Abstract
WHIM syndrome is a rare, autosomal dominant, immunodeficiency disorder so-named because it is characterized by warts, hypogammaglobulinemia, infections, and myelokathexis (defective neutrophil egress from the BM). Gain-of-function mutations that truncate the C-terminus of the chemokine receptor CXCR4 by 10-19 amino acids cause WHIM syndrome. We have identified a family with autosomal dominant inheritance of WHIM syndrome that is caused by a missense mutation in CXCR4, E343K (1027G → A). This mutation is also located in the C-terminal domain, a region responsible for negative regulation of the receptor. Accordingly, like CXCR4R334X, the most common truncation mutation in WHIM syndrome, CXCR4E343K mediated approximately 2-fold increased signaling in calcium flux and chemotaxis assays relative to wild-type CXCR4; however, CXCR4E343K had a reduced effect on blocking normal receptor down-regulation from the cell surface. Therefore, in addition to truncating mutations in the C-terminal domain of CXCR4, WHIM syndrome may be caused by a single charge-changing amino acid substitution in this domain, E343K, that results in increased receptor signaling.
Introduction
WHIM syndrome is a rare congenital immunodeficiency disorder so-named because it is characterized by warts, hypogammaglobulinemia, infections, and myelokathexis.1,2 Myelokathexis refers to neutropenia resulting from retention of mature neutrophils and increased neutrophil apoptosis in the BM.1 Many WHIM patients also have lymphopenia,3 monocytopenia,4 and deficiency of plasmacytoid dendritic cells in the circulation,5 and thus impairment in both innate and adaptive immunity. Warts develop after human papillomavirus (HPV) exposure (usually in late childhood) and are difficult to eradicate. Hypogammaglobulinemia is variable and, when present, is typically associated with low or absent B cells in the blood and impaired Ab responses after vaccination.6,7 The clinical course in WHIM syndrome is punctuated by recurrent bacterial infections,7 which can lead to chronic respiratory tract, joint, and hearing dysfunction and loss of dentition. Because neutrophils can be mobilized to the blood in response to acute infection, infected patients may present without neutropenia.8
Almost all cases of WHIM syndrome have an autosomal-dominant inheritance pattern.9,10 Rare de novo cases have been reported and a few cases remain genetically unexplained11 ; however, all previously reported, genetically defined cases have been caused by mutations changing the last 10-19 amino acids in the carboxy-tail (C-tail) of the chemokine receptor CXCR4.10,12,13 CXCR4 is a G-protein–coupled receptor specific for a single member of the chemokine family of leukocyte chemoattractants named CXCL12 (also known as SDF-1).14 It is highly conserved phylogenetically and is expressed on most mature leukocyte subtypes,15,16 as well as on hematopoietic progenitor cells,17 epithelial cells, keratinocytes, and cancer cells.10,14,18,19 After binding to CXCL12, CXCR4 initiates signal transduction by activating heterotrimeric Gi proteins, which activate downstream effectors such as calcium flux, AKT, and Erk1/2 to trigger cellular responses, including adhesion and cell migration.20 CXCR4 is critical for BM homing and egress of leukocytes (hematopoietic progenitor cells, peripheral blood neutrophils [PMNs], natural killer cells, and B cells),4,21-23 leukocyte trafficking in spleen and lymph nodes,15,24 and myeloid and B-cell development.25 Among the CXCR4-truncating mutations causing WHIM syndrome, the most common and best studied is R334X, which removes the C-terminal 19 amino acids.26 Neutropenia and myelokathexis have been recapitulated in mice and zebrafish genetically modified to express CXCR4R334X.27,28 Moreover, patients with WHIM syndrome due to CXCR4R334X have shown significant improvement in circulating leukocyte numbers after brief treatment with plerixafor, a CXCR4 antagonist approved by the US Food and Drug Administration for hematopoietic stem cell mobilization with G-CSF for transplantation of multiple myeloma or non-Hodgkin lymphoma patients after cytoreductive chemotherapy.4,29
The C-tail of CXCR4 has been shown by truncation mutagenesis studies to be important for ligand-induced receptor internalization, desensitization, ubiquitination, degradation, and endocytosis.30 Accordingly, truncations in this region of CXCR4 in WHIM syndrome cause increased signaling responses (eg, calcium flux and leukocyte chemotaxis), delayed desensitization, and defective receptor down-regulation.26,31 Additional structure-function studies involving site-directed mutagenesis have shown that several serines in the C-tail of CXCR4 are important phosphorylation sites,32-34 and 3 lysines are vital for degradation of the receptor,35 indicating that single-residue changes in this region may be functionally important; however, such changes have not been shown to cause disease. In the present study, we report a family with WHIM syndrome caused by such a mutation, E343K, affecting the charge of the C-terminus of CXCR4.
Methods
Patients
All research studies were performed after obtaining written informed consent consistent with the Declaration of Helsinki under a National Institutes of Health (NIH) institutional review board–approved protocol.
Genetic testing
Genomic DNA was isolated from peripheral blood, and candidate genes were sequenced using Sanger sequencing methods (ABI). Data were analyzed using Sequencher Version 4.9 software (Gene Codes). For ELANE genetic testing, blood samples were also sent to Gene DX in Gaithersburg, MD.
Microscopy
Microscopic images were taken with a Zeiss Axiovert 200M microscope equipped with a QImaging Retiga EXi camera using a 40× Zeiss oil-immersion objective for a total magnification of 400×. Image acquisition was performed using iVision-Mac Version 4.9 software (BioVision Technologies).
Construction of transfected cell lines
Construction of plasmids pcDNA3.1.CXCR4WT and pcDNA3.1.CXCR4R334X, encoding CXCR4WT or CXCR4R334X, respectively, has been described previously.31 Plasmid pcDNA3.1.CXCR4E343K was made by introducing the patient's single nucleotide (1027G → A) mutation into pcDNA3.1.CXCR4WT using the Quick Change II site-directed mutagenesis kit (Stratagene; for details, see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). K562 cells were transfected with plasmids as described previously (see supplemental Methods).31
K562 cells stably transfected with these plasmids were obtained by culture in 1.5 mg/mL of G418; cells were matched for receptor expression level by repeated selection with the CXCR4-specific Ab 12G5 (BD Biosciences), as described previously31 (also see supplemental Methods).
Flow cytometry
Calcium flux assay
Intracellular calcium flux assays were performed using transfected K562 cells and PBMCs as described previously31 with minor modifications (also see supplemental Methods).
Chemotaxis assay
Neutrophils were freshly isolated from the whole blood from an HD and from patients and chemotaxis was assessed as described previously (see supplemental Methods).31
Statistics
In vitro calcium-flux experiments and CXCR4 internalization assays with transfected K562 cells were performed in triplicate and repeated at least 3 times. Results are expressed as means ± SEM of summary data. Outliers in the data were detected and eliminated using the Grubb test (α = 0.05). Differences between WHIM patient and control subject data were tested for statistical significance using the Student t test. Dose-response curves were analyzed by sigmoidal regression using Prism Version 5 software (GraphPad).
Results
Phenotypic characterization of a family with WHIM syndrome
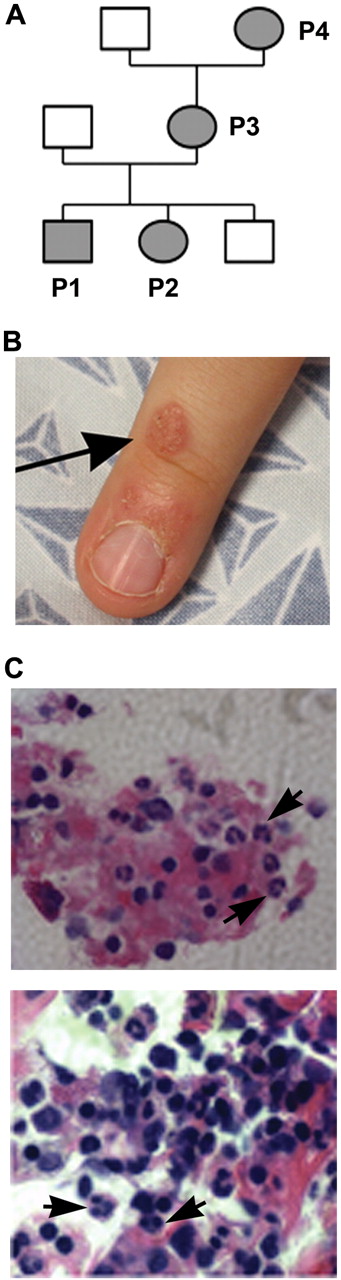
A white family from North Carolina was referred to the National Institutes of Health for evaluation of possible WHIM syndrome (Figure 1A). Patients P1 and P2 are 9-year-old male and 5-year-old female siblings; patient P3 is the 34-year-old mother of P1 and P2; and patient P4 is the 53-year-old grandmother of P1 and P2. Clinical features are summarized in Table 1. Blood for genetic testing was also obtained from a sibling brother and the father of P1 and P2, both of whom lacked a history consistent with congenital immunodeficiency.
Clinical manifestations in a family with WHIM syndrome. (A) Pedigree consistent with autosomal-dominant inheritance. (B) Representative area of skin warts on patient P1. Arrows point to clusters of representative warts on the dorsum of the right hand. (C) BM biopsy from P1 showing full myeloid maturation in the face of peripheral neutropenia, which is consistent with myelokathexis (400×). Arrows point to representative mature neutrophils and granulocyte precursors and an eyeglass-shaped neutrophil in which pyknotic nuclear lobes were connected by a long filament.
Clinical manifestations in a family with WHIM syndrome. (A) Pedigree consistent with autosomal-dominant inheritance. (B) Representative area of skin warts on patient P1. Arrows point to clusters of representative warts on the dorsum of the right hand. (C) BM biopsy from P1 showing full myeloid maturation in the face of peripheral neutropenia, which is consistent with myelokathexis (400×). Arrows point to representative mature neutrophils and granulocyte precursors and an eyeglass-shaped neutrophil in which pyknotic nuclear lobes were connected by a long filament.
Clinical manifestations in 4 patients with WHIM syndrome caused by missense mutation CXCR4E343K
| . | P1 . | P2 . | P3 . | P4 . | Normal range . |
|---|---|---|---|---|---|
| Age, y | 9 | 5 | 34 | 53 | NA |
| Sex | Male | Female | Female | Female | NA |
| Skin warts | + | − | + | − | NA |
| Genital warts | − | − | + | ? | NA |
| IgG, mg/dL | 694 | 791 | 705 | 725 | 572-1474 |
| IgA, mg/dL | 170 | 95 | 103 | 103 | 34-305 |
| IgM, mg/dL | 70 | 72 | 113 | 71 | 32-208 |
| IgE, IU/mL | 27 | 14 | 20 | 49 | 0-90 |
| WBCs, × 109/L | 2.57 | 3.39 | 3.07 | 2.93 | 4.31-11 |
| Neutrophils, × 109/L | 1.06 | 1.05 | 1.95 | 1.45 | 1.63-7.55 |
| Lymphocytes, × 109/L | 1.23 | 1.98 | 0.91 | 1.19 | 0.97-3.96 |
| Monocytes, × 109/L | 0.20 | 0.27 | 0.16 | 0.24 | 0.24-0.86 |
| Myelokathexis | + | Unknown | Unknown | Unknown | NA |
| Recurrent bacterial infections | + | + | + | − | NA |
| Recurrent yeast infections after antibiotics | − | + | − | − | NA |
| Hearing loss | + | + | − | − | NA |
| Delayed speech | + | + | − | − | NA |
| Infection despite vaccination | Influenza | Pneumococcal, influenza | Rubella | No | NA |
| . | P1 . | P2 . | P3 . | P4 . | Normal range . |
|---|---|---|---|---|---|
| Age, y | 9 | 5 | 34 | 53 | NA |
| Sex | Male | Female | Female | Female | NA |
| Skin warts | + | − | + | − | NA |
| Genital warts | − | − | + | ? | NA |
| IgG, mg/dL | 694 | 791 | 705 | 725 | 572-1474 |
| IgA, mg/dL | 170 | 95 | 103 | 103 | 34-305 |
| IgM, mg/dL | 70 | 72 | 113 | 71 | 32-208 |
| IgE, IU/mL | 27 | 14 | 20 | 49 | 0-90 |
| WBCs, × 109/L | 2.57 | 3.39 | 3.07 | 2.93 | 4.31-11 |
| Neutrophils, × 109/L | 1.06 | 1.05 | 1.95 | 1.45 | 1.63-7.55 |
| Lymphocytes, × 109/L | 1.23 | 1.98 | 0.91 | 1.19 | 0.97-3.96 |
| Monocytes, × 109/L | 0.20 | 0.27 | 0.16 | 0.24 | 0.24-0.86 |
| Myelokathexis | + | Unknown | Unknown | Unknown | NA |
| Recurrent bacterial infections | + | + | + | − | NA |
| Recurrent yeast infections after antibiotics | − | + | − | − | NA |
| Hearing loss | + | + | − | − | NA |
| Delayed speech | + | + | − | − | NA |
| Infection despite vaccination | Influenza | Pneumococcal, influenza | Rubella | No | NA |
All information shown is from the history, physical examination, and laboratory evaluation performed at the time of presentation to the National Institutes of Health.
NA indicates not applicable; +, present; and −, absent.
P1 had multiple clusters of warts on his hands near the finger tips (Figure 1B shows a representative lesion) and several warts on his face since age 3 that did not respond to topical salicylic acid or 5-fluorouracil. P2 did not have warts. P3 had abnormal Pap smears in 2000 and 2009 that were consistent with HPV infection. Although P1, P2, and P3 all had serum Ig levels above the lower limit of the normal range and a protective titer after diphtheria and tetanus vaccination, they also had a history of clinical failure of vaccination. In particular, P1 and P2 had virologically documented influenza twice despite receiving the annual seasonal vaccination more than 1 month before disease onset, and P3 had measles as a child despite vaccination with the rubella vaccine. All 3 patients had frequent bacterial infections involving the respiratory tract, including pharyngitis, sinusitis, pneumonia, and otitis. The latter eventually led to bilateral sensorineural hearing loss and speech problems in P1 and P2 with a need for hearing aids in P1. P2 also had recurrent vaginal yeast infections after treatment with antibiotics. Congenital neutropenia was diagnosed in P1 and P2 and their absolute neutrophil counts (ANCs) were noted to range from 0.4-1.3 × 109/L and to increase during infections. The mother, P3, had a history of mild neutropenia (twice documented recently as 1.4 and 1.95 × 109/L at the NIH). The grandmother, P4, had a history of neutropenia (1.4 × 109/L at the NIH) and a hysterectomy at age 20 for a diagnosis of unspecified malignancy, but did not have a history of recurrent infections, vaccine failure, or persistent warts. P1, P2, and P3 all had monocytopenia documented on multiple occasions, and P3 was consistently lymphocytopenic. At the NIH, the absolute monocyte count for P4 was at the lower limit of normal. At age 3, a BM biopsy performed on P1 showed myelokathexis (increased numbers of mature neutrophils in BM associated with neutropenia) with an eyeglass-shaped neutrophil in which the nuclear lobes were connected by a long filament (Figure 1C). Chromosomal analysis revealed a normal male karyotype with no observed clonal abnormalities. BM evaluation had not been performed on P2, P3, or P4. Because of recurrent infections despite vaccination and a daily prophylactic antibiotic, P2 was treated with G-CSF (Neupogen; Amgen) with an excellent response, including marked reduction in the incidence of infections. The available clinical history and laboratory values presented by this family suggested the possibility of WHIM syndrome.
Broad leukocytopenia in a family with WHIM syndrome
Because the patients had a history of leukopenia and because many patients with WHIM syndrome are panleukocytopenic,4 we characterized the leukocyte distribution in the blood of patients P1, P2, and P3 in greater detail using multicolor FACS analysis. We were unable to obtain blood from patient P4 for this analysis. As shown in Table 2, all 3 patients were broadly leukocytopenic. Of the 13 subsets with a reference range analyzed, all 3 patients were below the lower limit of normal for 3: CD8+ memory T cells, and classical and inflammatory monocytes; 2 of 3 patients were below the lower limit of normal for 7, including CD3+ T cells, total CD4+ naive and memory T cells, total CD8+ T cells and CD8+ naive T cells, NK cells, and CD27− B cells. P2 and P3 had normal total B cell and CD27+ memory B counts, but P1 had low B cell and CD27+ B-cell counts. In contrast, most other cases of WHIM syndrome that have been reported, including our previously reported WHIM patients with the CXCR4R334X mutation, have been severely deficient in circulating CD19+ B cells.4 In the monocyte population, P1, P2, and P3 had markedly reduced CD14+ CD16− classical and CD14+ CD16+ inflammatory monocytes, similar to our previous reported WHIM patients with R334X.4 We also checked the frequency of pDCs and mDCs in PBMCs from healthy donors (HDs), from P1, P2, P3, and P4, as well as from a WHIM syndrome patient with the R334X mutation (P1 from McDermott et al4 ). Consistent with a previous report,5 both cell types were severely deficient: 0.017% and 0.12%, respectively, in the WHIM patient with the R334X mutation. However, the average frequencies of pDCs (0.35 ± 0.05%) and mDCs (0.36 ± 0.06%) in P1-P4 were comparable to those in adult HDs (pDCs: 0.24 ± 0.04%; mDCs: 0.37 ± 0.05%).
Leukocyte subset analysis in the blood of 3 patients with WHIM syndrome caused by missense mutation CXCR4E343K
| Cell type . | P1 . | P2 . | P3 . | Reference range for children, cells/μL of blood, mean* . | Reference range for adults, cells/μL of blood, mean† . |
|---|---|---|---|---|---|
| CD3+ T cell | 943 | 1487 | 700 | 1200-2600 | 714-2266 |
| CD4+ T cell | 478 | 903 | 512 | 650-1500 | 359-1565 |
| CD4+ CD45RA+ naive T cell | 261 | 630 | 54 | 320-1000 | 454-733 |
| CD4+ CD45RO+ memory T cell | 149 | 147 | 392 | 230-630 | 219-1048 |
| CD4+ CD45RA− CD62L+ CCR7+ central memory T cell | 4 | 8 | 13 | ND | 16-49 |
| CD4+ CD45RA−CD62L− CCR7− effector memory T cell | 32 | 32 | 88 | ND | 57-130 |
| CD8+ T cell | 347 | 449 | 161 | 370-1100 | 178-853 |
| CD8+CD45RA+ naive T cell | 172 | 360 | 66 | 310-900 | 231-371 |
| CD8+CD45RO+ memory T cell | 26 | 20 | 30 | 70-390 | 50-352 |
| CD8+CD45RA−CD62L+CCR7+ central memory T cell | < 1 | < 1 | < 1 | ND | 0-12 |
| CD8+CD45RA−CD62L−CCR7− effector memory T cell | 7 | 21 | 10 | ND | 22-75 |
| CD3−CD56+ NK cell | 161 | 77 | 59 | 100-480 | 126-729 |
| CD19+ B cell | 127 | 414 | 152 | 270-860 | 61-329 |
| CD19+CD27+ memory B cell | 39 | 135 | 34 | 60-230 | 12-68 |
| CD19+CD27+CD38+IgD+ unswitched memory B cell | 1 | 1 | 2 | ND | 0-2 |
| CD19+CD27+CD38+IgD− switched memory B cell | 7 | 5 | 3 | ND | 4-21 |
| CD19+CD27− B cell | 53 | 71 | 118 | 140-470 | 90-176 |
| CD19+CD27−IgD+IgM+ transitional/naive B cell | 28 | 43 | 69 | ND | 42-85 |
| CD19+CD27−IgD−IgM+ immature B cell | < 1 | < 1 | 3 | ND | 2-10 |
| CD14+CD16− classical monocyte | 195 | 264 | 147 | 527-595 | 371-539 |
| CD14+CD16+ inflammatory monocyte | 5 | 6 | 13 | 80-106 | 14-30 |
| Cell type . | P1 . | P2 . | P3 . | Reference range for children, cells/μL of blood, mean* . | Reference range for adults, cells/μL of blood, mean† . |
|---|---|---|---|---|---|
| CD3+ T cell | 943 | 1487 | 700 | 1200-2600 | 714-2266 |
| CD4+ T cell | 478 | 903 | 512 | 650-1500 | 359-1565 |
| CD4+ CD45RA+ naive T cell | 261 | 630 | 54 | 320-1000 | 454-733 |
| CD4+ CD45RO+ memory T cell | 149 | 147 | 392 | 230-630 | 219-1048 |
| CD4+ CD45RA− CD62L+ CCR7+ central memory T cell | 4 | 8 | 13 | ND | 16-49 |
| CD4+ CD45RA−CD62L− CCR7− effector memory T cell | 32 | 32 | 88 | ND | 57-130 |
| CD8+ T cell | 347 | 449 | 161 | 370-1100 | 178-853 |
| CD8+CD45RA+ naive T cell | 172 | 360 | 66 | 310-900 | 231-371 |
| CD8+CD45RO+ memory T cell | 26 | 20 | 30 | 70-390 | 50-352 |
| CD8+CD45RA−CD62L+CCR7+ central memory T cell | < 1 | < 1 | < 1 | ND | 0-12 |
| CD8+CD45RA−CD62L−CCR7− effector memory T cell | 7 | 21 | 10 | ND | 22-75 |
| CD3−CD56+ NK cell | 161 | 77 | 59 | 100-480 | 126-729 |
| CD19+ B cell | 127 | 414 | 152 | 270-860 | 61-329 |
| CD19+CD27+ memory B cell | 39 | 135 | 34 | 60-230 | 12-68 |
| CD19+CD27+CD38+IgD+ unswitched memory B cell | 1 | 1 | 2 | ND | 0-2 |
| CD19+CD27+CD38+IgD− switched memory B cell | 7 | 5 | 3 | ND | 4-21 |
| CD19+CD27− B cell | 53 | 71 | 118 | 140-470 | 90-176 |
| CD19+CD27−IgD+IgM+ transitional/naive B cell | 28 | 43 | 69 | ND | 42-85 |
| CD19+CD27−IgD−IgM+ immature B cell | < 1 | < 1 | 3 | ND | 2-10 |
| CD14+CD16− classical monocyte | 195 | 264 | 147 | 527-595 | 371-539 |
| CD14+CD16+ inflammatory monocyte | 5 | 6 | 13 | 80-106 | 14-30 |
Values below the appropriate reference range are bolded and italicized.
Based on the reported values from healthy children.44-46
Based on values from 11-40 healthy adults in our clinical center.
ND indicates not defined.
Identification of missence mutation E343K in the C-terminus of CXCR4 in affected members of a family with WHIM syndrome
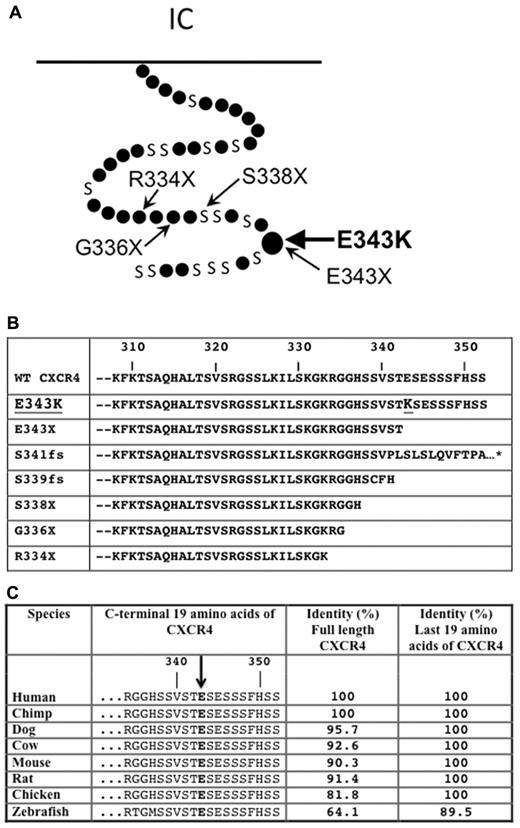
The multigenerational pattern of disease in this family was most consistent with autosomal dominant, Mendelian inheritance (Figure 1A). Since the clinical picture is most consistent with WHIM syndrome, we tested CXCR4 as a candidate for possible mutations. Previously it has been reported that several point mutaitons leading to a premature stop codon cause truncation of the last 10-19 amino acids of the C-tail of CXCR4 (Figure 2A) in patients with WHIM syndrome (eg, R334X, S338X, G336X, and E333X).12,36,37 Two additional mutations in patients with WHIM syndrome have been reported in the C-tail of CXCR4 that cause frame-shifts, and truncation of the receptor (Figure 2B).12,13,38 The predicted C-terminal amino acid sequences caused by all the reported mutations in WHIM syndrome are listed in Figure 2B. In the new family reported here, we found that the affected members were heterozygous for a novel 1027G→ A missense mutation in the portion of the open reading frame encoding the C-tail of CXCR4, which results in a single amino acid change, E343K, and a change in charge from negative to positive. We will refer to the mutant receptor hereafter as CXCR4E343K. This is the first missense mutation associated with WHIM syndrome. The brother of P1 and P2 and their father were both unaffected and homozygous for wild-type CXCR4 (CXCR4WT), suggesting that E343K is an autosomal-dominant mutation (Figure 1A). The grandmother, P4, was also heterozygous for the mutation, but had neutropenia as the only documented clinical manifestation of WHIM syndrome that we could identify. We were unable to find a previous report of this mutation in the medical literature, the National Center for Biotechnology Information single nucleotide polymorphism database (www.ncbi.nlm.nih.gov/projects/SNP), or data from the 1000 Genomes project (www.1000genomes.org), indicating that this is a bona fide rare mutation and not a polymorphism. Interestingly, the missense mutation was located at the exact amino acid that is truncated in the smallest truncation reported to cause WHIM syndrome (E343X), which was first identified by Hernandez et al12 and is in a highly conserved region in the receptor C-terminal region (Figure 2C). We also sequenced the neutrophil elastase gene ELANE in the patients and failed to find a mutation (data not shown).
CXCR4E343K is associated with WHIM syndrome. (A) Position of mutation E343K in the 7-transmembrane domain model of CXCR4. Small dots represent amino acids except serine (designated by S). The locations of other point mutations reported in WHIM syndrome are also highlighted. (B) Alignment of predicted amino acid sequences from the C-terminal domain of CXCR4WT and all reported CXCR4 mutants associated with WHIM syndrome. The position mutated to lysine (K) in CXCR4E343K is highlighted. *Missense mutation that extends the sequence by an additional 10 amino acids, not shown for clarity. (C) The glutamic acid changed to lysine in CXCR4E343K is highly conserved phylogenetically. Predicted amino acid sequences from the C-terminal domain of CXCRWT are aligned for the indicated species. The homology (percent amino acid identity) of human CXCR4 to CXCR4 from each of the other species listed is shown for both the full-length sequence and the C-terminal 19 amino acids.
CXCR4E343K is associated with WHIM syndrome. (A) Position of mutation E343K in the 7-transmembrane domain model of CXCR4. Small dots represent amino acids except serine (designated by S). The locations of other point mutations reported in WHIM syndrome are also highlighted. (B) Alignment of predicted amino acid sequences from the C-terminal domain of CXCR4WT and all reported CXCR4 mutants associated with WHIM syndrome. The position mutated to lysine (K) in CXCR4E343K is highlighted. *Missense mutation that extends the sequence by an additional 10 amino acids, not shown for clarity. (C) The glutamic acid changed to lysine in CXCR4E343K is highly conserved phylogenetically. Predicted amino acid sequences from the C-terminal domain of CXCRWT are aligned for the indicated species. The homology (percent amino acid identity) of human CXCR4 to CXCR4 from each of the other species listed is shown for both the full-length sequence and the C-terminal 19 amino acids.
CXCR4E343K is a gain-of-function mutation: analysis in transfected K562 cells
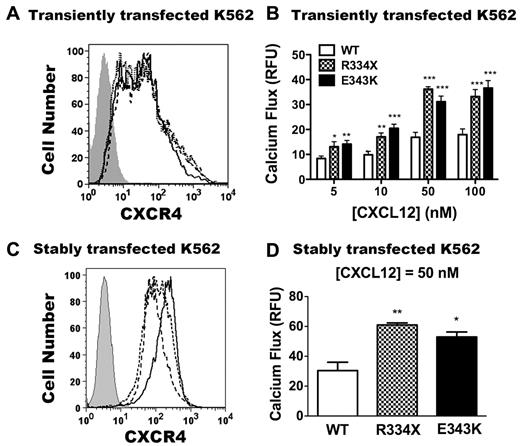
Because of its location in the carboxy terminus of the receptor, we hypothesized that CXCR4E343K may perturb receptor signaling and function. To test this, first we compared the agonist sensitivity of CXCR4E343K with CXCR4WT and the 19 amino acid–truncated receptor CXCR4R334X. We transiently transfected plasmids encoding CXCR4WT, CXCR4R334X, or CXCR4E343K into the cell line K562, which does not normally express native CXCR4 on the cell surface or respond to the CXCR4 agonist CXCL12.31 At 24 hours after transfection, we checked CXCR4 expression on all 3 transfectants by FACS using mAb 12G5 directed against the N-terminal extracellular domain of CXCR4. We verified that untransfected K562 cells had no CXCR4 expression, and found that cells transfected with all 3 CXCR4 variants exhibited similar levels of surface staining with 12G5 (Figure 3A), which allowed us to compare the signaling and function of these receptors quantitatively.
CXCR4E343K is a gain-of-function mutation: analysis in transfected K562 cells. (A,C) Expression of CXCR4 variants on untransfected K562 cells (gray area) or K562 cells transfected transiently (A) or stably (C) with CXCR4WT (solid line), CXCR4R334X (dashed line), and CXCR4E343K (dotted line). X-axis shows the mean fluorescence intensity of CXCR4 expression. (B) Peak calcium-flux responses of K562 cells transiently transfected with CXCR4WT, CXCR4R334X, and CXCR4E343K in response to the indicated doses of CXCL12. *P < .05; **P < .01; and ***P < .001 for the comparison between CXCR4WT and either CXCR4R334X or CXCR4E343K. Data are summarized as the means ± SEM from 4 independent experiments with 3 replicates each. (D) The peak calcium-flux response of K562 cells stably transfected with CXCR4WT, CXCR4R334X, and CXCR4E343K in response to 50nM CXCL12 stimulation. *P < .05 and **P < .01 for the comparison between CXCR4WT and CXCR4E343K or CXCR4R334X.
CXCR4E343K is a gain-of-function mutation: analysis in transfected K562 cells. (A,C) Expression of CXCR4 variants on untransfected K562 cells (gray area) or K562 cells transfected transiently (A) or stably (C) with CXCR4WT (solid line), CXCR4R334X (dashed line), and CXCR4E343K (dotted line). X-axis shows the mean fluorescence intensity of CXCR4 expression. (B) Peak calcium-flux responses of K562 cells transiently transfected with CXCR4WT, CXCR4R334X, and CXCR4E343K in response to the indicated doses of CXCL12. *P < .05; **P < .01; and ***P < .001 for the comparison between CXCR4WT and either CXCR4R334X or CXCR4E343K. Data are summarized as the means ± SEM from 4 independent experiments with 3 replicates each. (D) The peak calcium-flux response of K562 cells stably transfected with CXCR4WT, CXCR4R334X, and CXCR4E343K in response to 50nM CXCL12 stimulation. *P < .05 and **P < .01 for the comparison between CXCR4WT and CXCR4E343K or CXCR4R334X.
Consistent with our previous results, after stimulation with all doses of CXCL12 tested, no calcium flux response was observed in untransfected K562 cells (data not shown), and CXCR4R334X transiently transfected cells gave approximately 2-fold increased signals compared with CXCR4WT (Figure 3B).31 Interestingly, the cells transiently transfected with CXCR4E343K also had an approximately 2-fold increased signal compared with stimulated cells expressing CXCR4WT (Figure 3B). To confirm these results, we generated stably transfected K562 cells and selected cells matched for equivalent receptor expression on the cell surface. As shown in Figure 3C, CXCR4 was not detectable on untransfected K562 cells, whereas cells transfected with all 3 CXCR4 variants exhibited similar levels of surface staining, with slightly higher expression for the CXCR4WT transfectants. As described previously,31 responses of K562 cells stably expressing CXCR4R334X were approximately 2-fold greater than those of cells expressing CXCR4WT (Figure 3D) despite slightly increased receptor expression in the CXCR4WT cells. CXCL12-induced responses in cells stably expressing CXCR4E343K were equivalent to those for cells expressing CXCR4R334X. Therefore, like the R334X truncation mutation, E343K is a gain-of-function mutation in CXCR4, as shown by the calcium-flux assay in K562 cells.
CXCR4E343K is a gain-of-function mutation: analysis in WHIM patient leukocytes
To determine whether E343K also behaved as a gain-of-function mutation when expressed at natural abundance in primary leukocytes, we compared calcium flux and chemotaxis responses to CXCL12 stimulation for PBMCs and PMNs from the blood of WHIM patients and HDs.
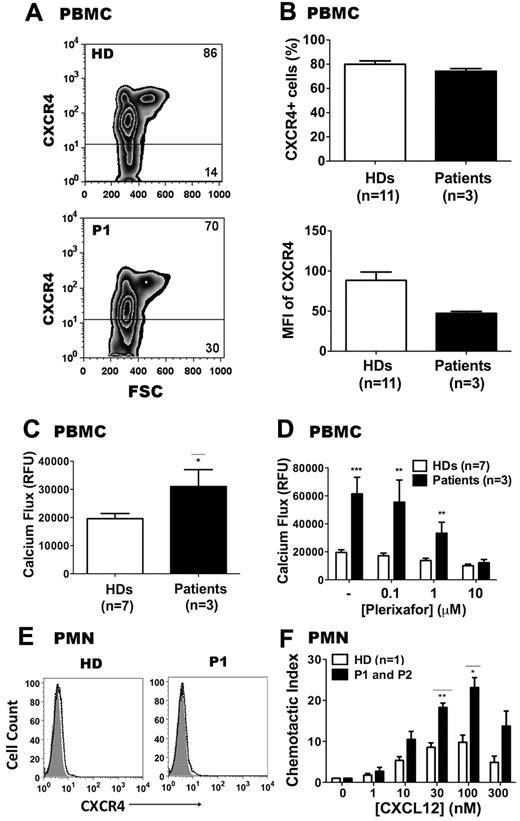
As shown in Figure 4A and B, total surface CXCR4 expression was lower on PBMCs from P1, P2, and P3 compared with HDs (n = 11), both for mean fluorescence intensity and for percentage of CXCR4+ cells. This is similar to what we have reported previously for a WHIM syndrome patient with the R334X mutation,31 and may be caused by preferential sequestration of CXCR4-expressing monocytes and T cells in the BM or lymphoid tissue. Despite this, after adjustment for CXCR4 expression, calcium flux responses to CXCL12 stimulation were higher for patient PBMCs compared with HD PBMCs (Figure 4C). To confirm that the increased calcium flux in response to CXCL12 in patient PBMCs was CXCR4 specific, PBMCs from HDs and from patients P1, P2, and P3 were pretreated with the specific CXCR4 inhibitor plerixafor (AMD3100) before being stimulated with 10nM CXCL12. As shown in Figure 4D, plerixafor could effectively block the CXCL12 response in cells from both HDs and from patients P1, P2, and P3. The IC50 (concentration at which half of the maximal signal was blocked) was approximately 1μM for PBMCs from both HDs, confirming our previously published results31 and extending them to these new patients.
CXCR4E343K is a gain-of-function mutation: analysis in primary leukocytes from patients with WHIM syndrome. (A-B) CXCR4 expression on PBMCs. Cells were gated on lymphocytes and monocytes. Representative zebra plots are shown in panel A and summary data in panel B. Percentages of CXCR4+ and CXCR4− cells in PBMCs are shown. (C) PBMC calcium-flux responses to 10nM CXCL12. (D) Patient and HD PBMC calcium-flux responses to CXCL12 can both be blocked by plerixafor. CXCL12 was added after 3 minutes of incubation with the indicated concentrations of plerixafor. (E) Representative CXCR4 expression on neutrophils from an HD and from P1. Shaded area is unstained; solid line, CXCR4; dashed line, isotype control. (F) Neutrophil chemotaxis. Comparisons are displayed of a HD versus P1 and P2 for different concentrations of CXCL12-induced chemotaxis. Chemotactic index was defined as 1 when no CXCL12 was added to the lower chamber. Results from both patients were averaged and graphed, with SEM displayed for each value. *P < .05; **P < .01; and ***P < .001 for the comparison between HDs and CXCR4E343K (E343K) patients, respectively.
CXCR4E343K is a gain-of-function mutation: analysis in primary leukocytes from patients with WHIM syndrome. (A-B) CXCR4 expression on PBMCs. Cells were gated on lymphocytes and monocytes. Representative zebra plots are shown in panel A and summary data in panel B. Percentages of CXCR4+ and CXCR4− cells in PBMCs are shown. (C) PBMC calcium-flux responses to 10nM CXCL12. (D) Patient and HD PBMC calcium-flux responses to CXCL12 can both be blocked by plerixafor. CXCL12 was added after 3 minutes of incubation with the indicated concentrations of plerixafor. (E) Representative CXCR4 expression on neutrophils from an HD and from P1. Shaded area is unstained; solid line, CXCR4; dashed line, isotype control. (F) Neutrophil chemotaxis. Comparisons are displayed of a HD versus P1 and P2 for different concentrations of CXCL12-induced chemotaxis. Chemotactic index was defined as 1 when no CXCL12 was added to the lower chamber. Results from both patients were averaged and graphed, with SEM displayed for each value. *P < .05; **P < .01; and ***P < .001 for the comparison between HDs and CXCR4E343K (E343K) patients, respectively.
To further examine whether the novel 1027G → A mutation (E343K) could augment CXCR4 function when expressed at natural abundance on primary cells from the patients, we compared chemotaxis responses of freshly isolated PMNs from the peripheral blood of an HD and from P1 and P2. PMNs from P1, P2, and P3 expressed very low levels of CXCR4 (Figure 4E), similar to HDs.39 Consistent with the PBMC calcium-flux results, the PMNs from P1 and P2 showed approximately 2-fold enhanced chemotaxis in response to CXCL12 across the 10-300nM dose range compared with PMNs from a HD tested at the same time (Figure 4F). Therefore, our findings demonstrate that the novel 1027G → A mutation (E343K) leads to increased CXCL12-induced and CXCR4-dependent responses when tested in either transfected cells or primary cells from patients with WHIM syndrome.
Impaired ligand-induced down-regulation of mutant receptor CXCR4E343K
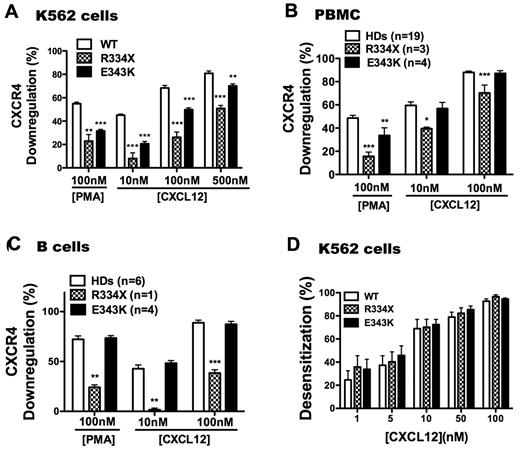
Normally, stimulation of G-protein–coupled receptors with agonists leads to a reduction in receptor levels detectable on the cell surface, a process known as down-regulation. Receptor levels and signaling capacity can be affected by many parameters, including retrograde trafficking (internalization), anterograde trafficking, and functional desensitization. Consistent with our previous findings,31,40 CXCR4WT was readily down-regulated in K562 cells by CXCL12 in a dose-dependent manner, as well as by phorbol 12-myristate 13-acetate (PMA); in contrast, CXCR4R334X down-regulation was reduced markedly for both stimuli. CXCR4E343K down-regulation was also reduced, but to a lesser degree than for CXCR4R334X (Figure 5A). When primary PBMCs and B cells from HDs were stimulated with PMA or CXCL12, the level of CXCR4 down-regulation was similar to what we observed for CXCR4WT-transfected K562 cells (Figure 5B-C). The down-regulation pattern for CXCR4R334X in primary cells was similar, but not identical, to transfected K562 cells. In particular, down-regulation of CXCR4R334X was reduced in response to both CXCL12 and PMA in PBMCs and B cells. In contrast, down-regulation was normal in both PBMCs and B cells from patients P1-P4 stimulated with CXCL12, and significantly reduced down-regulation of CXCR4 was only observed for PBMCs stimulated with PMA. Therefore, the increased signaling and function of CXCR4E343K may not be completely attributable to defective physical down-regulation of the receptor. Therefore, we also investigated whether impaired functional down-regulation might account for enhanced signaling by CXCR4E343K. In sequential stimulation experiments, CXCR4WT, CXCR4R334X, and CXCR4E343K signaling in K562 cells could be desensitized to a similar degree homologously, including complete desensitization at the highest dose tested, 100nM CXCL12 (Figure 5D). This suggests that impaired functional desensitization is not the sole cause of increased signaling by either CXCR4E343K or CXCR4R334X.
WHIM mutation CXCR4E343K attenuates CXCL12-induced receptor down-regulation but to a lesser degree than CXCR4R334X. Surface expression was measured with CXCR4-specific mAb 12G5 for the cell types indicated at the top of each panel after incubation in the presence or absence of the stimuli indicated on the x-axis. (A) Transiently transfected K562 cells. **P < .01 and ***P < .001 for the comparison between CXCR4WT (WT) and CXCR4E343K (E343K) or CXCR4R334X (R334X), respectively. Data are summarized as the means ± SEM from 4 experiments. (B) PBMCs. Data are pooled from multiple independent experiments with cells with the indicated CXCR4 genotypes (number of individuals studied for each genotype). P1-P3 were studied twice and P4 once; 1 CXCR4R334X patient was studied twice and the other 2 once, and each HD was studied once. One or 2 HDs were studied as controls in each of the 5 experiments involving WHIM patients, and the rest were studied in 8 additional independent experiments. Data are the means ± SEM of all values in each category. *P < .05; **P < .01; and ***P < .001 for the comparison between HDs and CXCR4R334X (R334X) or CXCR4E343K (E343K) patients, respectively. (C) B cells. After incubation with stimuli in duplicates, PBMCs were gated on CD19+ cells and analyzed for CXCR4 expression. Data are from 1 experiment that included WHIM patients P1-P4, 1 CXCR4R334X patient, and 1 HD, and 2 additional independent experiments for the other 4 HDs, and are the means ± SEM of all values in each category. **P < .01 and ***P < .001 for the comparison between HDs and CXCR4R334X (R334X) or CXCR4E343K (E343K) patients, respectively. (D) Normal homologous desensitization of WHIM variants CXCR4R334X (R334X) and CXCR4E343K (E343K) expressed in K562 cells. Data were pooled from 5 independent experiments done in triplicate with the indicated variants and are summarized as the means for each condition.
WHIM mutation CXCR4E343K attenuates CXCL12-induced receptor down-regulation but to a lesser degree than CXCR4R334X. Surface expression was measured with CXCR4-specific mAb 12G5 for the cell types indicated at the top of each panel after incubation in the presence or absence of the stimuli indicated on the x-axis. (A) Transiently transfected K562 cells. **P < .01 and ***P < .001 for the comparison between CXCR4WT (WT) and CXCR4E343K (E343K) or CXCR4R334X (R334X), respectively. Data are summarized as the means ± SEM from 4 experiments. (B) PBMCs. Data are pooled from multiple independent experiments with cells with the indicated CXCR4 genotypes (number of individuals studied for each genotype). P1-P3 were studied twice and P4 once; 1 CXCR4R334X patient was studied twice and the other 2 once, and each HD was studied once. One or 2 HDs were studied as controls in each of the 5 experiments involving WHIM patients, and the rest were studied in 8 additional independent experiments. Data are the means ± SEM of all values in each category. *P < .05; **P < .01; and ***P < .001 for the comparison between HDs and CXCR4R334X (R334X) or CXCR4E343K (E343K) patients, respectively. (C) B cells. After incubation with stimuli in duplicates, PBMCs were gated on CD19+ cells and analyzed for CXCR4 expression. Data are from 1 experiment that included WHIM patients P1-P4, 1 CXCR4R334X patient, and 1 HD, and 2 additional independent experiments for the other 4 HDs, and are the means ± SEM of all values in each category. **P < .01 and ***P < .001 for the comparison between HDs and CXCR4R334X (R334X) or CXCR4E343K (E343K) patients, respectively. (D) Normal homologous desensitization of WHIM variants CXCR4R334X (R334X) and CXCR4E343K (E343K) expressed in K562 cells. Data were pooled from 5 independent experiments done in triplicate with the indicated variants and are summarized as the means for each condition.
Discussion
In the present study, we provide genetic and biochemical evidence that WHIM syndrome can be caused by CXCR4E343K, the first missense mutation in CXCR4 identified in this disease. In particular, we found that: (1) clinical manifestations consistent with WHIM syndrome were found in 3 generations of a single family in a pattern consistent with Mendelian inheritance in an autosomal-dominant manner, the known inheritance pattern for classical WHIM syndrome; (2) mutation CXCR4E343K was found in heterozygous form in affected family members, but not in unaffected family members or in the general population (absent from both the National Center for Biotechnology Information single nucleotide polymorphism database and the 1000 Genomes Project database); (3) the mutation changes a negative charge to a positive one in the C-tail of CXCR4, where all other known WHIM mutations reside, including one that truncates the receptor at the same position, E343X; (4) the C-terminal domain of CXCR4 is normally responsible for negative regulation of the receptor; (5) CXCR4E343K results in gain-of-function, like other known WHIM syndrome mutations, and to a similar magnitude; and (6) exaggerated CXCR4 signaling as the cause of myelokathexis is consistent with the known role of normal CXCR4 signaling in regulating bone marrow egress of neutrophilis.41
None of the patients in the family had all four features of classical WHIM syndrome, consistent with the generally accepted notion that the syndrome is phenotypically heterogenous. P1, but not P2, has or has had skin warts, and P3 also has a history of abnormal Pap smears, while P4 had an early hysterectomy for a cancer of an unknown type. P2 is 5 years old and may simply not yet have been exposed to HPV. P1, P2, and P3 all have a history of recurrent bacterial infections. P1 and P2 had congenital neutropenia, but the ANC was documented to rise during episodes of acute infection, which is unusual for other types of congenital neutropenia and myelopoiesis defects. We were unable to obtain documentation of neutrophil counts for P3 and P4 from childhood, but the ANCs on presentation to NIH were low (P4) or at the lower limit of normal (P3). In addition, recent neutropenia was documented for P3. Bone marrow was directly examined only for P1, and this showed classical myelokathexis, without a defect in myeloid cell differentiation. Because of this and the gene findings, we could not justify bone marrow biopsies for the other patients. Nevertheless, we observed that low-dose G-CSF treatment resulted in a rapid increase in ANCs (within hours) to normal in P2, which would be expected in the setting of myelokathexis but not when neutropenia is caused by decreased neutrophil production.
Hypogammaglobulinemia is the least penetrant clinical feature of WHIM syndrome; approximately 10% of patients have normal IgG levels and most of the rest have only moderate deficiency.7 Therefore, the finding of normal IgG levels in all 4 patients with CXCR4E343K is consistent with the known clinical spectrum of the syndrome. Also unusual is the finding of normal absolute B-cell counts in the CXCR4E343K patients. Twelve of 15 reported WHIM patients with other genotypes have been reported to have abnormal B-cell counts,4,7 including our patients with the R334X mutation (n = 3), all of whom have severe deficiency.4 Nevertheless, as with other WHIM patients, vaccination failed to protect CXCR4E343K patients P1 and P2 from influenza and P3 from a measles infection, suggesting that their B-cell function may be at least partially impaired. It will be important to follow these children prospectively and determine whether more severe humoral immunodeficiency develops with time.
CXCR4E343K patients P1-P4 all had neutropenia; however, the ANC value was documented as < 0.5 × 109/L only in P1 and P2. To put this in perspective, neutropenia was also present in 28 of 28 previously reported WHIM patients7 and severe neutropenia (ANC < 0.5 × 109/L) has been observed in 18 of 24 (75%) WHIM patients with other mutations reported previously.7 In the present study, monocytopenia in P1, P2, and P3 was less severe compared with previously described patients with CXCR4R334X.4 CXCR4E343K was also associated with low levels of circulating memory T cells.
Because previously discovered mutations in WHIM syndrome all result in 10-19 amino acid truncations of the C-tail, it has not been possible to define the precise amino acid(s) critical for increased signaling and disease. Previous studies of the structural basis of CXCR4 down-regulation have identified specific functionally important amino acids, but this was outside of the context of disease.30,35,42 Therefore, the significance of the present study is in pinpointing the first amino acid that, when mutated, causes clinical manifestations consistent with WHIM syndrome. Additional evidence that there may be strong selective pressure for maintaining glutamate at this position in CXCR4 can be obtained from phylogenetic analysis, as shown in Figure 2C, which shows no variation from fish to humans. Although the full-length CXCR4 sequence is highly conserved phylogenetically, the C-tail region is even more highly conserved. All truncation mutants described in WHIM syndrome remove E343, and the gain of function observed for the longest truncation CXCR4R334X is quantitatively similar to that observed for CXCR4E343K. Therefore, it is formally possible that loss or mutation of E343 is the sole determinant of WHIM syndrome. Moreover, CXCR4R334X caused a greater defect than CXCR4E343K in ligand-induced receptor down-regulation, suggesting that E343 may cooperate with other C-tail residues to mediate this function. Three lysines in the C-tail of CXCR4 N-terminal to the R334X deletion site have been shown to be important for CXCR4 degradation30 ; the addition of a fourth by the glutamate-to-lysine change in CXCR4E343K may not change charge sufficiently to affect receptor down-regulation mechanisms, particularly because neither amino acid at position 343 can serve as a phosphorylation site for GRKs and is therefore unlikely to affect arrestin binding to a great extent. Nevertheless, weaker ligand-induced receptor down-regulation for CXCR4R334X than for CXCR4E343K did not translate into differences in the calcium flux and chemotaxis signaling assays. Calcium flux is a very rapid assay, measured in seconds, and requires low ligand concentrations, unlike receptor down-regulation, which occurs over minutes and requires high concentrations of ligand. Chemotaxis is measured in vitro using an artificial system and may not depend heavily on receptor down-regulation.43 Our present results are compatible with a previous study by Balabanian et al demonstrating enhanced activation of G-proteins by the R334X mutation in isolated cell membrane preparations in which receptor internalization does not occur.10 The significance of receptor down-regulation to the clinical, cellular, immunologic, and biochemical features in WHIM syndrome, and to the differences noted between patients with R334X versus E343K mutations in the present study, remains to be determined. We have also shown herein that the CXCR4 antagonist plerixafor (AMD3100) is equally effective at blocking CXCL12-induced calcium-flux responses in PBMCs from HDs and WHIM patients with the CXCR4E343K mutation (Figure 4D), similar to patients with the CXCR4R334X mutation.31 Because plerixafor treatment corrects panleukopenia and myelokathexis in WHIM patients with the R334X mutation in vivo,4 it may also be considered as a potential treatment for WHIM syndrome due to CXCR4E343K.
In conclusion, in the present study, we have identified a novel, autosomal-dominant, missense, nontruncating, charge-changing, gain-of-function mutation, CXCR4E343K, in the C-tail of CXCR4 that appears to cause WHIM syndrome. Increased receptor signaling does not appear to be fully explained by impairment of CXCR4 down-regulation, and plerixafor is a potential mechanism-directed treatment for these patients. Our results provide new knowledge about the genetic and biochemical basis of complex WHIM syndrome phenotypes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients with WHIM syndrome and their family members who volunteered to make this study possible; and Steve Tsang, PhD, National Institute of Allergy and Infectious Diseases (NIAID), for bioinformatics help in comparing the CXCR4 sequences from different species.
This work was supported by the Division of Intramural Research of the NIAID, and by an award from the Office of Rare Diseases Research through the Bench to Bedside Program of the Clinical Center, National Institutes of Health (NIH). Additional funding was provided by a grant from the National Cancer Institute, NIH (contract number HHSN261200800001E). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
National Institutes of Health
Authorship
Contribution: Q.L., H.C., T.O., and X.G. generated and analyzed the experimental data; S.A.-O., N.A.T., J.U., R.D., C.K., A.R.C., S.H.G., E.I.H., D.S.W., H.L.M., P.M.M., and D.H.M. provided patient recruitment and care; H.L.M., P.M.M., and D.H.M. supervised the experiments and analyzed the data; and Q.L., P.M.M. and D.H.M. wrote the manuscript with participation from all of the other authors.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for E.I.H. is Children's National Medical Center, Washington, DC.
Correspondence: David H. McDermott, 10 Center Dr, Bldg 10, Rm 11N107, Bethesda, MD 20892-1886; e-mail: dmcdermott@niaid.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal