Abstract
The European LeukemiaNet classification combines a heterogeneous group of aberrations as adverse-risk abnormalities. Our goal was to investigate the outcomes associated with distinct high-risk chromosomal abnormalities in acute myeloid leukemia (AML) after allogeneic hematopoietic stem cell transplantation (HSCT). We performed a retrospective cohort analysis in patients with high-risk AML who received first, HLA-compatible, allogeneic HSCT between January 2005 and December 2008. Data from 236 patients with a median age of 55 years were included. Because complex karyotype (CK), −5/5q−, and abnl(17p) are overlapping categories, a hierarchical classification system based on the presence or absence of abnl(17p) and −5/5q− was developed. Patients with abnl(17p) had a 2-year event-free survival (EFS) of 11% (95% confidence interval [CI], 0%-25%), patients with −5/5q− but no abnl(17p) a 2-year EFS of 29% (95% CI, 14%-44%), and patients with adverse-risk AML but neither of the 2 marker lesions a 2-year EFS of 49% (95% CI, 39%-59%). Notably, complex and monosomal karyotypes lost their prognostic value when these marker lesions were excluded. In conclusion, hierarchical classification of adverse-risk karyotypes by 2 marker lesions, abnl(17p) and −5/5q−, is effective in prognostication of the outcome of allogeneic HSCT in AML.
Introduction
As a result of more than a decade of clinical research and multiple large clinical trials, allogeneic hematopoietic cell transplantation (HSCT) has become established as the first-line consolidation therapy in acute myeloid leukemia (AML) defined by adverse-risk cytogenetic abnormalities.1-3 To date, this recommendation is valid without exception for all adverse-risk karyotypes.
Since the first cytogenetic classifications, modifications of the high-risk category have been proposed and accepted. Isolated trisomy 8, formerly considered a high-risk abnormality, failed to bear out its negative prognosis in larger series of patients.4 The classification of AML with translocations of chromosome band 11q23 has recently been refined depending on the fusion partner of the MLL gene.5 AML with t(9;11)(p22;q23), MLLT3-MLL is now a unique entity, classified as intermediate-risk II according to European LeukemiaNet (ELN), whereas balanced translocations other than that involving MLLT3 are grouped together as adverse-risk abnormalities. The most recent review of the cytogenetic classification by an international expert panel on behalf of the ELN has been published in 2010 and validated in large series.6,7 The adverse-risk group comprises the recurrent lesions inv(3)(q21q26.2) or t(3;3)(q21q26.2), t(6;9)(p23;q34), t(v;11)(v;q23), monosomy 5, del(5q), monosomy 7, abnl(17p), and complex karyotype.
Further efforts have been made to improve prognostication of AML. Breems et al proposed monosomal karyotype (MK+) defined as 2 or more monosomies or a single monosomy in the presence of structural abnormalities.8 The category MK+ AML helps to identify patients at extremely high risk of treatment failure in the context of induction and consolidation chemotherapy. A variety of subsequent reports have confirmed the poor prognosis of patients with MK+ AML even after allogeneic HSCT.9 Whether the category MK should replace that of complex karyotype (CK) is the subject of an ongoing debate in AML and myelodysplastic syndromes (MDS).10 Additional efforts have been made to determine the optimal cutoff for the number of abnormalities which define a CK or to refine the definition of complexity by requiring at least 1 structural abnormality.11,12 Neither MK nor CK, however, provides information about distinct genetic lesions that could stimulate the development of safer and more efficient therapies. Yet, the identification of molecular targets is the most promising way to improve outcomes for patients with high-risk AML.
Here, we present data on the prognostic value of distinct adverse chromosomal abnormalities in patients with AML treated with allogeneic HSCT. Because the largest subgroups of adverse karyotypes, −5, del(5q), −7, abnl(17p), and CKs overlap, we developed a hierarchical classification by presence or absence of −5/5q− or abnl(17p). We show data from a large cohort of patients with adverse-risk AML, who received their first allogeneic HSCT, where this hierarchical approach outperforms the heterogeneous classification by MK or CK.
Methods
Patient population
We performed a retrospective cohort analysis within the Cooperative German Transplant Study Group on patients with high-risk AML defined by cytogenetic abnormalities according to Cancer and Leukemia Group B (CALGB) 2002 criteria.13 The study was approved by the ethics committee of the Technical University Dresden and the institutional review board. Inclusion criteria were AML diagnosed according to the current WHO criteria and first allogeneic transplantation between January 2005 and December 2008. Patients with cord-blood, haploidentical, and second allogeneic HSCT were excluded from the analysis. Quality of the cytogenetic results was reviewed centrally and 64 patients with incomplete karyotype information were eliminated from the final analysis set.
Cytogenetic classification
The cytogenetic results were classified independently by scientists from different institutions (G.G., B.S., and B.M.) according to ELN, MK as introduced by Breems et al,8 and CK. The latter class was defined here as 3 or more abnormalities. Furthermore, we categorized by distinct lesions. Besides the recurrent genetic abnormalities t(6;9), inv(3)(q21q26.2) or t(3;3)(q21;q26.2), t(9;11)(p22;q23) and t(v;11)(v;q23) we analyzed the effects of abnormalities of 17p [abbreviated as abnl(17p)], −5/5q−, monosomy 7, and trisomy 8.
Definitions
AML in patients with a documented history of myelodysplasia was considered secondary AML (sAML) while therapy-related myeloid neoplasm (t-MN) indicated a history of exposure to chemotherapy or radiation therapy. Because HSCT had frequently been delivered during aplasia, we categorized remission status before the start of the conditioning regimen by the marrow blast count irrespective of the normalization of peripheral blood counts. Physical performance and comorbidity were assessed at HSCT and scored with the Karnofsky index and the hematopoietic cell transplantation-specific comorbidity index.14 Conditioning was divided into myeloablative regimens (> 10 mg/kg busulphan, > 150 mg/m2 melphalan, or > 8 Gy total-body irradiation [TBI]), reduced-intensity conditioning (RIC; ≤ 10 mg/kg busulphan, ≤ 150 mg/m2 melphalan, or ≤ 8 Gy TBI), FLAMSA-like (fludarabine 30 mg/m2, amsacrine 100 mg/m2, cytarabine 2 g/m2 for 4 days and RIC after a rest of 3 days),15 and nonmyeloablative regimens based on 2 Gy TBI.16
Statistical analysis
All time-dependent events were calculated from the time of transplantation. The log-rank test was used for univariate comparisons of overall survival (OS) and event-free survival (EFS), while competing-event statistics were used for cumulative incidence of relapse (CIR), nonrelapse mortality (NRM), and graft-versus-host disease (GVHD).17 In view of the dismal prognosis of patients with a relapse after allogeneic HSCT, we limit this report to EFS. Data for OS are shown in Table 1. Point estimates are given with approximate 95% confidence intervals (CIs). Age, Karnofsky performance status, hematopoietic cell transplantation-specific comorbidity index, type of AML (de novo vs sAML/t-MN), marrow blast count at HSCT (< 5% vs 5%-20% vs > 20% marrow blasts), donor type (HLA-identical sibling vs 8/8 alleles [HLA-A, -B, -C, and -DRB1]–matched unrelated donor [UD] vs partially mismatched UD), sex (female-male donor-recipient pairs vs any other combination), and CMV match (dichotomized negative–negative donor-recipient pairs vs any other combination) were selected a priori as potential confounding factors and included in multivariate Cox regression analyses. Ten thousand bootstrap samples of the same set of data were analyzed to assess the stability of the estimated regression coefficients for the cytogenetic classification.18
Outcome at 2 years after HSCT by cytogenetic classification
| . | Frequency, N . | OS% (95% CI) . | EFS% (95% CI) . | CIR% (95% CI) . | NRM% (95% CI) . |
|---|---|---|---|---|---|
| All patients | |||||
| ELN intermediate 2 | 70 | 71 (61-82) | 64 (53-76) | 22 (12-32) | 14 (5-22) |
| ELN adverse | 166 | 42 (34-50) | 37 (30-45) | 40 (32-48) | 23 (16-29) |
| P* | < .001 | < .001 | .007 | .1 | |
| Intermediate risk 2 | |||||
| Trisomy 8 alone | 31 | 67 (51-84) | 63 (46-81) | 20 (5-34) | 17 (3-31) |
| t(9;11)(p22;q23) | 8 | 70 (34-100) | 50 (15-85) | 50 (12-88) | 0 (0-0) |
| Other | 31 | 75 (59-91) | 69 (53-86) | 17 (3-30) | 14 (1-27) |
| P* | .9 | .6 | .1 | .5 | |
| AML with recurrent adverse abnormalities (WHO) | |||||
| Abnl (3) | 9 | 44 (12-77) | 33 (3-64) | 56 (19-92) | 11 (0-33) |
| t(6;9) | 14 | 71 (46-95) | 63 (38-89) | 15 (0-35) | 21 (0-44) |
| t(v;11)(v;q23) | 22 | 51 (28-73) | 41 (20-63) | 44 (21-66) | 15 (0-32) |
| P* | .3 | .2 | .05 | .8 | |
| ELN adverse by CK | |||||
| CK− | 69 | 53 (41-65) | 48 (36-60) | 30 (19-41) | 22 (12-33) |
| CK+ | 97 | 35 (24-45) | 29 (19-39) | 48 (37-58) | 23 (14-32) |
| P* | .03 | .03 | .03 | 1 | |
| Subclassification of CK | |||||
| 3-4 abnl | 35 | 62 (45-78) | 55 (37-72) | 28 (12-44) | 18 (5-31) |
| 5-9 abnl | 38 | 18 (4-33) | 12 (0-26) | 59 (39-79) | 29 (12-45) |
| ≥ 10 abnl | 24 | 21 (5-37) | 17 (2-32) | 62 (42-83) | 21 (4-38) |
| P* | < .001 | < .001 | .002 | .9 | |
| ELN adverse by MK | |||||
| MK− | 101 | 51 (40-61) | 46 (36-57) | 30 (21-39) | 24 (15-33) |
| MK+ | 65 | 30 (18-41) | 24 (14-35) | 55 (42-67) | 21 (10-31) |
| P* | .002 | .002 | .001 | .6 | |
| Hierarchical lesion-specific | |||||
| No abnl(17p) and no −5/5q− | 95 | 53 (43-64) | 49 (39-59) | 31 (22-41) | 20 (11-28) |
| No abnl(17p), but −5/5q− | 41 | 30 (15-45) | 29 (15-44) | 51 (35-67) | 20 (7-33) |
| abnl(17p) | 30 | 25 (8-41) | 11 (0-25) | 53 (32-73) | 36 (17-56) |
| P* | < .001 | < .001 | .05 | .1 |
| . | Frequency, N . | OS% (95% CI) . | EFS% (95% CI) . | CIR% (95% CI) . | NRM% (95% CI) . |
|---|---|---|---|---|---|
| All patients | |||||
| ELN intermediate 2 | 70 | 71 (61-82) | 64 (53-76) | 22 (12-32) | 14 (5-22) |
| ELN adverse | 166 | 42 (34-50) | 37 (30-45) | 40 (32-48) | 23 (16-29) |
| P* | < .001 | < .001 | .007 | .1 | |
| Intermediate risk 2 | |||||
| Trisomy 8 alone | 31 | 67 (51-84) | 63 (46-81) | 20 (5-34) | 17 (3-31) |
| t(9;11)(p22;q23) | 8 | 70 (34-100) | 50 (15-85) | 50 (12-88) | 0 (0-0) |
| Other | 31 | 75 (59-91) | 69 (53-86) | 17 (3-30) | 14 (1-27) |
| P* | .9 | .6 | .1 | .5 | |
| AML with recurrent adverse abnormalities (WHO) | |||||
| Abnl (3) | 9 | 44 (12-77) | 33 (3-64) | 56 (19-92) | 11 (0-33) |
| t(6;9) | 14 | 71 (46-95) | 63 (38-89) | 15 (0-35) | 21 (0-44) |
| t(v;11)(v;q23) | 22 | 51 (28-73) | 41 (20-63) | 44 (21-66) | 15 (0-32) |
| P* | .3 | .2 | .05 | .8 | |
| ELN adverse by CK | |||||
| CK− | 69 | 53 (41-65) | 48 (36-60) | 30 (19-41) | 22 (12-33) |
| CK+ | 97 | 35 (24-45) | 29 (19-39) | 48 (37-58) | 23 (14-32) |
| P* | .03 | .03 | .03 | 1 | |
| Subclassification of CK | |||||
| 3-4 abnl | 35 | 62 (45-78) | 55 (37-72) | 28 (12-44) | 18 (5-31) |
| 5-9 abnl | 38 | 18 (4-33) | 12 (0-26) | 59 (39-79) | 29 (12-45) |
| ≥ 10 abnl | 24 | 21 (5-37) | 17 (2-32) | 62 (42-83) | 21 (4-38) |
| P* | < .001 | < .001 | .002 | .9 | |
| ELN adverse by MK | |||||
| MK− | 101 | 51 (40-61) | 46 (36-57) | 30 (21-39) | 24 (15-33) |
| MK+ | 65 | 30 (18-41) | 24 (14-35) | 55 (42-67) | 21 (10-31) |
| P* | .002 | .002 | .001 | .6 | |
| Hierarchical lesion-specific | |||||
| No abnl(17p) and no −5/5q− | 95 | 53 (43-64) | 49 (39-59) | 31 (22-41) | 20 (11-28) |
| No abnl(17p), but −5/5q− | 41 | 30 (15-45) | 29 (15-44) | 51 (35-67) | 20 (7-33) |
| abnl(17p) | 30 | 25 (8-41) | 11 (0-25) | 53 (32-73) | 36 (17-56) |
| P* | < .001 | < .001 | .05 | .1 |
HSCT indicates hematopoietic stem cell transplantation; OS, overall survival; CI, confidence interval; EFS, event-free survival; CIR, cumulative incidence of relapse; NRM, nonrelapse mortality; ELN, European LeukemiaNet; AML, acute myeloid leukemia; and abnl, abnormality.
abnl(3), inv(3)(q21q26.2) or t (3;3)(q21;q26.2); t (v;11)(v;q23) indicate balanced translocations other than involving the MLLT3 gene on 9p22. Abnl(17p) indicates abnormality of chromosome 17. CK indicates complex karyotype defined by ≥ 3 independent clonal abnormalities. MK indicates monosomal karyotype defined by the presence of 2 or more distinct autosomal chromosome monosomies in the karyotype or a single autosomal monosomy in the presence of 1 or more structural chromosomal abnormalities.
P values were calculated using the Gray test for CIR and NRM and the log-rank test for OS and EFS.
Results
Patient characteristics
Data from 236 patients were analyzed. Baseline characteristics are shown in Table 2. The median age was 55 years with a wide range of 22 to 77 years. Twenty-two patients (9%) were older than 65. One hundred twenty-two patients (52%) were in first remission with a morphologically leukemia-free marrow while the remaining patients either suffered from relapse or exposed residual marrow blast before the start of the conditioning regimen. Patients with residual marrow blasts were frequently treated with sequential salvage chemotherapy and proceeded to RIC and allogeneic HSCT in aplasia without ever having achieved a remission. The FLAMSA-RIC regimen is one such sequential regimen which was administered in 27% of patients.15,19 Sixteen percent of patients were treated with standard myeloablative conditioning regimens; 49% of patients received RIC and 8% of patients nonmyeloablative conditioning regimens. The median interval from first diagnosis or relapse to HSCT was 106 days (range, 7-328 days).
Patient characteristics
| . | All, N = 236 (100%) . | Intermediate risk II (ELN), N = 70 (30%) . | Adverse risk (ELN), no abnl(17p)/no −5/5q−, N = 95 (40%) . | Adverse riks (ELN), no abnl(17p) but −5/5q−, N = 41 (17%) . | abnl(17p), N = 30 (13%) . |
|---|---|---|---|---|---|
| Median age (at time of diagnosis), y | 55 | 55 | 53 | 58 | 55 |
| Range | 21-77 | 22-75 | 21-77 | 28-69 | 30-71 |
| De novo AML/sAML/t-MN | |||||
| De novo | 142 (60) | 46 (66) | 60 (63) | 21 (51) | 15 (50) |
| sAML | 62 (26) | 15 (21) | 19 (20) | 17 (42) | 11 (37) |
| t-MN | 32 (14) | 9 (13) | 16 (17) | 3 (7) | 4 (13) |
| Treatment status | |||||
| Firstline | 187 (79) | 53 (76) | 72 (76) | 36 (88) | 26 (87) |
| Salvage | 49 (21) | 17 (24) | 23 (24) | 5 (12) | 4 (13) |
| Monosomal karyotype | |||||
| No | 171 (72) | 70 (100) | 76 (80) | 16 (39) | 9 (30) |
| Yes | 65 (28) | 19 (20) | 25 (61) | 21 (70) | |
| Karnofsky index* | |||||
| ≥ 80% | 213 (91) | 65 (94) | 83 (87) | 38 (95) | 27 (90) |
| < 80% | 21 (9) | 4 (6) | 12 (13) | 2 (5) | 3 (10) |
| Sorror index† | |||||
| Low risk | 55 (24) | 18 (27) | 20 (21) | 9 (22) | 8 (28) |
| Intermediate risk | 64 (28) | 16 (24) | 35 (37) | 8 (20) | 5 (17) |
| High risk | 113 (49) | 33 (49) | 40 (42) | 24 (59) | 16 (55) |
| Marrow blasts prior to HSCT‡ | |||||
| < 5% blasts | 141 (61) | 42 (61) | 61 (64) | 21 (53) | 17 (59) |
| 5%-20% blasts | 44 (19) | 14 (20) | 16 (17) | 8 (20) | 6 (21) |
| > 20% blasts | 48 (21) | 13 (19) | 18 (19) | 11 (28) | 6 (21) |
| Conditioning intensity | |||||
| Myeloablative | 38 (16) | 8 (11) | 18 (19) | 7 (17) | 5 (17) |
| Reduced intensity | 115 (49) | 33 (47) | 50 (53) | 19 (46) | 13 (43) |
| FLAMSA | 63 (27) | 19 (27) | 22 (23) | 13 (32) | 9 (30) |
| Flu/2 Gy | 20 (8) | 10 (14) | 5 (5) | 2 (5) | 3 (10) |
| Donor source | |||||
| Sibling | 62 (26) | 17 (24) | 29 (31) | 10 (24) | 6 (20) |
| Matched unrelated donor, 8/8 | 116 (49) | 33 (47) | 46 (48) | 22 (54) | 15 (50) |
| Partially unrelated donor | 58 (25) | 20 (29) | 20 (21) | 9 (22) | 9 (30) |
| Sex match | |||||
| Male recipient-female donor | 34 (14) | 8 (11) | 15 (16) | 7 (17) | 4 (13) |
| Any other | 202 (86) | 62 (89) | 80 (84) | 34 (83) | 26 (87) |
| CMV status§ | |||||
| Neg-neg | 50 (22) | 13 (19) | 25 (28) | 6 (15) | 6 (20) |
| Pos recipient | 146 (64) | 43 (63) | 53 (59) | 32 (78) | 18 (60) |
| Neg recipient–pos donor | 33 (14) | 12 (18) | 12 (13) | 3 (7) | 6 (20) |
| . | All, N = 236 (100%) . | Intermediate risk II (ELN), N = 70 (30%) . | Adverse risk (ELN), no abnl(17p)/no −5/5q−, N = 95 (40%) . | Adverse riks (ELN), no abnl(17p) but −5/5q−, N = 41 (17%) . | abnl(17p), N = 30 (13%) . |
|---|---|---|---|---|---|
| Median age (at time of diagnosis), y | 55 | 55 | 53 | 58 | 55 |
| Range | 21-77 | 22-75 | 21-77 | 28-69 | 30-71 |
| De novo AML/sAML/t-MN | |||||
| De novo | 142 (60) | 46 (66) | 60 (63) | 21 (51) | 15 (50) |
| sAML | 62 (26) | 15 (21) | 19 (20) | 17 (42) | 11 (37) |
| t-MN | 32 (14) | 9 (13) | 16 (17) | 3 (7) | 4 (13) |
| Treatment status | |||||
| Firstline | 187 (79) | 53 (76) | 72 (76) | 36 (88) | 26 (87) |
| Salvage | 49 (21) | 17 (24) | 23 (24) | 5 (12) | 4 (13) |
| Monosomal karyotype | |||||
| No | 171 (72) | 70 (100) | 76 (80) | 16 (39) | 9 (30) |
| Yes | 65 (28) | 19 (20) | 25 (61) | 21 (70) | |
| Karnofsky index* | |||||
| ≥ 80% | 213 (91) | 65 (94) | 83 (87) | 38 (95) | 27 (90) |
| < 80% | 21 (9) | 4 (6) | 12 (13) | 2 (5) | 3 (10) |
| Sorror index† | |||||
| Low risk | 55 (24) | 18 (27) | 20 (21) | 9 (22) | 8 (28) |
| Intermediate risk | 64 (28) | 16 (24) | 35 (37) | 8 (20) | 5 (17) |
| High risk | 113 (49) | 33 (49) | 40 (42) | 24 (59) | 16 (55) |
| Marrow blasts prior to HSCT‡ | |||||
| < 5% blasts | 141 (61) | 42 (61) | 61 (64) | 21 (53) | 17 (59) |
| 5%-20% blasts | 44 (19) | 14 (20) | 16 (17) | 8 (20) | 6 (21) |
| > 20% blasts | 48 (21) | 13 (19) | 18 (19) | 11 (28) | 6 (21) |
| Conditioning intensity | |||||
| Myeloablative | 38 (16) | 8 (11) | 18 (19) | 7 (17) | 5 (17) |
| Reduced intensity | 115 (49) | 33 (47) | 50 (53) | 19 (46) | 13 (43) |
| FLAMSA | 63 (27) | 19 (27) | 22 (23) | 13 (32) | 9 (30) |
| Flu/2 Gy | 20 (8) | 10 (14) | 5 (5) | 2 (5) | 3 (10) |
| Donor source | |||||
| Sibling | 62 (26) | 17 (24) | 29 (31) | 10 (24) | 6 (20) |
| Matched unrelated donor, 8/8 | 116 (49) | 33 (47) | 46 (48) | 22 (54) | 15 (50) |
| Partially unrelated donor | 58 (25) | 20 (29) | 20 (21) | 9 (22) | 9 (30) |
| Sex match | |||||
| Male recipient-female donor | 34 (14) | 8 (11) | 15 (16) | 7 (17) | 4 (13) |
| Any other | 202 (86) | 62 (89) | 80 (84) | 34 (83) | 26 (87) |
| CMV status§ | |||||
| Neg-neg | 50 (22) | 13 (19) | 25 (28) | 6 (15) | 6 (20) |
| Pos recipient | 146 (64) | 43 (63) | 53 (59) | 32 (78) | 18 (60) |
| Neg recipient–pos donor | 33 (14) | 12 (18) | 12 (13) | 3 (7) | 6 (20) |
ELN indicates European LeukemiaNet; abnl (17p), abnormality of chromosome 17p; AML, acute myeloid leukemia; sAML, secondary AML; t-MN, treatment-related AML; HSCT, hematopoietic stem cell transplantation; FLAMSA, chemotherapy with fludarabine, amsacrine, cytarabine, and subsequent reduced-intensity conditioning; Flu, fludarabine; CMV, cytomegalovirus; pos, positive; and neg, negative.
Data on *2 patients, †4 patients, ‡3 patients, and §7 patients are missing. Patients with available information add to 100%.
Outcome of the whole cohort
At the time of analysis, 110 patients were alive with a median follow-up of 28 months (range, 8 to 61 months). Two-year probabilities of OS, EFS, CIR, and NRM are shown in Table 1. The 4-year estimates for EFS, NRM, and CIR for all patients were 37% (95% CI, 30%-46%), 25% (95% CI, 18%-33%), and 37% (95% CI, 30%-44%), respectively.
NRM at day 30 and 6 months was 5% (95% CI, 2%-7%) and 11% (95% CI, 7%-15%), respectively. While early NRM was comparatively low, the hazard of relapse was highest in the first 6 months after transplantation. The overall probability of relapse within 6 months after HSCT was 22% (95% CI, 17%-27%). The incidence of acute GVHD grade 2-4 at day 100 was 36% (95% CI, 29%-42%) and grade 3-4 had been observed in 15% (95% CI, 10%-20%) of patients. At 1 year, the cumulative incidence of chronic GVHD (limited and extensive) was 54% (95% CI, 46%-61%).
Besides cytogenetic classification (as detailed in the following section), age (P = .05) and marrow blast count before HSCT (< 5% vs ≥ 5% bone marrow blasts, P = .05) were significantly associated with the end point EFS. Univariate analyses for EFS did not show a significant influence of type of AML, treatment status, time between diagnosis and HSCT, donor type, Karnofsky or hematopoietic cell transplantation-specific comorbidity index, sex match, CMV match, conditioning intensity, or transplantation center.
Survival in defined cytogenetic subgroups
The CALGB 2002 high-risk karyotypes were reclassified according to ELN 2010 criteria. Seventy patients were classified to intermediate-risk II and 166 patients to the adverse-risk category. EFS was significantly better in the intermediate-risk compared with the adverse-risk AML group (P < .001). The major reason for treatment failure was relapse in both groups. Both relapse incidence (P = .007) and NRM (P = .135) were lower in patients with intermediate-II compared with adverse-risk AML.
Complex karyotypes.
EFS at 2 years was worse in patients with CKs compared with patients with any other adverse karyotypes (P = .028). Increasing numbers of abnormalities were associated with shorter EFS (P < .001). The dismal outcome in patients with higher numbers of abnormalities was largely due to significantly higher relapse incidences (P = .002).
Monosomal karyotype.
The karyotypes of 65 patients (28%) met the criteria of MK. Patients with MK+ AML had a significantly lower EFS at 2 years than patients with MK− AML (P = .002). However, 4-year EFS was 19% (95% CI, 11%-34%) for MK+ patients compared with 39% (95% CI, 27%-51%) for MK− patients, indicating that a subgroup of MK+ patients achieved long-term EFS after HSCT.
Lesion-specific classification.
MK and CK comprise heterogeneous groups of abnormalities. The most frequent lesions were −5/5q− in 37%, losses of chromosome 7 in 22%, and abnl(17p) in 18% of the karyotypes under investigation. Marked overlap was observed between the 3 groups. Two-year EFS was poor in patients with −5/5q− (2-year EFS, 21%; 95% CI, 9%-33%), intermediate in patients with monosomy 7 (2-year EFS, 43%; 95% CI, 27%-59%), and very poor in patients with abnl(17p) (2-year EFS, 11%; 95% CI, 0%-24%). Of note, only 6 patients had an isolated del(5q), with only 1 patient being alive and in remission 50 months after HSCT.
Hierarchical classification.
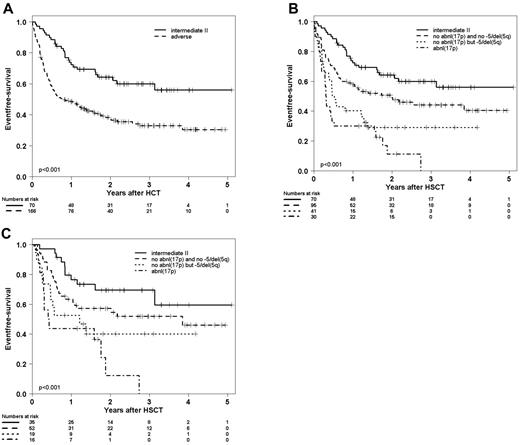
With the aim of specifying the genetic lesions conferring the poor prognoses of MK and CK, abnl(17p) and −5/5q− were subjected to further classification. To account for the overlap, karyotypes were classified in a hierarchical way. Abnl(17p) was assigned as ultra-high-risk, −5/5q− without abnl(17p) was assigned as high-risk, whereas the remaining adverse-risk ELN karyotypes were grouped together. This simple hierarchical system was used to reclassify (1) all adverse-risk karyotypes, (2) CKs, and (3) MKs. The results are shown in Figures 1 and 2. The classification led to better prediction of survival in all 3 populations.
EFS according to ELN and subclassification of adverse karyotypes. (A) EFS after HSCT of all patients according to classification into intermediate II and adverse risk. (B-C) Adverse-risk patients are classified hierarchically into 3 groups: (1) patients with abnl(17p) AML, (2) patients without abnl(17p) but with −5/5q−, and (3) patients with neither abnl(17p) nor −5/5q−. (B) EFS of all patients and (C) EFS of patients who received transplants in first remission with a morphologically leukemia-free marrow.
EFS according to ELN and subclassification of adverse karyotypes. (A) EFS after HSCT of all patients according to classification into intermediate II and adverse risk. (B-C) Adverse-risk patients are classified hierarchically into 3 groups: (1) patients with abnl(17p) AML, (2) patients without abnl(17p) but with −5/5q−, and (3) patients with neither abnl(17p) nor −5/5q−. (B) EFS of all patients and (C) EFS of patients who received transplants in first remission with a morphologically leukemia-free marrow.
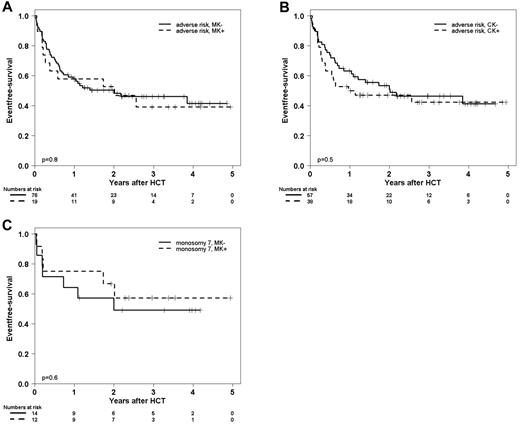
EFS in patients with adverse-risk karyotypes after exclusion of patients with either abnl(17p) or −5/5q−. EFS in patients with adverse-risk karyotypes classified by MK, CK, or monosomy 7 after exclusion of AML with either of the 2 marker lesions abnl(17p) or −5/5q−. (A) EFS at 2 years was 53% (95% CI, 30%-76%) in 19 patients with MK+ AML compared with 48% (95% CI, 36%-60%) in 76 patients with MK− AML. (B) EFS at 2 years was 47% (95% CI, 31%-63%) in 38 patients with CK+ AML compared with 51% (95% CI, 37%-75%) in 57 patients with CK− AML. (C) EFS at 2 years was 49% (95% CI, 22%-76%) in 14 patients with MK+ AML with monosomy 7 with compared with 57% (95% CI, 28%-86%) in 12 patients with MK− AML with monosomy 7.
EFS in patients with adverse-risk karyotypes after exclusion of patients with either abnl(17p) or −5/5q−. EFS in patients with adverse-risk karyotypes classified by MK, CK, or monosomy 7 after exclusion of AML with either of the 2 marker lesions abnl(17p) or −5/5q−. (A) EFS at 2 years was 53% (95% CI, 30%-76%) in 19 patients with MK+ AML compared with 48% (95% CI, 36%-60%) in 76 patients with MK− AML. (B) EFS at 2 years was 47% (95% CI, 31%-63%) in 38 patients with CK+ AML compared with 51% (95% CI, 37%-75%) in 57 patients with CK− AML. (C) EFS at 2 years was 49% (95% CI, 22%-76%) in 14 patients with MK+ AML with monosomy 7 with compared with 57% (95% CI, 28%-86%) in 12 patients with MK− AML with monosomy 7.
Among MK+ patients, −5/5q− and/or abnl(17p) were prevalent in 46 patients (71%). Most notably, after exclusion of these patients, the EFS of the remaining MK+ patients was comparable with that of MK− patients in the ELN adverse-risk category (Figure 2A). The same was true for CK+ AML (Figure 2B). Fifty-nine patients (61%) with CK+ AML displayed either abnl(17p) or −5/5q−.
The hierarchical classification further helped to predict EFS of 37 patients with a loss of chromosome 7 (log-rank test, P = .005 for overall comparison). Most remarkably, the 26 patients with monosomy 7 but without abnl(17p) or −5/5q− had a 2-year EFS of 57% (95% CI, 37%-77%) although 12 of these patients (46%) met the criteria of a monosomal karyotype.
As shown in Table 2, the rates of sAML/t-MN among patients with abnl(17p) or −5/5q− were slightly higher though not statistically significant (P = .07), and a trend to higher age was found (Mann-Whitney U test, P = .1). To adjust for different distributions of baseline variables, multivariate Cox regression modeling was performed. Age, performance status, hematopoietic cell transplantation-specific comorbidity index, type of AML, marrow blast count at HSCT, donor type, sex match, and CMV match were included in the regression model (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). With intermediate-II AML as reference category, the model-based hazard rate was 1.7 (95% CI, 1.05-2.9) for ELN adverse-risk patients without abnl(17p) or −5/5q−, while it was 3.1 (95% CI, 1.7-5.4) for patients with −5/5q− but no abnl(17p), and 5.2 (95% CI, 2.9-9.4) for patients with abnl(17p) AML. The bootstrapping coefficients and their CIs showed minimal differences to the original coefficients (data not shown).
Abnl (17p).
The analysis of abnl(17p) AML was driven by the hypothesis that patients with this abnormality do not benefit from allogeneic HSCT. This hypothesis was raised from a first cohort analysis of 47 patients with abnl(17p) AML who had a 3-year EFS of 6% after allogeneic HSCT (B.M., J.S., K. Schäfer-Eckart, N. Schmitz, M. Hänel, W. Rösler, N. Frickhofen, H. Link, A. Neubauer, U. Schuler, U. Platzbecker, J.M.M., G.E., M.B., M. Schaich, and F. Stölzel on behalf of the Study Alliance Leukemia (SAL), manuscript submitted, May 2012). Four patients belonged to both cohorts. In the dataset under investigation here, EFS in patients with abnl(17p) AML was 11% (95% CI, 0%-25%). At last follow-up, only 4 of 30 patients were alive and in remission with a maximum follow-up of 25 months. The poor outcome was due mainly to an increased incidence of relapse, 53% (95% CI, 32%-73%) at 2 years. NRM was significantly higher in this genetic subgroup than in patients with other cytogenetic high-risk abnormalities (NRM at 2 years: 36% vs 20%, P = .032). In the subgroup of 16 patients who received transplants in first remission, EFS at 2 years was 12% (95% CI, 0%-33%).
AML with recurrent adverse genetic abnormalities (WHO).
As expected, the numbers of patients with other recurrent aberrations were very small. Nine patients displayed a recurrent lesion of chromosome 3 [inv(3)(q21q26.2) or t(3;3)(q21;q26)] genetically defined by the RPN1-EVI1 fusion gene. Deregulated EVI1 expression leads to genomic instability and has been reported to be associated with monosomy 7, as was the case in 5 of 9 patients in this dataset. At last follow-up in March 2010, 2 of 9 patients had not relapsed or died from other causes 23 and 36 months after HSCT.
Fourteen patients harbored a t(6;9)(p23;q34) in their leukemic genome. The prognosis of t(6;9) AML has been linked to a second hit in the FLT-3 gene, a case in which increasing evidence suggests a beneficial role of allogeneic HSCT. Notably, this genetic subgroup had very good EFS at 2 years (63%, 95% CI, 38%-89%).
Thirty patients had an AML with abnormalities of chromosome band 11q23. EFS at 2 years was 50% (95% CI, 15%-85%) for patients with t(9;11)(p22;q23), and 41% (95% CI, 20%-63%) for patients with t(v;11)(v;q23) (log-rank test, P = .4). Kaplan-Meier plots for EFS in patients with adverse recurrent AML are shown in supplemental Figure 2.
Discussion
Adverse cytogenetic abnormalities in patients with AML establish the indication for allogeneic HSCT. This recommendation is based on results from studies using donor versus no-donor comparisons.1-3 Studies with this design generally face the problem of unbalanced treatment arms related to donor availability. The imbalance results in small-sized control groups, which lower the power of subgroup analyses. This obstacle partly contributes to the current situation, where outcomes for specific adverse genetic lesions after allogeneic HSCT have hitherto not been analyzed.
Although this large cohort study of cytogenetically defined high-risk AML is retrospective in nature, its strengths are that the patients received transplants over a short, recent period of time, comprehensive high-resolution HLA typing was performed uniformly, and baseline characteristics including comorbidity and high-quality cytogenetic data were available. Different myeloablative and RIC regimens have been administered before HSCT including FLAMSA-RIC.15 This regimen had been developed for patients with relapsed or refractory AML and is a sequential regimen of 4 days of high-dose Ara-C–based salvage chemotherapy followed by RIC and allogeneic HSCT after 3 days of rest. Because of the tight schedule, remission assessment between salvage chemotherapy and RIC is not possible. As a result, more patients with adverse karyotypes may proceed to allogeneic HSCT. This may be the case in our cohort of patients.
The type of conditioning regimen was not associated with EFS in univariate and multivariate analyses in this cohort of adverse-risk AML patients. This finding is in line with reports from a large retrospective and recently from a first randomized controlled trial.20,21 It is therefore highly unlikely that the use of different conditioning regimens accounted for the markedly different outcome in the genetic subgroups, especially since the regimens were equally distributed. Analyses of this dataset should thus permit assessment of the efficacy of allogeneic immunotherapy in AML in genetically defined subgroups.
With the use of 2 indicator lesions, abnl(17p) and −5/5q−, we were able to describe subgroups of cytogenetically adverse-risk AML with markedly different outcomes after allogeneic HSCT, ranging from 11%-49% EFS at 2 years (P < .001). Our results challenge the common perception that allogeneic immunotherapy can overcome any adverse cytogenetic abnormality. We defined 3 subgroups by hierarchical classification: (1) patients with abnl(17p) AML, (2) patients without abnl(17p) but with −5/5q−, and (3) patients with neither of the 2 marker lesions. In the context of allogeneic HSCT, this approach outperforms classification according to CK and MK as demonstrated in Figure 2.
While this classification will most likely be subject to further refinement, we argue in favor of a more general approach to differentiate between patients with adverse karyotypes: patients who are unlikely to experience sustained benefit from allogeneic HSCT, patients with an intermediate prognosis who are most likely to benefit from prophylactic strategies to prevent relapse, and patients with a reasonably good prognosis after allogeneic HSCT despite having an adverse karyotype associated with a poor prognosis after chemotherapy.
With respect to the extremely poor outcome of abnl(17p) AML, the data presented here are supported by evidence coming from small retrospective studies. In these retrospective studies, a detrimental outcome of patients with abnl(17p) AML after allogeneic HSCT has been reported uniformly.22,23 The reason for the detrimental effect of abnl(17p) on the outcome of patients with AML after allogeneic HSCT remains to be elucidated. TP53 lesions convey increased genetic instability allowing for the development of subclones which are resistant to graft-versus-leukemia (GvL) effects. Alternatively, TP53 lesions may entail primary resistance against GvL by reduced immunogenicity.
Of note, these results do not pertain for patients with abnl(17p) in the context of favorable recurrent lesions like t(8;21), inv(16), t(16;16), and t(15;17). Patients with these favorable recurrent abnormalities were not included into this cohort. Grimwade et al reported from patients treated with ATRA and anthracycline-based chemotherapy that those with t(15;17) and abnl(17p) exposed the same favorable outcome as patients without additional lesions.11
Currently, little is known about the outcome of −5/5q− AML after allogeneic HSCT. Pfeiffer et al reported data on conditioning with FLAMSA-RIC before allogeneic HSCT in high-risk AML.24 Four-year OS in 55 patients with −5/5q− was 30%, and this lesion was associated with inferior survival in the multivariate Cox regression analysis. The poor prognosis of patients with this lesion might be related to mutations in the TP53 gene (which are reported to occur in up to 86% of the patients with newly diagnosed AML who exposed −5/5q− in a CK) and lead to a poor prognosis after allogeneic HSCT.22 In Medical Research Council AML trials, TP53 mutations were present in 44% of patients with del(5q) and 66% in patients with a monosomy 5. Data on the TP53 mutations are not available for the cohort of patients presented here. However, one may hypothesize that patients with −5/5q− without concomitant cytogenetic abnl(17p) may be divided into patients with unmutated TP53 genes who have a reasonably good prognosis and a complementary group of patients with mutated TP53 genes with an outcome equivalent to that of the patients with abnl(17p) AML. This hypothesis is currently being investigated.
Despite robust effect estimates in an attempt to validate the results internally by bootstrap technique and external evidence which supports our approach, confirmatory analyses of independent datasets are absolutely necessary, and refinement and prospective validation should be the goal before this classification system is implemented into daily practice.
In this study, the number of patients with AML and inv(3)(q21q26.2), t(3;3)(q21q26.2), t(6;9)(p23;q34), or t(v;11)(v;q23) was too small to calculate reliable outcome estimates. Yet, future work will allow for an outcome prediction for rare recurrent adverse genetic lesions in the context of allogeneic HSCT. For example, in a large retrospective European Group for Blood and Marrow Transplantation (EBMT) analysis covering 172 patients with MLL-rearranged AML, a remarkable heterogeneity of the prognosis was observed for t(v;11)(v;q23) not involving MLLT3.25 Leukemia-free survival at 2 years was 16% (95% CI, 6%-26%) in 21 patients with t(6;11) compared with 68% (95% CI, 56%-80%) in 20 patients with t(11;19).
In conclusion, a hierarchical classification of adverse karyotypes according to distinct genetic lesions with respect to allogeneic HSCT is highly attractive. Two lesions, abnl(17p) and −5/5q−, indicate groups of AML patients who fare poorly after allogeneic HSCT. Notably, when patients with these 2 poor-risk markers were eliminated from the subset of patients with CK or MK, the remaining patients have a reasonably good prognosis after allogeneic HSCT. Two-year EFS was 53% in MK+ AML and 47% in CK+ AML without abnl(17p) or −5/5q−. Classification according to specific marker lesions rather than CK or MK may therefore be helpful.
In first remission, patients with −5/5q− AML without abnl(17p) had a 2-year EFS of 40% while patients with abnl(17p) AML had a 2-year EFS of only 12%. The role of allogeneic HSCT from an HLA-compatible donor in patients with abnl(17p) AML may therefore be questioned. For patients with abnl(17p) AML, alternative immunologic treatment strategies or the development of drugs which aim at restoring the TP53 function might be attractive.
Presented in part at the American Society of Hematology Annual Meeting, Orlando, FL, December 5, 2010; and at the European Group for Blood and Marrow Transplantation Annual Meeting, Paris, France, April 5, 2011.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the data managers C. Theuser, K. Davis, and E. Dammann for their extensive work and excellent contributions to this project.
This work was supported by the DKMS (German Bone Marrow Donor Center).
Authorship
Contribution: J.M.M. and J.S. designed the study; J.M.M., G.G., B.S., B.M., M.B., and J.S. analyzed and interpreted the data; D.B., M.S., H.B., G.B., F.B., S.B., R.S., H.M., U.H., G.E., M.B., and J.S. collected the cytogenetic and clinical data; G.E. provided financial and administrative support; and all authors contributed to the writing process and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Johannes Schetelig, MD, Medizinische KIinik und Poliklinik I, Universitätsklinikum Carl Gustav Carus, Fetscherstr. 74, 01307 Dresden, Germany; e-mail: johannes.schetelig@uniklinikum-dresden.de.