bars in the graph represent the α-values of the 4 amyloid proteins in each patient. α-value is the normalized patient over control ratio of each amyloid protein's spectral count (see supplemental Methods). Amyloidosis is attributed to the type whose corresponding amyloid protein has the highest α-value. Discontinuous line indicates the theoretical diagnostic threshold (α-value = 70; see supplemental Methods). Based on this algorithm, all patients were correctly classified (ie, amyloid-type definition was in accordance with IEM).
Typing of systemic amyloidosis through MS-based diagnostic algorithm in 26 patients. Bars in the graph represent the α-values of the 4 amyloid proteins in each patient. α-value is the normalized patient over control ratio of each amyloid protein's spectral count (see supplemental Methods). Amyloidosis is attributed to the type whose corresponding amyloid protein has the highest α-value. Discontinuous line indicates the theoretical diagnostic threshold (α-value = 70; see supplemental Methods). Based on this algorithm, all patients were correctly classified (ie, amyloid-type definition was in accordance with IEM).
Our technique was developed in patients with clear-cut amyloid deposits (Congo red score 3+/4+). In AL and AA samples with 1+/2+ amyloid, the MudPIT approach may not allow conclusive typing. In these cases, enrichment of amyloid areas by laser capture microdissection before MS, proposed by Vrana et al,8 may be necessary.
The described method is a novel approach for diagnostic amyloid typing in whole unfixed adipose tissue, coupling the advantages of using subcutaneous abdominal fat aspirates (ease of acquisition, wide applicability) with the automation and sensitivity of MudPIT. Analysis of whole tissue avoids fractionation steps, and the inclusion of the control group compensates for the background noise of carried-over plasma proteins. MS-based amyloid typing was concordant with IEM, validating the correctness of the results. Although thus far only implemented for the 4 described amyloid types, the approach can be expanded to the diagnosis of other forms. A limitation is that, in some types of systemic amyloidosis, particularly ATTR and ApoA-I, amyloid deposits may be scanty or absent in abdominal fat. In these cases, biopsy of involved organs, analyzed by laser capture microdissection and MS,8 is necessary. It has been argued that direct MS-based approaches for protein identification and quantification are less prone to biases than antibody-based methods, especially for proteins with altered conformation as amyloid ones. Our results indicate this method as a specific and informative novel potential diagnostic approach, granting its application on larger, independent patient sets, possibly through a multicenter collaboration.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Fondazione Cariplo Nobel project, Proteomic platform, Operational Network for Biomedicine Excellence in Lombardy, EURAMY project (Community's Sixth Framework Program), Fondazione Cariplo (N2009-2532), Ricerca Finalizzata Malattie Rare, Italian Ministry of Health, Istituto Superiore di Sanità (526D/63), Ministry of Research and University (2007AESFX2-003), and Associazione Italiana per la Ricerca sul Cancro Special Program Molecular Clinical Oncology (grant 9965).
Authorship
Contribution: F.B. wrote the manuscript, performed MudPIT analysis, and interpreted results; F.L. wrote the manuscript, processed tissue samples, collected clinical data, and interpreted results; D.D.S. and R.R. performed MudPIT analysis and interpreted results; V.V. processed tissue samples; G.P. and L.O. collected samples and clinical data; L.V. performed IEM; P.M. designed research, developed the algorithm for α-value, and revised the manuscript; and G.M. designed research, collected clinical data, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giampaolo Merlini, Amyloid Research and Treatment Center, Fondazione IRCCS Policlinico San Matteo and University of Pavia, Viale Golgi 19, 27100 Pavia, Italy; e-mail: gmerlini@smatteo.pv.it.
References
Author notes
F.B. and F.L. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal