Abstract
Skewing toward myeloid cell production is often observed in chronic inflammation and autoimmune diseases. Herein, we determined whether persistent myeloid activation and proinflammatory output occurring in pathologic conditions is at the level of hematopoietic stem and primitive progenitor cells (HSPPCs). By using a mouse arthritis model, we found that even though HSPPCs in arthritis still retained the capacity to differentiate into different lineages, they acquired enhanced in vitro and in vivo propensity in a disease-dependent manner to generate myeloid cells, the key perpetrators of tissue damage in arthritis. This myeloid skewing was cell intrinsic, as arthritic HSPPCs up-regulate myeloid-specific transcripts including S100a8. Exogenous S100a8 promoted myeloid cell output from wild-type HSPPCs, suggesting mechanistic involvement of this gene in the myeloid priming that occurs in arthritic HSPPCs. Therefore, our results indicate that in arthritic mice, HSPPCs adopt a pathologic state that favors disease persistence.
Introduction
Mature hematopoietic cells including cells of the innate and adaptive immune systems are derived from a small pool of progenitors that reside in the bone marrow. The most primitive compartment of this progenitor pool (herein, hematopoietic stem and primitive progenitor cells or HSPPCs) consists of self-renewing and non–self-renewing multipotential progenitors. These uncommitted HSPPCs differentiate into committed progenitors: granulocyte monocyte progenitors (GMPs), megakaryocyte erythrocyte progenitors (MEPs), and common lymphoid progenitors (CLPs).
In addition to homeostatically replenishing mature hematopoietic cells as these cells die, hematopoietic progenitors respond to various non–steady-state demands. Infectious or inflammatory states, for example, increase the demand for myeloid cells. Although infection and inflammation have classically been studied at the mature cell level, recent studies have indicated progenitor involvement as well. For example, inflammation induced by alum immunization or infection with Candida albicans increases GMP numbers or up-regulates C/EBPβ expression in these cells, which consequently promotes myeloid output.1,2
What is still unclear though is whether HSPPCs, the uncommitted precursors of committed progenitors, are actively involved in infectious and inflammatory states or whether they participate in the myeloid skewing associated with these states. HSPPCs, especially self-renewing hematopoietic stem cells (HSCs), are thought to be relatively protected from inflammatory signals by cell-intrinsic mechanisms like quiescence and cell-extrinsic mechanisms like presence of proximal regulatory T cells.3,4 Nevertheless, recent studies have documented that in vivo inflammatory insults including those of microbial origin may lead to HSPPC proliferation, expansion, and functional exhaustion.5 However, whether differentiation potential of these cells is also affected by inflammation has not been adequately addressed.
Examination of differentiation potential is key to understanding involvement of HSPPCs in disease. While it is easy to understand how expansion or proliferation of committed progenitors such as GMPs will consequently lead to increase in disease-relevant myeloid cells, it is not clear that expansion of HSPPCs, which are uncommitted, should lead to such specific mature cell outcome. Other lineages such as erythroid and lymphoid cells should increase as well, unless there are attendant mechanisms that skew differentiation potential of the HSPPCs. Furthermore, proliferation/expansion of HSPPCs is not specific to infection/inflammation as it is also seen in noninflammatory/noninfectious conditions such as blood loss or hypoxia where erythrocyte rather than myeloid cell demand increases.6,7 This indicates that HSPPC proliferation/expansion may be a nonspecific “knee-jerk” response to stress rather than a specific immune response. Therefore, unless the differentiation potential of HSPPCs is assessed prospectively, the question of whether inflammation-associated myeloid skewing occurs at this primitive level or only in more committed progenitors cannot be answered.
We have studied alterations in HSPPC homeostasis in inflammatory conditions using a mouse autoimmune arthritis model (KRNxG7).8,9 This mouse disease shares several clinical, histologic, and molecular features with human rheumatoid arthritis (RA).10 Although the arthritis is initiated by autoantibodies,11 myeloid cells are the direct effectors mediating joint and systemic pathologies of the disease. Not surprisingly, we previously documented a systemic increase in myeloid (Gr1+ or Mac1+) cells in all tissues of arthritic mice analyzed.9 Using an anti-Gr1–depleting antibody, Wipke and Allen have shown that myeloid cells are required for the joint inflammation (swelling) in arthritis.12 Subsequent studies have corroborated the indispensable role of neutrophils and monocyte lineages in arthritic joint inflammation.13-16 Lastly, osteoclasts, which are also of myeloid origin, are indispensable for bone erosions because they are the sole cells capable of bone resorption. In the absence of osteoclasts, joint swelling can proceed but joint bone erosions are spared in various arthritis models.17,18
In this study, we elucidate the developmental origins of the increased myeloid cells we previously observed in this KRNxG7 arthritic model and, in the process, uncover molecular changes at the HSC level that are induced by arthritis and prime these cells toward myeloid development.
Methods
Animals and arthritis induction
Generation of KRNxG7 arthritic mice and B6xG7, G7, or KRN controls has been previously described.9 For competitive experiments, we generated CD45 congenic B6xG7 or KRNxG7 by crossings involving C57BL/6, G7, and B6.SJL (CD45.1) and KRN transgenic mice. Serum transfer arthritis19 was induced by intraperitoneal injection with 250 μL of K/BxN arthritogenic serum with or without 200 μL of serum on day 2.
HSPPC sorting
Bone marrow cells were lineage depleted using MACS beads (Miltenyi Biotec), stained with fluorescent-conjugated antibodies including lineage marker antibodies and fluorescence-activated cell sorted to high purity using a MoFlo (DAKO) or FACSAria (BD Biosciences). Aberrant up-regulation of Sca1 surface marker on progenitors, which occurs in some inflammatory states and could confound KSL and other HSPPC analysis, did not occur in arthritic bone marrow (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
In vitro stromal cell–free competitive culture
Sorted KSL cells from arthritic and control mice were mixed at a 1:1 ratio (5000 cells each) and cultured in 24-well tissue-culture plates (TPP). Culture media consisted of StemSpan serum-free base medium (StemCell Technologies), 10% serum (Hyclone), KitL (1% supernatant), and Flt3L (PeproTech or e-Bioscience). Cells were cultured for 3.5-4 days. The experiment was performed in 2 sets (N = 3 in each set). Where indicated, Escherichia coli LPS (Sigma-Aldrich) was added to the culture.
Differentiation on OP9 cells
Sorted cells (1000-3000) were cultured on irradiated OP9 cells in 6-well or 24-well plates. Cells were grown in αMEM with 10% serum, KitL (1% of supernatant), 10 ng/mL IL-7 (PeproTech), and 20 ng/mL Flt3L (PeproTech). Analysis was performed after 5-8 days of culture. For competitive cultures, arthritic and control KSL cells were mixed at a 1:1 ratio before culture.
Osteoclast in vitro assay
Two thousand sorted cells were cultured noncompetitively in 96-well flat-bottom plates containing 200 μL of osteoclast-culturing media (α-MEM with 10% serum, 10% CMG supernatant [containing M-CSF] and 100 ng/mL RANKL). Plates were fixed on days 5-9 with 4% paraformaldehyde and stained with a histochemical TRAP staining kit (Sigma-Aldrich). Imaging was performed at room temperature with a Nikon Eclipse e400 microscope equipped with a PlanFluor lens and an optronics camera using Magnafire software (Meyer).
Transplantation
Recipient mice were lethally irradiated (10 Gy; single dose) a day before cell transplantation. For competitive transplantation, cell suspensions from 2 competing strains were mixed at the appropriate concentrations and injected in a 200-μL volume of HBSS (N = 4 recipients were analyzed at every time point with the exception of the 26-week time point for old recipients [N = 3]). All mice experiments were approved by the institutional animal care and use committee of the Washington University School of Medicine.
Quantitative real-time PCR
KSL cells (10 000-20 000) or CD150+CD48−CD34−KSL cells (4000-7000) sorted directly into TRIzol (Invitrogen) from 3 independent arthritic and age-matched control pools were used (12 independent pools in total). Standard TRIzol RNA extraction according to the manufacturer's instruction with inclusion of linear acrylamide (Ambion) as a carrier was performed. DNAse digestion, cDNA synthesis, and quantitative real-time PCR were done as described previously.9 Quantitative real-time PCR primers used in this study are listed in supplemental Table 3.
Murine S100A8 protein generation
Murine S100a8 gene C-terminally fused to the His-tag was cloned into pET21-a vector and overexpressed in BL21 (DE3) cells (Novagen). The recombinant S100a8 was purified from cell lysate using the arginine oxidative refolding method,20 followed by size-exclusion chromatography. The final protein was dialyzed into buffer containing 25mM HEPES (pH 7.5), 1mM CaCl2, and 150mM NaCl, concentrated (Amicon-Ultra; Millipore), 0.22-μm sterile filtered and quantified (BCA protein assay kit; Thermo Scientific).
Statistics
All tests of significance were performed using the Student t test.
Supplemental information
Additional information on the methods used in this study can be found in supplemental Methods.
Results
Arthritic KSL cells have increased myeloid potential in vitro
We determined whether the increased myeloid cells in arthritic mice9 could be traced to changes at the uncommitted HSPPC level. Kit+Sca1+Lin− (KSL) cells contain all HSPPCs. Our initial experiments indicated that this KSL phenotype identified a similar population of primitive cells in arthritic and control mice. Specifically, when KSL cells were cultured in clonal methylcellulose media replete with myelopoietic and/or erythropoietic cytokines, arthritic KSL cells formed colonies of similar number and size as control KSL cells did (supplemental Figure 1B-C). Furthermore, in noncompetitive culture on OP9 cells with lymphopoietic cytokines, KSL cells from arthritic and control mice generated similar cell output with similar B-cell frequency (supplemental Figure 1D). Importantly, we confirmed that arthritic KSL cells, like wild-type KSL cells, behaved very differently from committed myeloid progenitors (GMPs), which formed fewer and smaller colonies in methylcellulose culture and completely failed to thrive in lymphopoietic OP9 cultures (supplemental Figure 1B-D). Collectively, KSL cells from arthritic mice, like wild-type KSL cells, contained multipotential progenitors with similar capacity for multilineage differentiation, distinct from committed progenitors like GMPs.
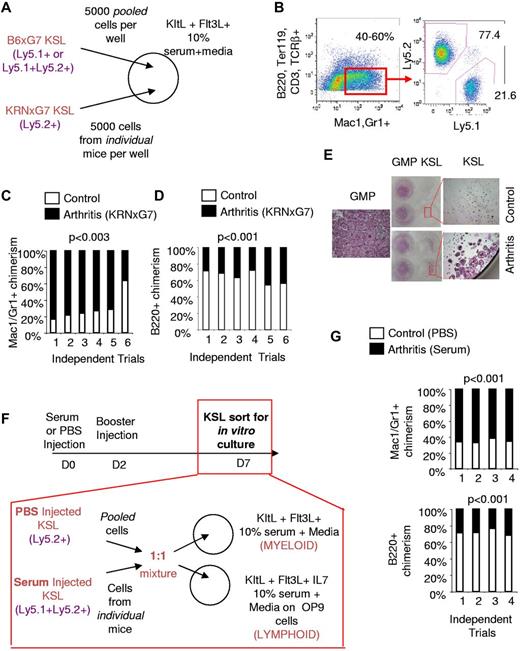
To directly compare the myeloid potential of arthritic and control KSL cells, we set up competitive in vitro cultures with limited cytokines. KSL cells sorted from KRNxG7 (arthritic) and B6xG7 (control) mice with different CD45 (Ly5) alleles were cultured in 1:1 ratio in stromal cell-free liquid culture without differentiation cytokines but with Kit ligand (KitL) and Flt3 ligand (Flt3L) to promote viability21,22 (Figure 1A). Under this permissive condition there was up to a 13-fold expansion of cells after 3-4 days of culture with approximately 20% of cells still having an immature Kit+Lin− phenotype (data not shown). More importantly, approximately 40%-60% of all cells generated were Mac1/Gr1+ but negative for other mature lineage markers (B220, CD3, TCRβ, Ter119) and, therefore, were myeloid cells (Figure 1B). Consistent with a greater myeloid potential of arthritic KSL cells, myeloid cells derived from arthritic KSL cells were approximately 3-fold more prevalent than those derived from control KSL cells (Figure 1B-C). In contrast, we found that when KSL cells from arthritic and control mice were cultured competitively on OP9 cells in the presence of lymphopoietic cytokines, B220+Gr1− (or B220+Mac1−CD11c−) lymphoid cells produced were preferentially (on average ∼ 2-fold) derived from control KSL cells (Figure 1D). Therefore, arthritic KSL cells, despite having similar multilineage differentiation capacity (supplemental Figure 1), have a specific higher propensity toward myeloid development (Figure 1A-D).
Arthritic KSL cells have increased in vitro myeloid potential. (A) Experimental scheme for in vitro competitive culture of arthritic and control KSL cells. Arthritic and control mice have different CD45 (Ly5) alleles. Arthritic and control KSL cells (5000 each) were mixed in the same well of a 24-well plate with survival cytokines, KitL+, Flt3L, and media containing 10% serum. (B left panel) FACS plot depicting myeloid differentiation of KSL cells in vitro after 3.5 days of culture. Myeloid cells (red gate) are Mac1+ and/or Gr1+ and negative for B-cell (B220), T-cell (CD3, TCRβ), and erythrocyte (Ter119) markers. Approximately 40%-60% of cells were present in this gate. (Right panel) FACS plot depicting analysis to determine arthritic KSL-derived (Ly5.2+) and control KSL-derived (Ly5.1+) myeloid cells. (C) Summary of in vitro competitive culture results. Each bar represents outcome of KSL cells from 1 independent arthritic mouse competed with control KSL cells. In 5 of 6 trials, arthritic KSL cells (black) contributed greater to myeloid cell output. (D) Outcome of culture of arthritic and control KSL cells on irradiated OP9 cells and media containing 10% serum, KitL, Flt3L, IL-7 to promote lymphoid cell formation. To ensure that lymphoid cells were being analyzed, B220+ cells lacking Gr1 or lacking Mac1 and CD11c were analyzed. Control and arthritic KSL-derived cells were distinguished based on CD45 (Ly5) allele expression. In 6 of 6 trials, arthritic KSL cells were inferior to control KSL cells in lymphoid cell output. (E) Tartrate-resistant acid phosphatase (TRAP) stained wells at least 6 days after culturing sorted KSL and GMPs (Kit+Sca1−Lin−CD34+FcγRIII/IIbhi) with M-CSF and RANKL to promote osteoclastogenesis. Low-magnification camera images and high-magnification (40×; 4× objective, 10× eyepiece) microscopy images of wells are shown. TRAP+ cells are pink/purple. The few TRAP+ cells in wells seeded with control KSL cells are not multinucleate unlike arthritic KSL- and GMP-derived cells. The single high-magnification image labeled GMP is representative of osteoclastogenesis from arthritic or control GMPs which gave similar results. (F) Experimental scheme for arthritis induction in naive mice (B6xG7) by injection of serum. Control mice were injected with PBS. Serum injection was administered on day 0 (D0) and day 2 and KSL cells sorted for competitive in vitro stromal cell–free and OP9 culture with indicated cytokines on day 7. (G) Relative contribution of KSL cells from arthritic (serum-injected B6xG7 mice) and control (PBS-injected B6xG7 mice) to Mac1/Gr1+ cells generated in stromal cell–free culture (top panel) or to B220+CD11c−Mac1− cells generated in OP9 culture (bottom panel). See panel F for experimental scheme. Each bar represents outcome of KSL cells from 1 independent arthritic mouse competed with control KSL cells.
Arthritic KSL cells have increased in vitro myeloid potential. (A) Experimental scheme for in vitro competitive culture of arthritic and control KSL cells. Arthritic and control mice have different CD45 (Ly5) alleles. Arthritic and control KSL cells (5000 each) were mixed in the same well of a 24-well plate with survival cytokines, KitL+, Flt3L, and media containing 10% serum. (B left panel) FACS plot depicting myeloid differentiation of KSL cells in vitro after 3.5 days of culture. Myeloid cells (red gate) are Mac1+ and/or Gr1+ and negative for B-cell (B220), T-cell (CD3, TCRβ), and erythrocyte (Ter119) markers. Approximately 40%-60% of cells were present in this gate. (Right panel) FACS plot depicting analysis to determine arthritic KSL-derived (Ly5.2+) and control KSL-derived (Ly5.1+) myeloid cells. (C) Summary of in vitro competitive culture results. Each bar represents outcome of KSL cells from 1 independent arthritic mouse competed with control KSL cells. In 5 of 6 trials, arthritic KSL cells (black) contributed greater to myeloid cell output. (D) Outcome of culture of arthritic and control KSL cells on irradiated OP9 cells and media containing 10% serum, KitL, Flt3L, IL-7 to promote lymphoid cell formation. To ensure that lymphoid cells were being analyzed, B220+ cells lacking Gr1 or lacking Mac1 and CD11c were analyzed. Control and arthritic KSL-derived cells were distinguished based on CD45 (Ly5) allele expression. In 6 of 6 trials, arthritic KSL cells were inferior to control KSL cells in lymphoid cell output. (E) Tartrate-resistant acid phosphatase (TRAP) stained wells at least 6 days after culturing sorted KSL and GMPs (Kit+Sca1−Lin−CD34+FcγRIII/IIbhi) with M-CSF and RANKL to promote osteoclastogenesis. Low-magnification camera images and high-magnification (40×; 4× objective, 10× eyepiece) microscopy images of wells are shown. TRAP+ cells are pink/purple. The few TRAP+ cells in wells seeded with control KSL cells are not multinucleate unlike arthritic KSL- and GMP-derived cells. The single high-magnification image labeled GMP is representative of osteoclastogenesis from arthritic or control GMPs which gave similar results. (F) Experimental scheme for arthritis induction in naive mice (B6xG7) by injection of serum. Control mice were injected with PBS. Serum injection was administered on day 0 (D0) and day 2 and KSL cells sorted for competitive in vitro stromal cell–free and OP9 culture with indicated cytokines on day 7. (G) Relative contribution of KSL cells from arthritic (serum-injected B6xG7 mice) and control (PBS-injected B6xG7 mice) to Mac1/Gr1+ cells generated in stromal cell–free culture (top panel) or to B220+CD11c−Mac1− cells generated in OP9 culture (bottom panel). See panel F for experimental scheme. Each bar represents outcome of KSL cells from 1 independent arthritic mouse competed with control KSL cells.
To confirm that the myeloid potential of arthritic KSL cells is indeed increased, we also examined the ability of KSL cells to generate osteoclasts, a specific myeloid lineage cell which mediates bone destruction at arthritic joints. Previous studies have shown that primitive progenitors fail to generate osteoclasts in vitro without prior activation of the progenitors such as with IL-3.23 Consistent with this report, sorted KSL cells from control (B6xG7) mice largely failed to appreciably generate osteoclasts (Tartrate resistant acid phosphatase [TRAP]+ multinucleate cells) when cultured with typical osteoclastogenic cytokines—M-CSF and RANKL only—without prior activation (Figure 1E). Under these same conditions, however, arthritic KSL-containing wells displayed osteoclast potential approximately 80% (15 of 19) of the time. On the rare occasion that control KSL cells formed osteoclasts, these cells were invariably smaller and/or fewer than those formed from arthritic KSL cells (Figure 1E and data not shown). Taken together, arthritic KSL cells have increased in vitro myeloid, including osteoclast, generation potential.
To determine whether myeloid skewing of arthritic KSL cells is triggered by the disease state, we transferred acute arthritic disease to wild-type mice as previously reported.19 Specifically, we induced transient arthritis in wild-type (B6xG7) mice by injecting them with serum derived from chronic arthritic mice and sorted KSL cells from these mice during robust disease (7 days after injection; Figure 1F). These cells were then cultured competitively in stromal cell free and on OP9 cells with KSL cells from CD45 congenic B6xG7 mice that only received PBS (Figure 1F). In multiple trials, KSL cells from serum-transferred mice preferentially contributed to Mac1/Gr1+ myeloid output (Figure 1G) while the very same KSL cells contributed less to B220+CD11c−Mac1− lymphoid output (Figure 1G). This supports the notion that the increased in vitro myeloid and decreased lymphoid potential are indeed arthritis dependent.
Arthritic KSL cells including long term HSCs have increased myeloid potential in vivo
The increased in vitro myeloid potential of arthritic KSL cells correlates not only with a systemic in vivo increase in Gr1+ myeloid cells,9 but also with a developmental skewing in favor of myeloid precursors in arthritic bone marrow (supplemental Figure 2). At the committed progenitor level, GMPs are increased relative to MEPs (supplemental Figure 2A) and CLPs.9 Downstream of these committed progenitors, immature Gr1lo myeloid cells are more prominently increased (supplemental Figure 2B). This correlative analysis was suggestive of increased myeloid development in arthritic bone marrow because of increased myeloid potential of KSL cells.
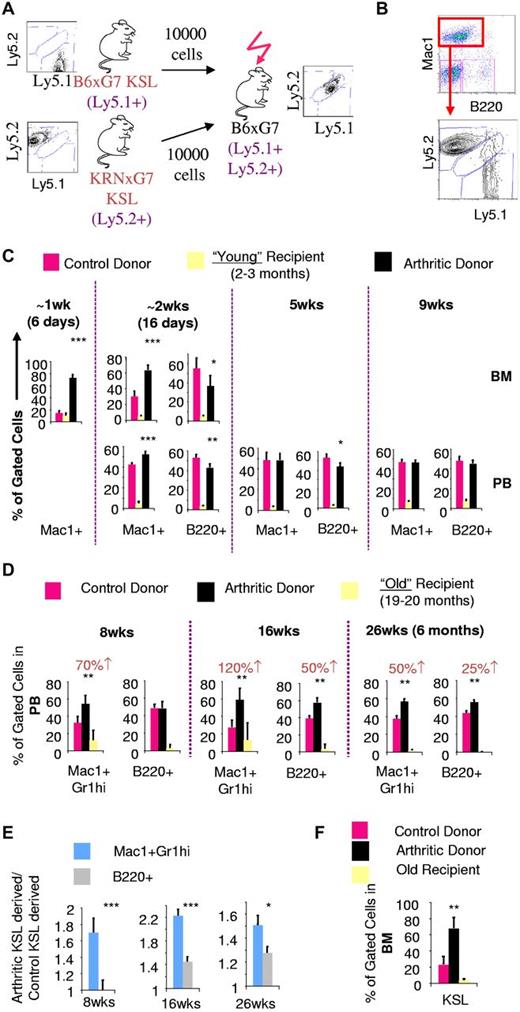
To prospectively determine whether arthritic KSL cells, with increased in vitro myeloid potential, generated increased myeloid cells in vivo, we performed competitive transplantation experiments. Equal numbers of sorted KSL cells from arthritic and control mice with different Ly5 alleles were then transplanted into lethally irradiated young (2-3 months old) recipients (Figure 2A). Myeloid and B-lymphoid cell output were tracked using Mac1 and B220, respectively. Costaining with Gr1 and CD19, respectively, confirmed the myeloid and B-lymphoid status of cells analyzed (data not shown).
Increased in vivo myeloid potential of arthritic LT-HSCs is revealed by old recipient transplantation. (A) Experimental scheme for competitive in vivo transplantation. Donors and recipients have distinct Ly5 alleles. Recipients were generated to be histocompatible with both donors and also express a different Ly5 allele. (B) FACS plot depicting analysis of Ly5 chimerism of bone marrow Mac1+ cells in recipient mice 6 days after transplantation. At an early time point (< 10 days after transplantation), peripheral blood was virtually devoid of leukocytes and hence only bone marrow analysis could be performed. (C) Quantification of arthritic and control KSL contribution to Mac1+ and B220+ cells in bone marrow (BM) and peripheral blood (PB) of young recipients based on Ly5 chimerism. (D) PB chimerism 8 weeks (wks), 16 weeks, and 26 weeks (6 months) after transplantation into old recipients gated on Mac1+Gr1hi myeloid cells or B220+ cells. Numbers in red indicate absolute percentage increase in contribution to Mac1+Gr1hi or B220+ lineages by arthritic KSL cells compared to contribution by control KSL cells. (E) Statistical analysis to compare the fold contribution to different lineages of arthritic KSL cells relative to control KSL. (F) BM KSL chimerism 26 weeks after transplantation into old recipients; *P < .05, **P < .01, ***P < .001 relative to control KSL-derived cells (see also supplemental Figure 3).
Increased in vivo myeloid potential of arthritic LT-HSCs is revealed by old recipient transplantation. (A) Experimental scheme for competitive in vivo transplantation. Donors and recipients have distinct Ly5 alleles. Recipients were generated to be histocompatible with both donors and also express a different Ly5 allele. (B) FACS plot depicting analysis of Ly5 chimerism of bone marrow Mac1+ cells in recipient mice 6 days after transplantation. At an early time point (< 10 days after transplantation), peripheral blood was virtually devoid of leukocytes and hence only bone marrow analysis could be performed. (C) Quantification of arthritic and control KSL contribution to Mac1+ and B220+ cells in bone marrow (BM) and peripheral blood (PB) of young recipients based on Ly5 chimerism. (D) PB chimerism 8 weeks (wks), 16 weeks, and 26 weeks (6 months) after transplantation into old recipients gated on Mac1+Gr1hi myeloid cells or B220+ cells. Numbers in red indicate absolute percentage increase in contribution to Mac1+Gr1hi or B220+ lineages by arthritic KSL cells compared to contribution by control KSL cells. (E) Statistical analysis to compare the fold contribution to different lineages of arthritic KSL cells relative to control KSL. (F) BM KSL chimerism 26 weeks after transplantation into old recipients; *P < .05, **P < .01, ***P < .001 relative to control KSL-derived cells (see also supplemental Figure 3).
Six days after transplantation, we found that myeloid cells were preferentially derived from arthritic KSL cells (∼ 5-fold; Figure 2B-C). Sixteen days after transplantation, myeloid cells were still preferentially derived from arthritic KSL cells although less so (∼ 2-fold). However, at 5 weeks and beyond, myeloid cells were equally derived from arthritic and control KSL cells. In contrast, B-lymphoid contribution was greater from control mice with the difference disappearing with increased time after transplantation.
The detection of preferential myeloid contribution of arthritic KSL cells only at early time points (< 5 weeks; Figure 2C) suggested that myeloid priming or arthritic KSL cells might not exist in the self-renewing long-term hematopoietic stem cell (LT-HSC) fraction. Alternatively, it was possible that LT-HSCs are indeed affected but in an environment-dependent manner. Functional outcomes because of cell-autonomous genetic modifications are easily detected several months after transplantation because genetic modifications are stable. On the other hand, because epigenetic changes including environmentally induced perturbations may be reversible, their functional outcome would only be detectable in the long-term if the change is inherently stable or if the recipient environment maintains the change. We surmised that the environment of young B6xG7 recipients could erase/reverse myeloid priming of arthritic LT-HSCs and was hence unsuitable to test myeloid priming of arthritic LT-HSCs.
Similar to arthritic mice, aged wild-type mice have an increase in myeloid cells at the expense of other lineages and this myeloid skewing is at the HSC level.24 This indicates that the environment of old mice is permissive to the sustenance and myeloid output of myeloid primed HSCs. We competitively transplanted equal numbers of arthritic (KRNxG7) and control (B6xG7) KSL cells from 2- to 3-month-old mice into old (20 months old) B6xG7 recipients (supplemental Figure 3A).
Eight weeks after transplantation, myeloid cell frequency in peripheral blood was approximately 40%, higher than myeloid cell frequency of young recipients, indicating that myeloid-sustaining environmental effects of the aged recipient is preserved despite lethal irradiation and transplantation (supplemental Figure 3B). At this time point (8 weeks), myeloid (Mac1+Gr1hi) cells in the old recipient mice were preferentially derived from arthritic KSL cells (1.7-fold) while in contrast B-lymphoid (B220+) cells were equally derived from donor arthritic and control KSL cells (Figure 2D, supplemental Figure 3C). In fact, relative contribution to Mac1+Gr1hi myeloid cells by arthritic KSL cells was still greater than relative contribution to B220+ cells at 16 weeks and 6 months after transplantation (Figure 2D-E), time points reconstituted solely by LT-HSCs.25 Therefore, LT-HSCs within the arthritic KSL cell population have greater in vivo myeloid potential than control LT-HSCs. Furthermore, these primitive progenitors from arthritic mice engraft old recipients better than control cells as evidenced by bone marrow KSL chimerism analysis 6 months after transplantation (Figure 2F), and increase in overall arthritic KSL-derived cells at 16 weeks and 6 months (Figure 2D).
KSL cells from arthritic mice up-regulate a myeloid inflammatory gene signature
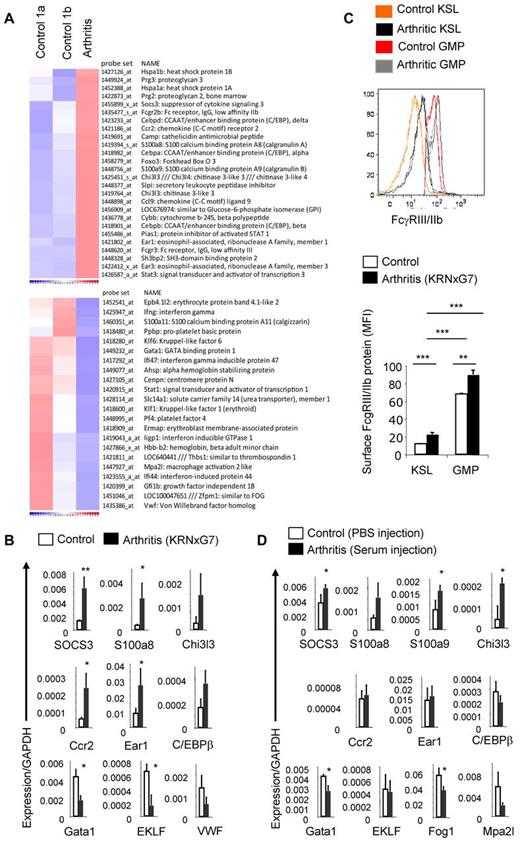
To determine the molecular basis of the increased myeloid potential of arthritic HSPPCs, we comprehensively compared the transcriptomes of KSL cells from arthritic (KRNxG7) and control (both B6xG7 and KRN) mice using mouse Affymetrix 430 2.0 microarrays. Gene ontology–based clustering analysis of enriched genes26,27 confirmed that on a global level, arthritic KSL cells were similar to KSL cells from B6xG7 and KRN control mice as well as HSC-enriched populations from previous studies (supplemental Figure 4A). In fact, of more than 45 000 probesets in the array, only approximately 700 (1%-2%) detected transcripts that were differentially regulated in arthritic KSL cells versus both control KSL cell pools. All microarray data are available at the Gene Expression Omnibus under accession no. GSE35458 (http://www.ncbi.nlm.nih.gov/geo; deposited July 2012).
We found that the C/EBP family of myeloid transcription factors—C/EBP α, β, and δ—were up-regulated while, on the other hand, key erythroid transcription factors like GATA1, EKLF (Klf1), Fog1 (Zfpm1), and Gfi1b were down-regulated in arthritic KSL cells (Figure 3A, supplemental Figure 4B). The Foxo3a transcription factor which plays a crucial role in arthritis by promoting survival of neutrophils15 was also up-regulated in arthritic KSL cells. Most notably, several myeloid-specific genes including Chi3l3, SOCS3, S100a8, and S100a9 were up-regulated in arthritic KSL cells; S100A8 and S100A9 are in fact the most abundant protein in myeloid cells.28 Expression of genes implicated in osteoclastogenesis (including Ccr2,29 Ccl9,30 and Sh3bp231 in addition to Chi3l332 and S100a833 ) was also increased (Figure 3A). On the other hand, expression of Ifnγ, which suppresses osteoclastogenesis,34 and several IFNγ-regulated genes were reduced in arthritic KSL cells while expression of Adar1, a negative regulator of interferon signaling35 was increased (Figure 3A and data not shown).
Arthritic KSL cells show molecular evidence of myeloid priming. (A) Microarray-based heat map depicting selected genes up-regulated (top) or down-regulated (bottom) in arthritic (KRNxG7) KSL cells relative to both B6xG7 (control 1a) and KRN (control 1b) KSL populations. KSL cells from 7-9 mice per strain was used. (B) Quantitative real-time PCR validation of selected differentially regulated genes. Three independent KSL pools each from KRNxG7 arthritic and B6xG7 control mice were used. KSL cells were also independent from pools used for microarray. (C) Top panel, FcγRIII/IIb protein expression on KSL cells and GMP determined by FACS. Histograms from 2 arthritic mice (KRNxG7) and 2 control mice (KRN) are shown. Bottom panel, Median fluorescence intensity (MFI) of anti-FcγRIII/IIb–PE signal on gated KSL cells and GMP as a surrogate quantification of FcγRIII/IIb surface protein level. Representative of at least 2 experiments with at least 3 arthritic and control mice per experimental set up. (D) Quantitative real-time PCR of selected genes in KSL cells from B6 mice that received PBS (control) or serum (developed arthritis). *P < .05, **P < .01, ***P < .001 (see also supplemental Figure 4).
Arthritic KSL cells show molecular evidence of myeloid priming. (A) Microarray-based heat map depicting selected genes up-regulated (top) or down-regulated (bottom) in arthritic (KRNxG7) KSL cells relative to both B6xG7 (control 1a) and KRN (control 1b) KSL populations. KSL cells from 7-9 mice per strain was used. (B) Quantitative real-time PCR validation of selected differentially regulated genes. Three independent KSL pools each from KRNxG7 arthritic and B6xG7 control mice were used. KSL cells were also independent from pools used for microarray. (C) Top panel, FcγRIII/IIb protein expression on KSL cells and GMP determined by FACS. Histograms from 2 arthritic mice (KRNxG7) and 2 control mice (KRN) are shown. Bottom panel, Median fluorescence intensity (MFI) of anti-FcγRIII/IIb–PE signal on gated KSL cells and GMP as a surrogate quantification of FcγRIII/IIb surface protein level. Representative of at least 2 experiments with at least 3 arthritic and control mice per experimental set up. (D) Quantitative real-time PCR of selected genes in KSL cells from B6 mice that received PBS (control) or serum (developed arthritis). *P < .05, **P < .01, ***P < .001 (see also supplemental Figure 4).
To validate these microarray results, we analyzed expression of 24 genes, including some up-regulated and down-regulated genes from the microarray, using quantitative real-time PCR. Of 24 genes assessed, expression of 18 was changed in a manner consistent with the microarray experiment (Figure 3B and data not shown). Notably, we were able to validate the increased expression of highly myeloid-specific genes like S100a8 and Chi3l3 as well as decreased expression of key erythropoietic transcription factors like Gata1. We also detected increased cell-surface protein expression of FcγR III/IIb on arthritic KSL cells (Figure 3C) consistent with up-regulation of FcγRIII and FcγRIIb transcripts in arthritic KSL cells from the microarray analysis (Figure 3A). During normal myelopoiesis, FcγR III/IIb cell-surface expression is up-regulated at the GMP level and is sustained in mature cells. Importantly, the level of FcγRIII/IIb cell-surface protein on arthritic KSL cells was much less than on GMPs (Figure 3C), thus implying that arthritic KSL cells are not misidentified GMPs but rather bona fide KSL cells on a path toward myeloid differentiation.
Lastly, we determined whether these gene expression changes were indeed disease dependent. We induced transient arthritis in wild-type B6 mice using the serum transfer model and determined mRNA expression of several genes in KSL cells of these mice during active disease compared with mice injected with PBS. Indeed, most of the gene expression changes observed in KRNxG7 arthritic KSL could be recapitulated in wild-type (B6) mice induced to develop arthritis (Figure 3D). Based on consistent up-regulation of the myeloid-specific genes S100a8, S100a9, Chi3l3, and SOCS3 in KSL cells from KRNxG7 arthritic mice and from serum-transferred wild-type arthritic mice, we define these genes a “myeloid inflammatory signature.” We rechecked the expression of S100a8 and S100a9 approximately 4 weeks after arthritis induction in the serum transfer model when disease had resolved/was resolving and found that expression of these genes was down-regulated at this time point (supplemental Figure 5).
In summary, a molecular program of activated myeloid genes (myeloid priming) at the expense of erythroid genes is triggered in KSL cells of arthritic mice by the arthritic environment.
Myeloid inflammatory signature genes are up-regulated in most primitive HSCs
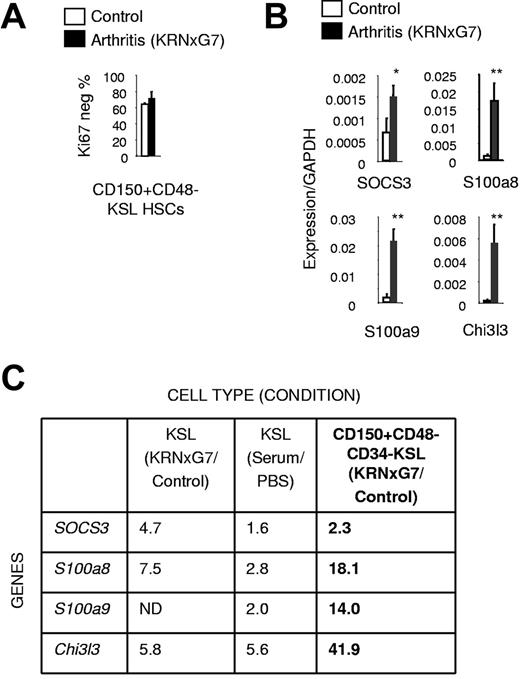
To confirm myeloid priming of LT-HSCs at the molecular level, we analyzed CD150+CD48−CD34−KSL cells representing less than 5% of KSL cells. Previous studies have shown that all LT-HSC activity is contained within this CD150+CD48−CD34− fraction of KSL cells.36,37 Specifically, 40%-50% of CD150+CD48−KSL37 and 40% of CD34−KSL cells38 have LT-HSC activity in single-cell transplantation experiments implying that CD150+CD48−CD34−KSL represents a highly enriched LT-HSC population. We initially verified that just like control HSCs,39 phenotypically defined HSCs (CD150+CD48−KSL cells) from arthritic mice were mostly quiescent with approximately 70% lacking expression of the cell-cycle marker, Ki67 (Figure 4A, supplemental Figure 6A-C). Because cell-cycle state exquisitely correlates with differentiation state of progenitors,39,40 this result indicated that the same very immature population was being identified by this surface phenotype as in control mice. We then prospectively sorted CD150+CD48−CD34−KSL cells from arthritic and control mice and performed a gene expression analysis.
Molecular myeloid priming exists in fractionated LT-HSCs from arthritic mice. (A) Quantification of Ki67-negative (quiescent) fraction of HSCs. (B) Quantitative real-time PCR of myeloid inflammatory signature genes (see Figure 3B,D) in arthritic and control HSCs (CD150+CD48−CD34−KSL IL7Rα−). (C) Table comparing fold increase in expression of myeloid inflammatory signature genes in arthritic KSL cells and HSCs relative to controls (based on Figures 3B,D, 4B); *P < .05, **P < .01 relative to control KSL or control KSL-derived cells (see also supplemental Figure 6).
Molecular myeloid priming exists in fractionated LT-HSCs from arthritic mice. (A) Quantification of Ki67-negative (quiescent) fraction of HSCs. (B) Quantitative real-time PCR of myeloid inflammatory signature genes (see Figure 3B,D) in arthritic and control HSCs (CD150+CD48−CD34−KSL IL7Rα−). (C) Table comparing fold increase in expression of myeloid inflammatory signature genes in arthritic KSL cells and HSCs relative to controls (based on Figures 3B,D, 4B); *P < .05, **P < .01 relative to control KSL or control KSL-derived cells (see also supplemental Figure 6).
We found that although the expression of several transcription factors was unchanged (supplemental Figure 6D), expression of all the myeloid genes constituting the myeloid inflammatory signature identified earlier—SOCS3, S100a8, S100a9, and Chi3l3—was increased in arthritic HSCs compared with control HSCs (Figure 4B). In fact, the magnitude of the increase at the HSC level was greater than the fold increase at the bulk KSL level (Figure 4C). Therefore, activation of a myeloid program by the arthritic environment is initiated at the HSC level.
S100a8, which is up-regulated in arthritic HSPPCs, promotes myeloid skewing
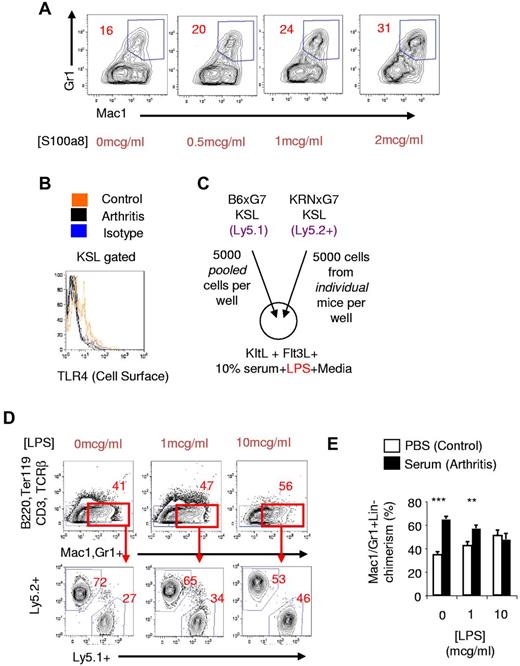
We considered the possibility that these myeloid inflammatory signature genes could be mechanistically involved in the myeloid priming of arthritic HSPPCs. In particular, S100a8, which is highly up-regulated in arthritic HSPPCs (Figures 3–4) and is abundant in the serum and joints of arthritis patients,41,42 has been shown to be an endogenous TLR4 agonist.43 This is significant because exogenous stimulation of TLR4 using LPS drives myeloid differentiation of KSL cells.21,44 Furthermore, S100a8, acting through TLR4, has been shown to promote osteoclastogenesis33 and up-regulation of FcγRIII/IIb45 ; we have shown that both phenomena occur in arthritic KSL cells (Figures 1E, 3C).
To directly determine whether S100a8 could promote myeloid development from HSPPCs, we cultured KSL cells from naive mice with recombinant murine S100a8 protein. We excluded any LPS effect by adding polymyxin B to these culture media. We found that S100a8 promoted a dose-dependent increase in myeloid cell output from naive KSL cells: a 100% increase at a dose of 2 mcg/mL S100a8 (Figure 5A). The magnitude of this effect was lower than could be attained with LPS (supplemental Figure 7A). Nevertheless, our results indicated that S100a8 could be mechanistically involved in promoting the myeloid skewed output of arthritic KSL cells.
S100a8 promotes myeloid development of KSL cells. (A) Naive KSL cells from B6 mice were cultured on OP9 cells with KitL, Flt3L, IL-7, and different concentrations of S100a8. Polymyxin B (100 μg/mL) was also added to inhibit any LPS that may be present. (B) Cell-surface TLR4 protein expression in arthritic (KRNxG7) and control KSL cells using clone UT41. Isotype control staining is shown in blue. Diminished cell-surface TLR4 protein was also seen with MTS510 clone. (C) Experimental scheme for in vitro competitive culture of arthritic and control KSL cells in the presence of LPS. (D top panel) FACS plot depicting myeloid generation (red gate) of KSL cells in response to different doses of LPS. KRNxG7 arthritic mice were used. (Bottom panel) FACS analysis to determine myeloid cells derived from arthritic KSL cells (Ly5.2+) or control KSL (Ly5.1+). (E) Relative contribution of KSL cells to Mac1/Gr1+B220−CD3−TCRβ−Ter119− cells after in vitro competitive culture of arthritic KSL cells (serum-injected B6xG7 mice) and control KSL cells (PBS-injected B6xG7 mice); **P < .01, ***P < .001.
S100a8 promotes myeloid development of KSL cells. (A) Naive KSL cells from B6 mice were cultured on OP9 cells with KitL, Flt3L, IL-7, and different concentrations of S100a8. Polymyxin B (100 μg/mL) was also added to inhibit any LPS that may be present. (B) Cell-surface TLR4 protein expression in arthritic (KRNxG7) and control KSL cells using clone UT41. Isotype control staining is shown in blue. Diminished cell-surface TLR4 protein was also seen with MTS510 clone. (C) Experimental scheme for in vitro competitive culture of arthritic and control KSL cells in the presence of LPS. (D top panel) FACS plot depicting myeloid generation (red gate) of KSL cells in response to different doses of LPS. KRNxG7 arthritic mice were used. (Bottom panel) FACS analysis to determine myeloid cells derived from arthritic KSL cells (Ly5.2+) or control KSL (Ly5.1+). (E) Relative contribution of KSL cells to Mac1/Gr1+B220−CD3−TCRβ−Ter119− cells after in vitro competitive culture of arthritic KSL cells (serum-injected B6xG7 mice) and control KSL cells (PBS-injected B6xG7 mice); **P < .01, ***P < .001.
We then assessed for in vivo evidence of S100a8 stimulation of arthritic KSL cells in vivo by examining cell-surface protein expression of TLR4, a known murine S100a8 receptor.33,43,45 In vivo or in vitro TLR4 stimulation leads to diminution of the cell-surface TLR4 protein signal by receptor internalization or altered conformation.21,46,47 We found that while TLR4 was detectable on KSL cells from nonarthritic mice as previously reported,21 the TLR4 signal on the surface of KRNxG7 arthritic KSL cells was reduced to close to background levels (Figure 5B).
To confirm that functional TLR4 was diminished on the surface of arthritic KSL cells, we competitively cultured sorted KSL cells from KRNxG7 arthritic mice and CD45 congenic B6xG7 mice in the presence of LPS (Figure 5C). We found that while arthritic KSL cells, generated almost 3-fold more myeloid cells than control KSL cells did in the absence of LPS, the myeloid contribution from arthritic and control KSL cells virtually equalized in the presence of 10 μg/mL LPS (Figure 5D). Therefore, arthritic KSL cells exhibited diminished responsiveness to de novo in vitro LPS. This refractoriness to ex vivo LPS stimulation could be established in KSL cells from wild type (B6xG7) mice induced to develop arthritis by arthritogenic serum transfer (Figure 5E).
Therefore, even though the KRNxG7 and serum transfer arthritis models do not involve LPS administration, KSL cells from these mice exhibit diminished cell-surface TLR4 and are consequently refractory to ex vivo de novo LPS stimulation. This suggests endogenous stimulation of TLR4, which is consistent with increased activity of S100a8 in vivo, at the KSL cell surface.
Discussion
Recent studies have explored the impact of individual cytokines and inflammatory molecules on the homeostasis of HSPPCs. However, the extrapolation of some of these findings to actual disease has been called into question. For example, although the Gram-negative bacterial antigen LPS induces expansion of HSPPCs in a TLR4-dependent manner,21,48 HSPPC expansion induced by actual Gram-negative bacterial infection of mice is independent of TLR signaling.48 Disease phenomena are usually more complex and difficult to predict because they involve multiple cytokines and molecules that individually may have opposing, redundant, additive, or synergistic effects. For instance, while treatment of mice with several individual cytokines triggers changes in cell cycling of HSPPCs,5 we have found that cycling of arthritic HSPPCs is unchanged relative to controls (Ma et al9 and this study). Therefore, studying HSPPCs in actual disease contexts provides the most direct means of obtaining disease-relevant changes that occur in these cells.
Using in vivo transplantation, in vitro differentiation, and gene expression analyses of sorted cells, we have shown in this study that uncommitted HSPPCs (including the most primitive HSCs from arthritic mice) are a rudimental contributor to enhanced myeloid production in arthritis. Specifically, we have shown that arthritic HSPPCs acquire molecular changes, myeloid priming, which underlie an increased myeloid potential demonstrable in in vitro and in vivo functional assays. The ability to induce arthritis in wild-type mice injected with arthritogenic serum allowed us to determine that the myeloid priming and increased myeloid potential of HSPPCs is indeed arthritis dependent.
Myeloid cells including neutrophils, monocyte lineage cells, and osteoclasts are crucial for the joint-specific inflammation and destruction as well as the systemic inflammation that occur in arthritis.12-18 Therefore, it appears there is a positive feedback loop whereby the inflammatory environment triggers myeloid priming and increased myeloid output from HSPPCs that in turn perpetuates the inflammation in a vicious cycle. This could serve as a potentially powerful mechanism for sustaining a chronic inflammatory disease. Whether myeloid-primed HSPPCs are also recruited to arthritic joint where they may generate pathologic myeloid effector cells in situ should be addressed in the future. HSPPC proliferation and differentiation potential in other non–steady-state conditions need also be examined to determine whether the HSPPC properties we found in this study can be generalized to other chronic diseases. Understanding such inflammatory stem cells would provide novel cellular targets for therapeutic intervention in chronic inflammatory diseases.
Similar to the KRNxG7 arthritic mice, physiologic aging is also associated with myeloid skewing at the expense of lymphoid and erythroid lineages. Even though this myeloid skewing in aged mice is also at the stem cell level, there are important differences between aged and KRNxG7 arthritic HSPPCs. Recent studies have suggested that normal aging is associated with the expansion of myeloid-primed HSC clones, identifiable by high expression of CD150, at the expense of lymphoid-primed and balanced HSC clones.49 However, we do not see significant differences in the level of CD150 expression between arthritic and control CD34−CD48−KSL HSCs (supplemental Figure 8). Therefore, it is unlikely that myeloid priming of arthritic HSCs is a result of expansion of particular HSC clones, like that which occurs in aging. This might also explain our finding that arthritic HSC myeloid priming is rapidly reversed after transplantation into young recipients unlike aged HSCs whose preferential myeloid output is sustained several months after transplantation.24 At the molecular level, aged Flk2−CD34−KSL cells up-regulate myeloid transcripts at the expense of lymphoid transcripts,24 while arthritic HSPPCs appear to up-regulate myeloid transcripts at the expense of erythroid transcripts. Most prominently, genes that we have demonstrated to be strikingly and consistently up-regulated in arthritic KSL cells and HSCs including S100a8, S100a9, and Chi3l3 were all down-regulated in the normal aging of stem cells.24 It will be interesting to investigate whether HSCs and primitive progenitors of RA patients also exhibit myeloid priming or if the myeloid skewing that occurs in physiologic aging partly accounts for the preferential arthritis incidence in aged individuals. Future work will also need to address the molecular and cellular mechanism by which the aged environment allows better engraftment of arthritic KSL cells than control KSL cells and sustains their preferential myeloid output.
The molecular mechanism by which myeloid priming occurs in arthritic HSPPCs is not completely clear at the moment. However, our data indicate that S100a8 could be involved. S100a8 and its stabilizing heterodimer S100a9 were consistently up-regulated in arthritic mice KSL cells and even more prominently at the HSC level. Furthermore, recombinant murine S100a8 protein promoted increased myeloid output from naive KSL cells in vitro. In vivo, arthritic KSL cells have down-regulated TLR4 expression on the cell surface, reminiscent of myeloid-skewing LPS stimulation of HSPPCs,21 and are rendered resistant to ex vivo LPS stimulation. In addition, we have shown that arthritic KSL cells have increased osteoclastogenic activity and increased cell-surface FcγRIII/IIb, both previously described effects of S100a8 on less primitive hematopoietic cells.33,45 Therefore, it is possible that endogenous S100a8 produced by arthritic HSCs promote the myeloid priming of these cells in an autocrine manner. This might occur through TLR4 but could also occur via RAGE, the other signaling receptor for S100a8. Interestingly, it has been shown that another endogenous TLR4 agonist, HMGB1, could induce LPS tolerance of monocyte lineage cells in a RAGE-dependent manner.50 Therefore, there is intersection between RAGE and TLR4 pathways downstream of the S100a8 signaling. Future work to elucidate the molecular mechanism of arthritic HSC myeloid priming should address questions such as: which receptor—TLR4 or RAGE—is required specifically for the functional outcome of increased myeloid output, the role of other up-regulated genes such as S100a9, and the molecular factors in the arthritic environment that induce myeloid priming in arthritic HSCs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Changwon Park, Yunglin Ma, Wei Zhou, Deborah Novack, Paul Allen, and Steven Teitelbaum for materials or technical advice, and the Alvin J. Siteman Cancer Center for cell sorting and gene expression analysis.
This work was supported by National Institutes of Health grants HL55337 and HL63736 (K.C.). The Alvin J. Siteman Cancer Center is supported by National Cancer Institute grant P30 CA91842.
K.A.O.J. is the recipient of American Heart Association predoctoral fellowship 09PRE2300025. F.L. is the recipient of American Heart Association postdoctoral fellowship 10POST4570022.
National Institutes of Health
Authorship
Contribution: K.A.O.J. and K.C. planned experiments, analyzed data, and wrote the manuscript; K.A.O.J., with the help of F.L., Q.T., and C.-K.K., performed experiments; O.L. and D.F. made murine S100A8 protein; and J.C.M. helped with microarray data analyses.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kyunghee Choi, Department of Pathology and Immunology, 660 South Euclid Ave, Box 8118, Washington University School of Medicine, St Louis, MO 63110; e-mail: kchoi@wustl.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal