Abstract
Multiple myeloma (MM) is a clonal plasma cell disorder frequently accompanied by hematopoietic impairment. We show that hematopoietic stem and progenitor cells (HSPCs), in particular megakaryocyte-erythrocyte progenitors, are diminished in the BM of MM patients. Genomic profiling of HSPC subsets revealed deregulations of signaling cascades, most notably TGFβ signaling, and pathways involved in cytoskeletal organization, migration, adhesion, and cell-cycle regulation in the patients. Functionally, proliferation, colony formation, and long-term self-renewal were impaired as a consequence of activated TGFβ signaling. In accordance, TGFβ levels in the BM extracellular fluid were elevated and mesenchymal stromal cells (MSCs) had a reduced capacity to support long-term hematopoiesis of HSPCs that completely recovered on blockade of TGFβ signaling. Furthermore, we found defective actin assembly and down-regulation of the adhesion receptor CD44 in MM HSPCs functionally reflected by impaired migration and adhesion. Still, transplantation into myeloma-free NOG mice revealed even enhanced engraftment and normal differentiation capacities of MM HSPCs, which underlines that functional impairment of HSPCs depends on MM-related microenvironmental cues and is reversible. Taken together, these data implicate that hematopoietic suppression in MM emerges from the HSPCs as a result of MM-related microenvironmental alterations.
Introduction
The hallmark of multiple myeloma (MM) is a BM infiltration by a clonal population of malignant plasma cells (PCs).1 At diagnosis, the prevalent clinical symptom is a normochromic-normocytic anemia in ∼ 75% of patients, while leukopenia and thrombocytopenia are less frequently encountered.2 The underlying mechanisms of the hematopoietic suppression are incompletely understood. Among the mechanisms unraveled is cleavage of the erythroid transcription factor GATA-1 in combination with a direct cytotoxic effect of Fas-L and TRAIL expressing MM cells on immature erythroblasts,3,4 and induction of hepcidin, the principal iron-regulatory hormone and a pathogenic factor in anemia of inflammation (AI).5 Still, marrow replacement by malignant PCs is widely considered as mechanistic rationale.2 However, suppression of normal hematopoiesis can occur in cases with a relatively low extent of malignant infiltration and does not necessarily reflect a merely physical “crowding out” of benign cells.6 It is by now well established that malignant PC infiltration induces a disruption of the BM homeostasis between the highly organized cellular and extracellular compartments.6 The release of cytokines and growth factors from the malignant clone and innocent bystander cells leads to alterations of the BM cytokine milieu that are supportive to the neoplastic cells.7,8 During this process, compounds with enzymatic activity act on the composition of the extracellular matrix,9 while activation of osteoclasts and inhibition of osteoblasts can cause osteolytic bone lesions as the disease progresses.10 Based on the enhanced understanding of the functional importance of the BM microenvironment for hematopoietic stem and progenitor cells (HSPCs) by providing, for example, structural orientation or growth factors for self-renewal and orchestrated differentiation,11,12 involvement of HSPCs in patients with MM seems likely. In this study, we performed a detailed quantitative, molecular, and functional assessment of distinct HSPC subsets in patients with de novo MM.
Methods
Patients' samples
Between July 2007 and December 2011, 71 patients with de novo MM were entered into this study. Patients were diagnosed according to the criteria of the International Myeloma Working Group13 and BM aspirates were obtained by a standard operating procedure including defined aspiration volume, heparin dose, and anatomical location of the puncture. Control samples were donated by 52 age-matched healthy volunteers. Supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) shows detailed patient characteristics. The entire investigation was approved by the institutional review board of the Heinrich-Heine-University (reference number 2832).
Flow cytometry and FACS
Lin−, CD34+, CD38− HSCs and Lin−, CD34+, CD38+ progenitor subsets including IL-3Rαlo, CD45RA− common myeloid progenitors (CMPs), IL-3Rαlo, CD45RA+ granulocyte-macrophage progenitors (GMPs), IL-3Rα−, CD45RA− megakaryocyte-erythrocyte progenitors (MEPs), and CD10+, IL-7Rα+ common lymphoid progenitors (CLP) were isolated from BM HSPCs of MM patients and healthy donors by FACS as described.14 CD3−, CD11c−, CD14−, CD20−, CD123+, CD303+, CD304+ plasmacytoid dendritic cells (pDCs), CD19+, CD38+, CD34+ pro-B cells, CD19+, CD38+, CD34− pre-B cells, CD19+, CD20+ CD38− mature B cells and CD27−, CD10+, CD38+ immature B cells were quantified by flow cytometry (FCM) within mononuclear cells (MNCs) as described.15,16
RNA isolation, amplification, and hybridization to microarrays
RNA was isolated from sorted HSPC subsets using the RNeasy pico kit and the RNase-Free DNase kit as recommended by the manufacturer (QIAGEN). Ten nanograms of total RNA were used for linear amplification with the WT-Ovation Pico RNA Amplification System (NuGen Technologies Inc). Five micrograms of cRNA were biotin-labeled using the FL-Ovation cDNA Biotin Module Version 2 (NuGen Technologies Inc). Quality control of RNA and cRNA was performed using a bioanalyzer (Agilent 2001 Biosizing; Agilent Technologies). After fragmentation, labeled cRNA of each individual sample was hybridized to Affymetrix U133 2.0 GeneChips and stained according to the manufacturer's instructions. Array data has been stored in the Gene Expression Omnibus database (www.ncbi.nlm.nih.gov/geo/; accession no. GSE24870) according to MIAME standards.
Quantification, normalization, and statistical analysis of gene expression data
Array data were normalized by the variance stabilizing method17 and summarized by median polish. To identify differential expression between HSPC subsets of patients and donors, the rank product test was used. Genes were considered to be regulated between 2 HSPC subsets if the rank product showed a false-positive rate below 0.05. Significantly regulated genes were subsequently used to extract pathways from the Biocarta database (http://pid.nci.nih.gov/browse_pathways.shtml#biocarta). To assess the relevance of the resulting pathways, the Globaltest was performed and significance was determined by Cochran Q test.18 Statistical and numerical analyses were done in the R statistical programming environment using the Bioconductor toolset (http://www.bioconductor.org). For database queries and restructuring custom scripts, the PERL scripting language were used.
Immunohistochemistry
Paraffin-mounted BM sections from MM patients and healthy donors were deparaffinized and hydrated. After rinsing 3 times in PBS, sections were immersed in 3% hydrogen peroxide for 20 minutes at room temperature. Ag unmasking solution (Vector Laboratories Inc) was used for Ag retrieval. Primary Ab staining was carried out overnight at 4°C using a 1:1000 dilution of a polyclonal rabbit phosphorylated (p)–smad2 Ab (Cell Signaling Technology) followed by staining with the VECTOR NovaRED substrate kit (Vector Laboratories Inc) and counterstaining with hematoxylin. Sections were mounted with VECTAMount permanent mounting medium (Vector Laboratories Inc) and imaged with an AxioPlan 2 microscope equipped with an AxioCam HR using a 20× objective (PlanNeofluar). p-smad2 staining was quantified by counting the total number of positively stained cells in three hot fields (area of high density of p-smad2 staining) aided by Motic Image Plus software.
Cell lysis and immunoblotting
Protein lysates of HSPCs extracted by guanidine isothiocyanate–phenol-chloroform were subjected to SDS-PAGE. Immunoblotting and band detection was performed as described.19
Semisolid clonogenic assays
Purified HSPCs were seeded in semisolid ready-to-use methylcellulose growth medium (MethoCult H4436; StemCell Technologies) according to the manufacturer's instructions.
Long-term culture-initiating cell assays
Six thousand HSPCs were sorted into 5 mL of IMDM (Sigma-Aldrich) containing 12.5% FBS, 12.5% horse serum (both StemCell Technologies), 100 IU/mL pen/strep, and L-glutamine (both Sigma-Aldrich), plated in limiting dilutions onto irradiated confluent AFT024 feeder layers subcultured in 96-well plates (Costar), and maintained with weekly medium changes. After 5 weeks, medium was replaced by clonogenic methylcellulose medium H4534 (StemCell Technologies) containing 10 IU/mL erythropoietin (NeoRecormon 1000 IU; Roche), and cells were cultured for 2 more weeks at 37°C, 5% CO2 under humidified conditions.
Proliferation and cell-cycle analysis of HSPCs
HSPCs were cultured in fully supplemented IMDM containing 20% FBS (Biochrom Ltd) and cytokines (10 ng/mL thrombopoietine [TPO] and FLT-3L, 25 ng/mL SCF) at 37°C, 5% CO2 under humidified conditions. Viable cell concentrations were determined using a Casy Cell Counter (Roche Diagnostics GmbH). Cell-cycle analysis was either performed with the FITC BrdU Flow Kit (BD Biosciences) according to the manufacturer's instructions (BD Biosciences) or by intracellular Ki67 and Hoechst 33342 staining as described.20
Immunofluorescence imaging
For immunofluorescence imaging, HSPCs were fixated on adhesive microscope slides (HistoBond; Paul Marienfeld GmbH) by cytocentrifugation at 277g for 5 minutes with a Cytospin III (Shandon). Fixated cells were stained with a murine Ab against the hyaluronic acid (HA) receptor CD44 (clone L178; BD Pharmingen) at 1:1000 dilution and an Alexa Fluor 488–conjugated secondary goat anti–mouse Ab at a 1:2000 dilution (Molecular Probes). The cell nucleus and actin filaments of the cytoskeleton cells were stained with Hoechst 33342 at a 1:5000 dilution (Molecular Probes) and phalloidin-rhodamin-complex at a 1:1000 dilution (Sigma-Aldrich), respectively. Slides were analyzed with an AxioPlan 2 epifluorescence microscope equipped with an AxioCam HR and AxioVision software (all Carl Zeiss) using a 100× objective (Plan Neofluar).
In vitro adhesion and migration assays
Chemotaxis assays were performed using a 96-transwell plate with a pore size of 5 μm (NeuroProbe Inc). Sorted HSPC subsets (2.5 × 103 cells each) in fully supplemented IMDM were added to the upper well. Chemotaxis toward 100 ng/mL human stromal-derived factor 1 (SDF-1; PeproTech GmbH) in the lower chamber was allowed to continue for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. For adhesion analysis, sorted HSPC subsets (5 × 103 cells each) were plated on HA (300 ng/mL)– and fibronectin (FN; 50 ng/mL)–coated slides in fully supplemented IMDM and incubated for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. Nonadherent cells were removed by 3 washes with PBS and adherent cells were quantified by light microscopy.
Generation of MSCs
A total of 1-3 × 107 BM MNCs were seeded in DMEM low glucose (Sigma-Aldrich) containing 30% FBS, 100 IU/mL pen/strep and L-glutamine and incubated at 37°C, 5% CO2 under humidified conditions for 14-21 days. Nonadherent cells were removed weekly and fresh medium was added. After the incubation period, colonies > 50 cells (CFU-F) were counted by light microscopy as described.21 The mesenchymal stromal cell (MSC) character of the cells was determined by FACS (≥ 95% expression of CD73, CD90, CD105 while lacking CD34 and CD45) and differentiation assays. Osteogenic differentiation of the MSCs was induced by dexamethasone (10−7M), ascorbic acid (50 μg/mL), and β-glycerolphosphate (10mM; all Sigma-Aldrich) and visualized by Alizarin Red, van Kossa, and alkaline phosphatase (ALP) staining after 14 days. For induction of adipogenic and chondrogenic differentiation, cells were plated in DMEM high glucose (Sigma-Aldrich), and the medium was supplemented with indomethacin (0.2mM), IBMX (1mM), insulin (0.1 mg/mL), and dexamethasone (10−6M or ITS + 1 (1%), dexamethasone (10−7M), L-Proline (40 μg/mL), ascorbate-2-phosphate (50 μg/mL), and TGFβ3 (10 ng/mL, all Sigma-Aldrich), respectively. Adipogenic and chondrogenic differentiation was visualized by Oil Red O or Safranin O staining, respectively, after 21 days. Images were captured with an Axiovert 25 microscope (Carl Zeiss) using the 5× objective (Zeiss CP-Achromat 5× Ph0) for native and osteogenic differentiated MSCs and the 10× objective (Zeiss CP-Achromat 10× Ph1) for chondrogenic differentiated MSCs in combination with the Spot Software (Diagnostic Instruments Inc).
ELISA
Human TGFβ1 concentrations were measured using a Platinum TGFβ1 ELISA kit as recommended by the manufacturer (eBioscience). To activate TGFβ1, samples and standards were acidified with hydrochloride (HCl) 1 hour before the assay. To determine IL-1β, TNFα, and MIP-1α concentrations the following ELISA kits were used, according to the manufacturers' instructions: IL-1β and TNFα PeliKine compact ELISA kits (Sanquin), and the MIP-1 α ELISA kit (Boster Biologic Technology).
PCR
RNA was purified using the RNeasy mini kit in combination with the RNase-Free DNase kit as recommended by the manufacturer (both QIAGEN). For quantification of mRNA levels, a StepOne Plus real-time cycler (Applied Biosystems) was used. PCRs were performed using the Power SYBR Green PCR MasterMix (Applied Biosystems) according to the manufacturer's instructions. Primer sequences are provided in supplemental Methods. For detection of TGFβ1, the QuantiTect Primer Assay HS-TGFB1_1_SG (QIAGEN) was used. Fold changes were calculated by the ΔΔCt method.
Xenotransplantation assay
For xenotransplantations, 8- to 12-week-old NOG22 mice from in-house breedings at the pathogen-free animal facility of the German Cancer Research Center (DKFZ) were sublethally irradiated with a single dose (200 cGy) and received purified human BM HSPCs in limiting dilutions (1 × 103-1 × 106 cells) by intrafemoral injections within 24 hours after irradiation. Subsequently, mice were provided with antibiotic-supplemented water for 2 weeks posttransplantation and monitored for human cell engraftments in the peripheral blood and BM by FACS. Cells were washed with staining media, blocked with rat IgG, and stained with fluorochrome-conjugated Abs against CD45, CD19, CD33, CD11b, and CD71 (eBioscience).
Statistical analyses
Parameters influencing hemoglobin (Hgb) concentration, white blood cell (WBC), and platelet (PLT) counts in MM patients were analyzed by a multiple regression model. Significant factors identified by univariate analysis (P ≤ .05) were included into the multivariate analysis. Calculations were performed using SPSS (IBM).
Results
The megakaryocyte-erythrocyte progenitors subset is particularly diminished in MM patients
The majority of patients examined in this study (82%) were anemic (< 12 g/dL in women and < 14 g/dL in men), whereas leukopenia (< 4.000/μL) and thrombocytopenia (< 150.000/μL) were observed in 15% and 23% of patients, respectively. In an attempt to identify clinical parameters associated with this hematopoietic suppression in MM, we found that Hgb concentration and PLT count of the patients inversely correlated with the degree of MM infiltration in the BM. Detailed results of this evaluation are shown in Table 1. We reasoned that the pattern of hematopoietic suppression in MM patients is reflected by the quantitative distribution of HSPCs in the BM and therefore analyzed HSPC subsets of MM patients and healthy donors by flow cytometry (Figure 1A). We found that the proportion of MEP within the HSPCs was smaller (P < .01) in MM patients. No difference was observed in the proportion of HSCs in MM patients and healthy donors. Alike, GMP did not differ proportionally. Looking at CMP, we found a slightly greater percentage in MM patients compared with healthy donors (P < .05; Figure 1B). Again, in terms of absolute cell counts, the greatest difference was observed in MEP with a ∼ 5-fold lower cell number in comparison to healthy donors (P < .001). We also found significantly lower numbers of HSCs (∼ 4-fold), CMPs (∼ 2-fold), and GMPs (∼ 3-fold) in the BM of MM patients (Figure 1C). In addition, CLPs, pro-B and pre-B cells as well as pDCs were all significantly diminished in MM patients (supplemental Figure 1).
Univariate and multivariate analyses
| Factor . | Univariate . | Multivariate . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hgb . | WBC . | PLT . | Hgb . | WBC . | PLT . | |||||||
| P . | r . | P . | r . | P . | r . | P . | r . | P . | r . | P . | r . | |
| Age, y | NS | NA | NS | NA | .01 | −0.3 | NS | NA | NS | NA | .002 | −0.4 |
| Sex | .013 | −0.3 | NS | NA | NS | NA | .001 | −0.3 | NS | NA | NS | NA |
| ISS | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA |
| BM infiltration | .001 | −0.6 | NS | NA | NS | NA | .003 | −0.3 | NS | NA | .0004 | −0.4 |
| Kidney function | .04 | NA | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA |
| Factor . | Univariate . | Multivariate . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hgb . | WBC . | PLT . | Hgb . | WBC . | PLT . | |||||||
| P . | r . | P . | r . | P . | r . | P . | r . | P . | r . | P . | r . | |
| Age, y | NS | NA | NS | NA | .01 | −0.3 | NS | NA | NS | NA | .002 | −0.4 |
| Sex | .013 | −0.3 | NS | NA | NS | NA | .001 | −0.3 | NS | NA | NS | NA |
| ISS | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA |
| BM infiltration | .001 | −0.6 | NS | NA | NS | NA | .003 | −0.3 | NS | NA | .0004 | −0.4 |
| Kidney function | .04 | NA | NS | NA | NS | NA | NS | NA | NS | NA | NS | NA |
Hgb indicates hemoglobin; WBC, white blood cell; PLT, platelet; ISS, International Staging System; NS, not significant; and NA, not applicable.
HSPC subsets are diminished in the BM of MM patients. The subset composition of HSPCs including HSC (Lin−, CD34+, CD38−), CMP (Lin−, CD34+, CD38+, IL-3αlo, CD45RA−), GMP (Lin−, CD34+, CD38+, IL-3Rαlo, CD45RA+), and MEP (Lin−, CD34+, CD38+, IL-3Rα−, CD45RA−) was determined in BM samples of 5 healthy donors and 12 MM patients. (A) Contour plots of representative experiments are shown. HSPC subset concentrations were calculated based on the number of total nucleated cells (TNC) per microliter of BM aspirate. Bar charts represent the (B) mean proportions and (C) absolute cell counts of HSPC subsets in healthy donors (□) and MM patients (■). Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001. (D) Computer simulation of HSPC subset composition in (left) healthy donors and (right) MM patients based on microarray datasets.
HSPC subsets are diminished in the BM of MM patients. The subset composition of HSPCs including HSC (Lin−, CD34+, CD38−), CMP (Lin−, CD34+, CD38+, IL-3αlo, CD45RA−), GMP (Lin−, CD34+, CD38+, IL-3Rαlo, CD45RA+), and MEP (Lin−, CD34+, CD38+, IL-3Rα−, CD45RA−) was determined in BM samples of 5 healthy donors and 12 MM patients. (A) Contour plots of representative experiments are shown. HSPC subset concentrations were calculated based on the number of total nucleated cells (TNC) per microliter of BM aspirate. Bar charts represent the (B) mean proportions and (C) absolute cell counts of HSPC subsets in healthy donors (□) and MM patients (■). Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001. (D) Computer simulation of HSPC subset composition in (left) healthy donors and (right) MM patients based on microarray datasets.
Computer simulation indicates normal hierarchical development of HSPCs in MM patients
We then asked whether the altered subset composition of HSPCs in MM patients is due to a deregulation of the developmental program in HSPC. Therefore, we developed a computational algorithm to simulate the cellular organization within the HSPCs compartment based on gene-expression profiles of HSCs, CMPs, GMPs, and MEPs (not shown) and found that the HSPC subset conformation in MM patients and healthy donors did not differ and was in accordance with the current model for hierarchical development of HSPCs (Figure 1D). A brief description of the algorithm is provided in supplemental Methods.
Colony formation and long-term self-renewal are reduced in BM HSPCs of MM patients
To assess colony-forming capacity, 2000 BM HSPCs from MM patients and healthy donors were plated into semisolid medium. The patients' cells gave rise to fewer CFU-erythroid (CFU-E) and burst-forming units-erythroid (BFU-E) reflecting a ∼ 19- and 13-fold reduction in comparison to healthy donors (P < .01 and P < .05, respectively). There was no significant difference as far as CFU granulocyte-macrophage (CFU-G, CFU-M, CFU-GM) and multipotent CFU granulocyte, erythrocyte, macrophage, megakaryocyte (CFU-GEMM) were concerned (Figure 2 A). Looking at proportional differences, CFU-E and BFU-E were ∼ 3- and 2-fold decreased (P < .01 and P < .05, respectively) in MM patients (Figure 2B). Assessment of the long-term self-renewal potential of HSPCs of MM patients and healthy donors in vitro revealed a ∼ 3-fold lower long-term culture initiating cell frequency (P < .05) among the patients' cells (Figure 2C).
Colony formation and long-term self-renewal of HSPCs is impaired in MM patients. Equal numbers of HSPCs from the BM of 4 healthy donors and 8 MM patients were seeded in semisolid ready-to-use methylcellulose growth medium, and colonies (CFU-E, BFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) were counted after 14 days. □ and ■ represent data for healthy donors and MM patients, respectively. Means of (A) absolute numbers and (B) proportions of colonies are shown. For LTC-IC assays, HSPCs from the BM of 6 healthy donors and 6 MM patients were plated in limiting dilutions (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto irradiated confluent AFT024 feeder layers and maintained in liquid medium. After 5 weeks, the liquid medium was replaced by methylcellulose medium and wells were scored for the presence of secondary colony-forming cells after 2 weeks. (C) The absolute frequency of LTC-ICs was calculated using Lda STAT software. Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01.
Colony formation and long-term self-renewal of HSPCs is impaired in MM patients. Equal numbers of HSPCs from the BM of 4 healthy donors and 8 MM patients were seeded in semisolid ready-to-use methylcellulose growth medium, and colonies (CFU-E, BFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) were counted after 14 days. □ and ■ represent data for healthy donors and MM patients, respectively. Means of (A) absolute numbers and (B) proportions of colonies are shown. For LTC-IC assays, HSPCs from the BM of 6 healthy donors and 6 MM patients were plated in limiting dilutions (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto irradiated confluent AFT024 feeder layers and maintained in liquid medium. After 5 weeks, the liquid medium was replaced by methylcellulose medium and wells were scored for the presence of secondary colony-forming cells after 2 weeks. (C) The absolute frequency of LTC-ICs was calculated using Lda STAT software. Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01.
Gene expression analysis reveals consistent molecular deregulations in distinct HSPC subsets of MM patients
To get a molecular understanding of this functional impairment, single gene and pathway-based analyses were performed. Forty-three percent of the significantly deregulated pathways were found in all 4 HSPC subsets analyzed and 83% were observed in at least 3 subsets reflecting a relatively consistent pattern within the distinct HSPC subsets. Among the deregulated pathways were those involved in cell motility, migration, adhesion, and cell-cycle regulation including cell-to-cell adhesion signaling, the role of p85 in regulation of actin organization and cell migration, the integrin signaling pathway as well as cyclins and cell-cycle regulation and cell cycle: G1/S checkpoint. Furthermore, several signaling pathways, among them TGFβ signaling, p38 MAPK signaling, and NF-κB signaling, were significantly altered (Table 2). We mapped the relative expression levels of all genes in a given pathway to identify changes within their molecular interaction and reaction networks that contribute to the deregulations. As the most prominent example, the TGFβ signaling pathway is illustrated in supplemental Figure 2. A list of the top 10 up- and down-regulated single genes is given in Table 3, including CD44 and essential components of the TGFβ signaling pathway.
Selection of consistently deregulated pathways in HSC, CMP, MEP, and GMP
| Functional group/pathway |
| Organization of the cytoskeleton, adhesion, and motility |
| Y branching of actin filaments |
| Role of PI3K subunit p85 in regulation of actin organization and cell migration |
| Role of MAL in Rho-mediated activation of srf |
| Signaling pathway from G-protein families |
| M-calpain and friends in cell motility |
| Integrin signaling pathway |
| Cell-to-cell adhesion signaling |
| Cell-cycle regulation and proliferation |
| Cell cycle: G1/S check point |
| Cell-cycle G2/M checkpoint |
| Cyclins and cell-cycle regulation |
| Influence of Ras and Rho proteins on G1 to S transition |
| Inhibition of cellular proliferation by gleevec |
| Regulation of cell-cycle progression by Plk3 |
| Cell-signaling cascades |
| AKT signaling pathway |
| ERK1/2 MAPK signaling pathway |
| Growth hormone signaling pathway |
| NF-κB signaling pathway |
| p38 MAPK signaling pathway |
| Ras signaling pathway |
| TGF beta signaling pathway |
| TNFR 1 signaling pathway |
| Miscellaneous |
| Bone remodeling |
| EPO signaling pathway |
| Hypoxia and p53 in the cardiovascular system |
| HIF in the cardiovascular system |
| Oxidative stress-induced gene expression via Nrf2 |
| Telomeres, telomerase cellular aging, and immortality |
| VEGF hypoxia and angiogenesis |
| Functional group/pathway |
| Organization of the cytoskeleton, adhesion, and motility |
| Y branching of actin filaments |
| Role of PI3K subunit p85 in regulation of actin organization and cell migration |
| Role of MAL in Rho-mediated activation of srf |
| Signaling pathway from G-protein families |
| M-calpain and friends in cell motility |
| Integrin signaling pathway |
| Cell-to-cell adhesion signaling |
| Cell-cycle regulation and proliferation |
| Cell cycle: G1/S check point |
| Cell-cycle G2/M checkpoint |
| Cyclins and cell-cycle regulation |
| Influence of Ras and Rho proteins on G1 to S transition |
| Inhibition of cellular proliferation by gleevec |
| Regulation of cell-cycle progression by Plk3 |
| Cell-signaling cascades |
| AKT signaling pathway |
| ERK1/2 MAPK signaling pathway |
| Growth hormone signaling pathway |
| NF-κB signaling pathway |
| p38 MAPK signaling pathway |
| Ras signaling pathway |
| TGF beta signaling pathway |
| TNFR 1 signaling pathway |
| Miscellaneous |
| Bone remodeling |
| EPO signaling pathway |
| Hypoxia and p53 in the cardiovascular system |
| HIF in the cardiovascular system |
| Oxidative stress-induced gene expression via Nrf2 |
| Telomeres, telomerase cellular aging, and immortality |
| VEGF hypoxia and angiogenesis |
All pathways shown were significantly deregulated in HSC, CMP, GMP, and MEP. Pathways were considered significantly deregulated with a P value < .05.
HSC indicates hematopoietic stem cell; MEP, megakaryocyte-erythroid progenitor; CMP, common myeloid progenitor; and GMP, granulocyte-monocyte progenitor.
Top 10 up- and down-regulated genes per subset
| Top 10 genes up-regulated in: . | Top 10 genes down-regulated in: . | ||||||
|---|---|---|---|---|---|---|---|
| Gene . | Description . | FC . | P . | Gene . | Description . | FC . | P . |
| HSC | |||||||
| TGFB1I1 | TGF beta 1–induced transcript 1 | 3.00 | .0001 | SKIL | SKI-like oncogene | −8.13 | .000008 |
| MEX3D | mex-3 homolog D (Caenorhabditis elegans) | 2.87 | .0004 | RAD23B | Homolog (Saccharomyces cerevisiae) | −4.46 | .0000009 |
| SOCS3 | Suppressor of cytokine signaling 3 | 2.77 | .0006 | CD44 | CD44 (Indian blood group) | −3.98 | .00001 |
| VWF | von Willebrand factor | 2.70 | .0004 | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −3.10 | .00008 |
| UCHL1 | Ubiquitn thiolesterase L1 | 2.56 | .0001 | PSPH | Phosphoserine phosphatase | −3.09 | .0005 |
| PRR16 | Proline-rich 16 | 2.47 | .000005 | ARL17P1 | ADP-ribosylation factor-like 17 pseudogene 1 | −3.00 | .0001 |
| CLU | Clusterin | 2.38 | .00005 | LRRC37A | Leucine-rich repeat contain 37A | −2.86 | .0006 |
| VANGL1 | vang-like 1 (Drosophila) | 2.37 | .0002 | RUFY2 | RUN and FYVE dom. containing 2 | −2.84 | .00003 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | 2.30 | .00007 | SMEK2 | SMEK homolog 2, suppressor of mek1 (Dictyostelium) | −2.78 | .00008 |
| FLOT2 | flotillin 2 | 2.28 | .0003 | LRP5L | Low-density lipoprotein receptor-related protein 5-like | −2.72 | .0006 |
| CMP | |||||||
| FMO5 | Flavin-containing monooxygenase 5 | 3.06 | .00001 | MME | Membrane metalloendopeptidase | −9.83 | .000008 |
| TSPAN6 | Tetraspanin 6 | 2.77 | .00004 | CD24 | CD24 molecule | −7.70 | .00001 |
| HSPB11 | Heat shock protein family B (small), member 11 | 2.66 | .00006 | AKAP12 | A kinase (PRKA) anchor protein 12 | −6.44 | .000001 |
| MAT2A | Methionine adenosyltransferase II, alpha | 2.48 | .00008 | POU2AF1 | POU class 2–associating factor 1 | −5.93 | .00003 |
| EIF1 | Eukaryotic translation initiation factor 1 | 2.40 | .00005 | RAG2 | Recombination activating gene 2 | −5.19 | .0001 |
| CFH | Complement factor H | 2.34 | .0003 | DNTT | Deoxynucleotidyltransferase, terminal | −4.27 | .0007 |
| TUBD1 | Tubulin, delta 1 | 2.22 | .0003 | RAG1 | Recombination activating gene 1 | −4.24 | .00005 |
| ADORA2B | Adenosine A2b receptor | 2.17 | .0008 | IER3 | Immediate early response 3 | −4.03 | .0006 |
| EGLN3 | egl nine homolog 3 (C elegans) | 2.12 | .00007 | BLNK | B-cell linker | −3.79 | .0007 |
| TXNIP | Thioredoxin-interacting protein | 2.11 | .0005 | S1PR1 | Sphingosine-1-phosphate receptor 1 | −3.29 | .00006 |
| GMP | |||||||
| CHRM3 | Cholinergic receptor, muscarinic 3 | 3.36 | .0004 | MTMR6 | Myotubularin-related protein 6 | −3.68 | .0003 |
| KCNK5 | Potassium channel, subfamily K, member 5 | 3.26 | .0002 | PELI1 | pellino homolog 1 (Drosophila) | −3.31 | .0001 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | 2.78 | .0002 | RAB11FIP1 | RAB11 family–interacting protein 1 (class I) | −3.22 | .00003 |
| EIF3B | Eukaryotic translation initiation factor 3, subunit B | 2.74 | .0002 | KIF1A | Kinesin family member 1A | −3.15 | .0008 |
| BCL2L11 | BCL2-like 11 (apoptosis facilitator) | 2.73 | .0006 | PTPRE | Protein tyrosine phosphatase, receptor type E | −3.00 | .0005 |
| TAL1 | T-cell acute lymphocytic leukemia 1 | 2.39 | .00009 | DUSP10 | Dual specificity phosphatase 10 | −2.88 | .0001 |
| LEPR | Leptin receptor | 2.39 | .00007 | YT healthy donors C1 | YTH domain containing 1 | −2.80 | .0003 |
| CLU | Clusterin | 2.38 | .00008 | JMJD1C | jumonji domain containing 1C | −2.79 | .00007 |
| PTPN7 | Protein tyrosine phosphatase, nonreceptor type 7 | 2.23 | .0004 | B3GALT2 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransfer., polypeptide 2 | −2.71 | .0003 |
| UBAP2L | Ubiquitin-associated protein 2-like | 2.20 | .0003 | CDC73 | cell divis. cycle 73, Paf1/RNA pol. II compl. homolog (S cerevisiae) | −2.67 | .0009 |
| MEP | |||||||
| TXNIP | Thioredoxin-interacting protein | 3.95 | .00002 | NIACR2 | Niacin receptor 2 | −5.79 | .0002 |
| TLR3 | Toll-like receptor 3 | 3.87 | .0004 | BLNK | B-cell linker | −4.55 | .0004 |
| VWF | von Willebrand factor | 3.64 | .0002 | ARL4C | ADP-ribosylation factor-like 4C | −4.09 | .00008 |
| STXBP6 | Syntaxin-binding protein 6 | 3.48 | .0004 | BCL6 | B-cell CLL/lymphoma 6 | −4.00 | .0003 |
| CCNF | Cyclin F | 3.22 | .0006 | KIF1A | Kinesin family member 1A | −3.99 | .00001 |
| DLK1 | Delta-like 1 homolog (Drosophila) | 3.16 | .0009 | HERPUD1 | Homocyst.-inducible, ER stress-induce, ubiquitin-like d. member 1 | −3.92 | .0001 |
| VEGFA | Vascular endothelial growth factor A | 3.00 | .00008 | CD44 | CD44 (Indian blood group) | −3.85 | .0001 |
| ZKSCAN1 | Zinc finger with KRAB and SCAN domains 1 | 2.94 | .00005 | DDIT3 | DNA damage–inducible transcript 3 | −3.67 | .0006 |
| MTMR3 | Myotubularin-related protein 3 | 2.91 | .0009 | RBM34 | RNA-binding motif protein 34 | −3.62 | .0001 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 2.85 | .0002 | SMEK1 | SMEK homolog 1, suppressor of mek1 (Dictyostelium) | −3.42 | .00002 |
| Top 10 genes up-regulated in: . | Top 10 genes down-regulated in: . | ||||||
|---|---|---|---|---|---|---|---|
| Gene . | Description . | FC . | P . | Gene . | Description . | FC . | P . |
| HSC | |||||||
| TGFB1I1 | TGF beta 1–induced transcript 1 | 3.00 | .0001 | SKIL | SKI-like oncogene | −8.13 | .000008 |
| MEX3D | mex-3 homolog D (Caenorhabditis elegans) | 2.87 | .0004 | RAD23B | Homolog (Saccharomyces cerevisiae) | −4.46 | .0000009 |
| SOCS3 | Suppressor of cytokine signaling 3 | 2.77 | .0006 | CD44 | CD44 (Indian blood group) | −3.98 | .00001 |
| VWF | von Willebrand factor | 2.70 | .0004 | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −3.10 | .00008 |
| UCHL1 | Ubiquitn thiolesterase L1 | 2.56 | .0001 | PSPH | Phosphoserine phosphatase | −3.09 | .0005 |
| PRR16 | Proline-rich 16 | 2.47 | .000005 | ARL17P1 | ADP-ribosylation factor-like 17 pseudogene 1 | −3.00 | .0001 |
| CLU | Clusterin | 2.38 | .00005 | LRRC37A | Leucine-rich repeat contain 37A | −2.86 | .0006 |
| VANGL1 | vang-like 1 (Drosophila) | 2.37 | .0002 | RUFY2 | RUN and FYVE dom. containing 2 | −2.84 | .00003 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | 2.30 | .00007 | SMEK2 | SMEK homolog 2, suppressor of mek1 (Dictyostelium) | −2.78 | .00008 |
| FLOT2 | flotillin 2 | 2.28 | .0003 | LRP5L | Low-density lipoprotein receptor-related protein 5-like | −2.72 | .0006 |
| CMP | |||||||
| FMO5 | Flavin-containing monooxygenase 5 | 3.06 | .00001 | MME | Membrane metalloendopeptidase | −9.83 | .000008 |
| TSPAN6 | Tetraspanin 6 | 2.77 | .00004 | CD24 | CD24 molecule | −7.70 | .00001 |
| HSPB11 | Heat shock protein family B (small), member 11 | 2.66 | .00006 | AKAP12 | A kinase (PRKA) anchor protein 12 | −6.44 | .000001 |
| MAT2A | Methionine adenosyltransferase II, alpha | 2.48 | .00008 | POU2AF1 | POU class 2–associating factor 1 | −5.93 | .00003 |
| EIF1 | Eukaryotic translation initiation factor 1 | 2.40 | .00005 | RAG2 | Recombination activating gene 2 | −5.19 | .0001 |
| CFH | Complement factor H | 2.34 | .0003 | DNTT | Deoxynucleotidyltransferase, terminal | −4.27 | .0007 |
| TUBD1 | Tubulin, delta 1 | 2.22 | .0003 | RAG1 | Recombination activating gene 1 | −4.24 | .00005 |
| ADORA2B | Adenosine A2b receptor | 2.17 | .0008 | IER3 | Immediate early response 3 | −4.03 | .0006 |
| EGLN3 | egl nine homolog 3 (C elegans) | 2.12 | .00007 | BLNK | B-cell linker | −3.79 | .0007 |
| TXNIP | Thioredoxin-interacting protein | 2.11 | .0005 | S1PR1 | Sphingosine-1-phosphate receptor 1 | −3.29 | .00006 |
| GMP | |||||||
| CHRM3 | Cholinergic receptor, muscarinic 3 | 3.36 | .0004 | MTMR6 | Myotubularin-related protein 6 | −3.68 | .0003 |
| KCNK5 | Potassium channel, subfamily K, member 5 | 3.26 | .0002 | PELI1 | pellino homolog 1 (Drosophila) | −3.31 | .0001 |
| NBL1 | Neuroblastoma, suppression of tumorigenicity 1 | 2.78 | .0002 | RAB11FIP1 | RAB11 family–interacting protein 1 (class I) | −3.22 | .00003 |
| EIF3B | Eukaryotic translation initiation factor 3, subunit B | 2.74 | .0002 | KIF1A | Kinesin family member 1A | −3.15 | .0008 |
| BCL2L11 | BCL2-like 11 (apoptosis facilitator) | 2.73 | .0006 | PTPRE | Protein tyrosine phosphatase, receptor type E | −3.00 | .0005 |
| TAL1 | T-cell acute lymphocytic leukemia 1 | 2.39 | .00009 | DUSP10 | Dual specificity phosphatase 10 | −2.88 | .0001 |
| LEPR | Leptin receptor | 2.39 | .00007 | YT healthy donors C1 | YTH domain containing 1 | −2.80 | .0003 |
| CLU | Clusterin | 2.38 | .00008 | JMJD1C | jumonji domain containing 1C | −2.79 | .00007 |
| PTPN7 | Protein tyrosine phosphatase, nonreceptor type 7 | 2.23 | .0004 | B3GALT2 | UDP-Gal:betaGlcNAc beta 1,3-galactosyltransfer., polypeptide 2 | −2.71 | .0003 |
| UBAP2L | Ubiquitin-associated protein 2-like | 2.20 | .0003 | CDC73 | cell divis. cycle 73, Paf1/RNA pol. II compl. homolog (S cerevisiae) | −2.67 | .0009 |
| MEP | |||||||
| TXNIP | Thioredoxin-interacting protein | 3.95 | .00002 | NIACR2 | Niacin receptor 2 | −5.79 | .0002 |
| TLR3 | Toll-like receptor 3 | 3.87 | .0004 | BLNK | B-cell linker | −4.55 | .0004 |
| VWF | von Willebrand factor | 3.64 | .0002 | ARL4C | ADP-ribosylation factor-like 4C | −4.09 | .00008 |
| STXBP6 | Syntaxin-binding protein 6 | 3.48 | .0004 | BCL6 | B-cell CLL/lymphoma 6 | −4.00 | .0003 |
| CCNF | Cyclin F | 3.22 | .0006 | KIF1A | Kinesin family member 1A | −3.99 | .00001 |
| DLK1 | Delta-like 1 homolog (Drosophila) | 3.16 | .0009 | HERPUD1 | Homocyst.-inducible, ER stress-induce, ubiquitin-like d. member 1 | −3.92 | .0001 |
| VEGFA | Vascular endothelial growth factor A | 3.00 | .00008 | CD44 | CD44 (Indian blood group) | −3.85 | .0001 |
| ZKSCAN1 | Zinc finger with KRAB and SCAN domains 1 | 2.94 | .00005 | DDIT3 | DNA damage–inducible transcript 3 | −3.67 | .0006 |
| MTMR3 | Myotubularin-related protein 3 | 2.91 | .0009 | RBM34 | RNA-binding motif protein 34 | −3.62 | .0001 |
| TNFSF10 | Tumor necrosis factor (ligand) superfamily, member 10 | 2.85 | .0002 | SMEK1 | SMEK homolog 1, suppressor of mek1 (Dictyostelium) | −3.42 | .00002 |
FC indicates fold change.
HSPCs of MM patients show disturbed actin polymerization and down-regulation of the HA receptor CD44
Because the transcriptional analysis of HSPC subsets suggested molecular deregulations of pathways related to cytoskeletal organization, MM patients' and healthy donors' HSPCs were stained with phalloidine to visualize the actin skeleton (Figure 3A). A significantly higher percentage of HSPCs obtained from MM patients showed defective actin organization whereas a normal actin ring structure was predominant in HSPCs of healthy donors (Figure 3B). The transcriptional ∼ 4-fold down-regulation of the cellular adhesion receptor CD44 prompted us to analyze CD44 expression in HSC samples of MM patients and their healthy counterparts by fluorescence microscopy which showed reduced receptor expression in the patients' cells (Figure 3Aiii,vii). Accordingly, FCM revealed a significantly lower CD44 expression in HSCs, CMPs, GMPs, and MEPs (Figure 3C-D).
MM HSPCs have disturbed migratory and adhesive properties. (A) Fluorescence micrographs show staining of (i,v) the nucleus, (ii,vi) the actin structure, (iii,vii) CD44, and (iv,viii) a merge in a representative HSC from a healthy donor and an MM patient, respectively. One representative of 4 experiments is shown. (B) For quantification of normal and defective actin assembly, 100 cells were counted and the mean percentages of cells with a normal and defective actin ring structure were determined. (C) Mean fluorescence intensity (MFI) of CD44 was determined by staining with an FITC-conjugated anti-CD44 Ab and flow cytometric analysis. Representative histogram plots of 6 experiments are shown for each subset indicated. Gray line indicates isotype control; black line, healthy donors; and red line, MM patients. (D) Bar charts represent the mean MFI of CD44 in healthy donors (□) and MM patients (■) for each HSPC subset indicated. Error bars represent SEM. (E) Chemotaxis toward stromal-derived factor 1 (SDF-1) in the lower chamber was allowed to continue for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. The relative transmigration of distinct HSPC subsets of MM patients (■) compared with their healthy counterparts (□) is shown. Highly purified HSPC subsets (5 × 103 cells each) of 4 healthy donors (□) and 4 MM patients (■) were allowed to adhere on hyaluronic acid (HA)– and fibronectin (FN)–coated glass slides for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. Mean percentages of adherent cells on (F) HA- and (G) FN-coated glass slides are shown. (H) Equal numbers of BM HSPCs of 6 MM patients and 6 healthy donors were grown in liquid medium supplemented with SCF, TPO, and FLT-3L. On days 0, 2, 3, 6, and 7, viable cell concentrations were determined. Cell-cycle distribution of total BM HSPCs and HSPC subsets was determined ex vivo by staining with FITC-conjugated Ki-67 Ab (intracellular) and Hoechst 33 342 and flow cytometric analysis. The bar charts show the mean percentage of (I) HSPCs and (J) MEP in G0 and G1/G2/S/M phases obtained from the BM of healthy donors (□) and MM patients (■). Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001.
MM HSPCs have disturbed migratory and adhesive properties. (A) Fluorescence micrographs show staining of (i,v) the nucleus, (ii,vi) the actin structure, (iii,vii) CD44, and (iv,viii) a merge in a representative HSC from a healthy donor and an MM patient, respectively. One representative of 4 experiments is shown. (B) For quantification of normal and defective actin assembly, 100 cells were counted and the mean percentages of cells with a normal and defective actin ring structure were determined. (C) Mean fluorescence intensity (MFI) of CD44 was determined by staining with an FITC-conjugated anti-CD44 Ab and flow cytometric analysis. Representative histogram plots of 6 experiments are shown for each subset indicated. Gray line indicates isotype control; black line, healthy donors; and red line, MM patients. (D) Bar charts represent the mean MFI of CD44 in healthy donors (□) and MM patients (■) for each HSPC subset indicated. Error bars represent SEM. (E) Chemotaxis toward stromal-derived factor 1 (SDF-1) in the lower chamber was allowed to continue for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. The relative transmigration of distinct HSPC subsets of MM patients (■) compared with their healthy counterparts (□) is shown. Highly purified HSPC subsets (5 × 103 cells each) of 4 healthy donors (□) and 4 MM patients (■) were allowed to adhere on hyaluronic acid (HA)– and fibronectin (FN)–coated glass slides for 3 hours at 37°C, 5% CO2 in a humidified atmosphere. Mean percentages of adherent cells on (F) HA- and (G) FN-coated glass slides are shown. (H) Equal numbers of BM HSPCs of 6 MM patients and 6 healthy donors were grown in liquid medium supplemented with SCF, TPO, and FLT-3L. On days 0, 2, 3, 6, and 7, viable cell concentrations were determined. Cell-cycle distribution of total BM HSPCs and HSPC subsets was determined ex vivo by staining with FITC-conjugated Ki-67 Ab (intracellular) and Hoechst 33 342 and flow cytometric analysis. The bar charts show the mean percentage of (I) HSPCs and (J) MEP in G0 and G1/G2/S/M phases obtained from the BM of healthy donors (□) and MM patients (■). Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001.
HSPC subsets of MM patients have impaired migratory and adhesive capacities
For functional corroboration of these findings, we assessed the migratory and adhesive capacities of HSPC subsets of MM patients and healthy donors using modified Boyden chamber assays as well HA and FN detachment assays, respectively. We found that the proportions of migrating HSCs, CMPs, GMPs, and MEPs were significantly reduced by ∼ 2- to 4-fold in the patients' samples (Figure 3E). In addition, the fractions of adherent cells on HA-coated and, to a lesser extent, FN-coated coverslips were significantly lower in HSCs, CMPs, GMPs, and MEPs of MM patients compared with their healthy counterparts (Figure 3F-G).
Proliferation and cell cycling is impaired in BM HSPCs of MM patients
As suggested by the results of the gene expression analysis, we wondered whether the reduction of HSPCs in the BM of MM patients is due to reduced proliferative and cell cycling activities. We found that proliferation was decreased in HSPCs of MM patients (Figure 3H) and a significantly smaller percentage of HSPCs was actively cycling (Figure 3I). Cell-cycle analysis of the distinct HSPC subsets using Ki-67 and Hoechst33342 dye staining showed a greater fraction of MEP from MM patients in the inactive G0 phase and a smaller fraction in G1/S/G2/M phases compared with those of healthy donors (G0P < .05; G1/S/G2/M P < .01; Figure 3J). No difference in cell-cycle activity was found in the other HSPC subsets.
TGFβ signaling is overactivated in BM nucleated cells and HSPCs of MM patients
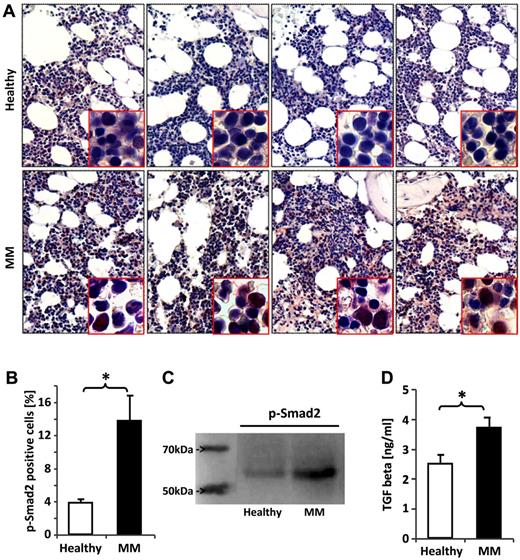
Given the particularly broad transcriptional deregulation of the TGFβ signaling pathway in HSPCs of MM patients, we analyzed BM sections by immunohistochemical staining for p-smad2, the activated form of the downstream mediator of TGFβ receptor I kinase (TBRI). Activation of smad2 was seen in nucleated BM cells in the BM sections of MM patients showing more intense staining (Figure 4A) and a significantly greater number of positively stained cells compared with those of healthy donors (Figure 4B). To assess smad2 activation specifically in HSPCs, immunoblotting was performed and revealed increased smad2 activation in the patients' cells (Figure 4C). Consistent with the activation of TGFβ signaling in BM cells, we detected a significantly elevated mean TGFβ concentration in the BM extracellular fluid (BMEF) of MM patients determined by ELISA (Figure 4D). The levels of TNFα, MIP-1α, and IL-1β were not increased (supplemental Figure 3). No differences in the gene expression of TGFβ and TBRI were observed in HSPCs, suggesting that activation of TGFβ signaling is not autocrine, whereas expression of smad2, which is transcriptionally up-regulated after sustained TGFβ stimulation, was significantly increased (supplemental Figure 4).
Smad2 is overactivated in BM cells and HSPCs of MM patients. BM biopsy sections from patients with MM and healthy controls were fixed and immunostained with an Ab against phospho-smad2, the activated form of the downstream mediator of TGFβ receptor I kinase (TBRI) activation. (A) Four representative samples of healthy donors and MM patients, respectively. P-smad2 staining was quantified by counting the total number of positively stained cells in 3 hot fields for each patient sample under ×200 magnification aided by Motic Image Plus software. (B) The bar chart shows the mean percentage of phospho-smad2–positive BM cells in healthy donors and MM patients. HSPCs from the BM of healthy donors and MM patients were assessed for smad2 phosphorylation by immunoblotting. (C) One representative of 4 experiments is shown. TGFβ1 concentrations were assessed by ELISA. (D) The bar charts display the mean TGFβ1 concentration in the BMEF. The Student t test was used to detect statistically significant differences. *P < .05.
Smad2 is overactivated in BM cells and HSPCs of MM patients. BM biopsy sections from patients with MM and healthy controls were fixed and immunostained with an Ab against phospho-smad2, the activated form of the downstream mediator of TGFβ receptor I kinase (TBRI) activation. (A) Four representative samples of healthy donors and MM patients, respectively. P-smad2 staining was quantified by counting the total number of positively stained cells in 3 hot fields for each patient sample under ×200 magnification aided by Motic Image Plus software. (B) The bar chart shows the mean percentage of phospho-smad2–positive BM cells in healthy donors and MM patients. HSPCs from the BM of healthy donors and MM patients were assessed for smad2 phosphorylation by immunoblotting. (C) One representative of 4 experiments is shown. TGFβ1 concentrations were assessed by ELISA. (D) The bar charts display the mean TGFβ1 concentration in the BMEF. The Student t test was used to detect statistically significant differences. *P < .05.
Inhibition of the TGFβ signaling pathway in MM HSPCs stimulates proliferation, self-renewal, and erythroid colony growth
Based on these findings we wondered whether HSPC function could be restored by blockade of the TGFβ signaling pathway. In fact, SD-208, an inhibitor of TBRI kinase-mediated smad2 phosphorylation, significantly increased proliferation and erythroid colony formation (Figure 5A,C) as well as the LTC-IC frequency of MM HSPCs (Figure 5B). This is reflected by a significant increase of the proportion of cells in S phase and a significantly decreased cell fraction in G0/1 phase shown in cell-cycle assays using BrdU incorporation and 7-AAD staining (Figure 5D). In contrast, inhibition of p38 MAPK signaling by SCIO-469 and NFKB signaling by TPCA-1 did not enhance erythroid colony formation of HSPCs of MM patients (supplemental Figure 5).
Inhibition of TBRI restores self-renewal and clonogenic capacities of MM HSPCs. (A) BM HSPCs of 6 MM patients were preincubated for 1 hour with a vehicle, SD-208 (0.5μM) or TGFβ in combination with SD-208 and grown in the presence of SCF, TPO, and FLT-3L. On days 0, 2, 3, 6, and 7, cell aliquots were taken and viable cell concentrations were determined and compared with BM HSPCs of healthy donors (P < .05). To determine the frequency of long-term culture-initiating cells 6 × 103 HSPCs from the BM of 5 MM patients were preincubated with vehicle or SD-208 and plated (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto irradiated confluent AFT024 feeder layers subcultured in 96-well plates and maintained in liquid medium. After 5 weeks, the liquid medium was replaced by methylcellulose medium and secondary CFC were scored after 2 weeks. The absolute frequency of LTC-ICs was calculated using Lda STAT software. (B) LTC-IC frequency of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients ( ) is shown. Purified HSPCs from the BM of 6 MM patients were seeded into semisolid ready-to-use methylcellulose growth medium at a concentration of 5 × 102 cells/mL after preincubation with vehicle or SD-208. (C) Colony numbers were counted after 2 weeks under an inverted light microscope. Proportions of colonies (BFU-E/CFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) derived from HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs (
) is shown. Purified HSPCs from the BM of 6 MM patients were seeded into semisolid ready-to-use methylcellulose growth medium at a concentration of 5 × 102 cells/mL after preincubation with vehicle or SD-208. (C) Colony numbers were counted after 2 weeks under an inverted light microscope. Proportions of colonies (BFU-E/CFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) derived from HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs ( ) are shown. Cell-cycle analyses of HSPCs obtained from the BM of 4 MM patients were performed after 7 days in liquid medium supplemented with SCF, TPO, and FLT-3L and treatment with either vehicle or SD-208 using the PE BrdU Flow Kit and flow cytometric analysis. (D) The bar chart displays the mean percentages of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients (
) are shown. Cell-cycle analyses of HSPCs obtained from the BM of 4 MM patients were performed after 7 days in liquid medium supplemented with SCF, TPO, and FLT-3L and treatment with either vehicle or SD-208 using the PE BrdU Flow Kit and flow cytometric analysis. (D) The bar chart displays the mean percentages of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients ( ) in G0/1 phase and S phase. Error bars represent SEM. The Student t test or one-way ANOVA was used to detect statistically significant differences. *P < .05; **P < .01.
) in G0/1 phase and S phase. Error bars represent SEM. The Student t test or one-way ANOVA was used to detect statistically significant differences. *P < .05; **P < .01.
Inhibition of TBRI restores self-renewal and clonogenic capacities of MM HSPCs. (A) BM HSPCs of 6 MM patients were preincubated for 1 hour with a vehicle, SD-208 (0.5μM) or TGFβ in combination with SD-208 and grown in the presence of SCF, TPO, and FLT-3L. On days 0, 2, 3, 6, and 7, cell aliquots were taken and viable cell concentrations were determined and compared with BM HSPCs of healthy donors (P < .05). To determine the frequency of long-term culture-initiating cells 6 × 103 HSPCs from the BM of 5 MM patients were preincubated with vehicle or SD-208 and plated (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto irradiated confluent AFT024 feeder layers subcultured in 96-well plates and maintained in liquid medium. After 5 weeks, the liquid medium was replaced by methylcellulose medium and secondary CFC were scored after 2 weeks. The absolute frequency of LTC-ICs was calculated using Lda STAT software. (B) LTC-IC frequency of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients ( ) is shown. Purified HSPCs from the BM of 6 MM patients were seeded into semisolid ready-to-use methylcellulose growth medium at a concentration of 5 × 102 cells/mL after preincubation with vehicle or SD-208. (C) Colony numbers were counted after 2 weeks under an inverted light microscope. Proportions of colonies (BFU-E/CFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) derived from HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs (
) is shown. Purified HSPCs from the BM of 6 MM patients were seeded into semisolid ready-to-use methylcellulose growth medium at a concentration of 5 × 102 cells/mL after preincubation with vehicle or SD-208. (C) Colony numbers were counted after 2 weeks under an inverted light microscope. Proportions of colonies (BFU-E/CFU-E, CFU-G/CFU-M/CFU-GM, CFU-GEMM) derived from HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs ( ) are shown. Cell-cycle analyses of HSPCs obtained from the BM of 4 MM patients were performed after 7 days in liquid medium supplemented with SCF, TPO, and FLT-3L and treatment with either vehicle or SD-208 using the PE BrdU Flow Kit and flow cytometric analysis. (D) The bar chart displays the mean percentages of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients (
) are shown. Cell-cycle analyses of HSPCs obtained from the BM of 4 MM patients were performed after 7 days in liquid medium supplemented with SCF, TPO, and FLT-3L and treatment with either vehicle or SD-208 using the PE BrdU Flow Kit and flow cytometric analysis. (D) The bar chart displays the mean percentages of HSPCs of healthy donors (□), vehicle-treated HSPCs of MM patients (■), and SD-208–treated HSPCs of MM patients ( ) in G0/1 phase and S phase. Error bars represent SEM. The Student t test or one-way ANOVA was used to detect statistically significant differences. *P < .05; **P < .01.
) in G0/1 phase and S phase. Error bars represent SEM. The Student t test or one-way ANOVA was used to detect statistically significant differences. *P < .05; **P < .01.
MSCs of MM patients are functionally impaired and have a decreased stem cell–supporting capacity
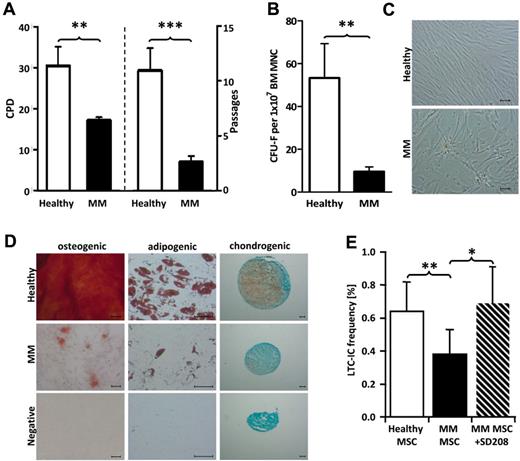
MM patient-derived MSCs proliferated slower, as indicated by lower cumulative population doublings (CPD; P < .01), and entered growth arrest after fewer culture passages (P < .001; Figure 6 A). The clonogenic capacity of patient-derived MSCs was reduced (P < .01; Figure 6B) and the cells were large and flat (Figure 6C). Although trilineage differentiation capacity was retained in the patients' MSC, adipogenic, osteogenic, and chondrogenic differentiation were quantitatively impaired (Figure 6D). To decipher whether these MSC alterations were relevant for the functional impairment of HSPCs in MM patients, normal HSPCs were cultured on healthy donor– and patient-derived MSCs. Hereby, we found that the long-term ability of patient-derived MSCs to support HSPCs was significantly reduced, but reached the level of healthy donor–derived MSCs on blockade of TGFβ signaling (Figure 6E).
MSCs of MM patients are functionally altered and have an impaired long-term ability to support HSPCs. (A) Bar charts show cumulative population doublings (CPD; left) and number of passages (right) of healthy donors and MM MSCs. (B) At the end of the primary incubation period, colonies > 50 cells (CFU-F) were counted by light microscopy. Bar chart displays the number of CFU-F per 1 × 107 MNCs for healthy donors and MM patients. (C) Micrographs depict morphology of healthy donors and MM patients' MSCs. For assessment of the differentiation capacity, MSCs were differentiated into osteogenic, adipogenic, and chondrogenic lineages. (D) Representative micrographs of the osteogenic, adipogenic, and chondrogenic differentiation of healthy donors' and patients MSCs (each one of 5 experiments performed). To determine the long-term ability of MSCs to support HSPCs, 34+ HSPCs from the BM of healthy donors were plated (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto MSCs from healthy donors and MM patients (with or without SD-208) subcultured in 96-well plates and maintained for 5 weeks. (E) After 5 weeks, the liquid medium was replaced by methylcellulose medium and secondary CFCs were scored after 2 weeks. The absolute frequency of LTC-ICs was calculated using Lda STAT software. LTC-IC frequency of healthy donor BM HSPCs maintained on MSCs of 5 healthy donors or 8 MM patients with or without SD-208 is shown. Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001.
MSCs of MM patients are functionally altered and have an impaired long-term ability to support HSPCs. (A) Bar charts show cumulative population doublings (CPD; left) and number of passages (right) of healthy donors and MM MSCs. (B) At the end of the primary incubation period, colonies > 50 cells (CFU-F) were counted by light microscopy. Bar chart displays the number of CFU-F per 1 × 107 MNCs for healthy donors and MM patients. (C) Micrographs depict morphology of healthy donors and MM patients' MSCs. For assessment of the differentiation capacity, MSCs were differentiated into osteogenic, adipogenic, and chondrogenic lineages. (D) Representative micrographs of the osteogenic, adipogenic, and chondrogenic differentiation of healthy donors' and patients MSCs (each one of 5 experiments performed). To determine the long-term ability of MSCs to support HSPCs, 34+ HSPCs from the BM of healthy donors were plated (22 replicates per concentration: 180, 60, 20, 7.5 cells/well) onto MSCs from healthy donors and MM patients (with or without SD-208) subcultured in 96-well plates and maintained for 5 weeks. (E) After 5 weeks, the liquid medium was replaced by methylcellulose medium and secondary CFCs were scored after 2 weeks. The absolute frequency of LTC-ICs was calculated using Lda STAT software. LTC-IC frequency of healthy donor BM HSPCs maintained on MSCs of 5 healthy donors or 8 MM patients with or without SD-208 is shown. Error bars represent SEM. The Student t test was used to detect statistically significant differences. *P < .05; **P < .01; ***P < .001.
Xenotransplantation assays revealed normal differentiation capacity and even enhanced engraftment of MM HSPCs
To assess in vivo whether the functional impairment of HSPCs in MM patients depends on the continuous exposure to the MM-related BM microenvironmental conditions or persists in their absence and affects BM engraftment and repopulation capacities, BM HSPCs obtained from MM patients or age-matched healthy donors were transplanted into NOG mice. We found no differences in the short-term engraftment of HSPCs of MM patients and healthy donors after 4 weeks (Figure 7A). In addition, the differentiation capacities did not show significant variations at this time point (Figure 7Bi-iii). Interestingly, the long-term engraftment of BM HSPCs of MM patients was even higher 30 weeks after transplantation (P < .001; Figure 7C-D). Analysis of the engrafted human CD45+ cells showed an increased percentage of CD33+ myeloid cells among the MM patient-derived cells (P < .05) while there was no significant difference in the percentage of CD19+, CD11b+, and CD71+ cells between patient- and donor-derived cells, indicating intact differentiation capacities of the MM patients' HSPCs (Figure 7Ei-iv).
Engraftment of MM HSPCs is enhanced in xenotransplantation assays. A total of 1-3 × 106 HSPCs obtained from the BM of 4 healthy donors and 4 MM patients were transplanted into sublethally irradiated NOG mice by intrafemoral injection. Peripheral blood (PB) was analyzed 4 weeks after transplantation and analyzed by flow cytometry. (A) Mean proportions of human CD45+ hematopoietic cells within PB. (Bi-iii) Mean proportions of CD19+, CD33+, and CD71+ cells are shown. (C) Total BM (TBM) cells were harvested 30 weeks after transplantation and analyzed by flow cytometry. Representative FACS plots show the proportions of human CD45+ hematopoietic cells within TBM after transplantation of HSPCs obtained from the BM of a healthy donors and a MM patient, respectively. (D) Bar chart shows the mean proportions of the human CD45+ cell population within TBM cells for all experiments. Human hematopoietic differentiation markers were analyzed in human CD45+ cells 30 weeks after transplantation. (Ei-iv) Bar charts show mean proportions of CD19+, CD11b, CD33+, and CD71+ cells. Error bars represent SD. Paired 2-tailed t test was used to detect statistically significant differences. *P < .05; ***P < .001.
Engraftment of MM HSPCs is enhanced in xenotransplantation assays. A total of 1-3 × 106 HSPCs obtained from the BM of 4 healthy donors and 4 MM patients were transplanted into sublethally irradiated NOG mice by intrafemoral injection. Peripheral blood (PB) was analyzed 4 weeks after transplantation and analyzed by flow cytometry. (A) Mean proportions of human CD45+ hematopoietic cells within PB. (Bi-iii) Mean proportions of CD19+, CD33+, and CD71+ cells are shown. (C) Total BM (TBM) cells were harvested 30 weeks after transplantation and analyzed by flow cytometry. Representative FACS plots show the proportions of human CD45+ hematopoietic cells within TBM after transplantation of HSPCs obtained from the BM of a healthy donors and a MM patient, respectively. (D) Bar chart shows the mean proportions of the human CD45+ cell population within TBM cells for all experiments. Human hematopoietic differentiation markers were analyzed in human CD45+ cells 30 weeks after transplantation. (Ei-iv) Bar charts show mean proportions of CD19+, CD11b, CD33+, and CD71+ cells. Error bars represent SD. Paired 2-tailed t test was used to detect statistically significant differences. *P < .05; ***P < .001.
Discussion
MM patients frequently suffer from hematopoietic suppression at the time of diagnosis with anemia as a prevailing symptom,1,2 while relatively few studies have addressed the underlying pathophysiology.3-5 In the BM of MM patients, we found a significant reduction of HSPC, in particular of MEPs, which were functionally impaired with regard to clonogenic and long-term self-renewal capacity as well as proliferative activity. Thus, the depletion of BM HSPCs in MM patients is not only due to a pure anatomic crowding out, but also results from functional impairment. Microarray-based computer simulation of the cellular organization within the total HSPC population resembles normal hierarchical development23 in MM patients, suggesting that hematopoietic suppression is not the result of an intrinsic deregulation of the developmental program. Instead, the pattern of transcriptional deregulations in the HSPC subsets supports their origin from extrinsic MM-related perturbations of the BM. As the most prominent we focused on the TGFβ signaling pathway, which was transcriptionally activated in the HSPCs of MM patients. Consequently, the expression of p-smad2, a biomarker for activated TGFβ signaling,24 was increased in BM HSPCs and BM cells in MM patients. We detected elevated TGFβ concentrations in the BMEF of MM patients which suggests that overactivation of TGFβ signaling in BM cells is due to abundant release of TGFβ by PCs and MSCs in MM patients as previously described.25
Activation of TGFβ signaling suppresses erythropoiesis via cell-cycle inhibition of erythroid precursors and has a strong antiproliferative effect on HSPCs leading to reduced colony formation and long-term self-renewal.26-31 Inhibition of smad2 phosphorylation by SD208, a selective TGFβ receptor I kinase inhibitor,32 recovered, at least in part, colony formation and long-term self-renewal and increased the proliferation and cell-cycle activity in MM HSPCs. Similar findings have recently been reported for progenitor cells of patients with myelodysplastic syndromes (MDS).24 At this point, it is worth mentioning that thalidomide, a drug with an inhibitory effect on TGFβ signaling33 and high effectiveness in the treatment of MM,34 led to a significant raise of Hgb concentrations in patients with MM34 and MDS.35 The rise of the Hgb concentration was associated with a decline in serum TGFβ levels.35 In addition to the canonical pathway, TGFβ activates several noncanonical pathways including p38 and other MAPK pathways24,31,36,37 that were also transcriptionally deregulated in the patients' HSPCs. As another potential mechanism of myelosuppression, we found (albeit not significant) increased levels of TNFα, a suppressor of normal HSC activity,38 in the BMEF of MM patients that has been shown to inhibit erythroid colony formation of HSPCs via activation of NFKB signaling.39 It is therefore conceivable that activation of these pathways contributes to the observed deregulation of HSPC, although inhibition of p38 MAPK and NF-κB signaling did not restore clonogenic capacities of the MM HSPCs examined.
In further support of the role of TGFβ for the functional impairment of HSPCs in MM patients, we found a significantly reduced long-term ability of MM patient-derived MSCs to maintain HSPCs that reached the level of healthy donor–derived MSCs when TGFβ signaling was blocked during the coculture. Patient-derived MSCs showed an altered phenotype including a slower rate of proliferation and differentiation that was not enhanced by TGFβ signaling blockade and, at the beginning of MSC cultivation, even further slowed cell growth (data not shown) reflecting the importance of TGFβ signaling for growth and differentiation of MSC.40 Activation of TGFβ signaling, which enhances the secretion of IL-6 and VEGF by MSC,25 does not trigger the release of other cytokines with known inhibitory effects on HSPC.41 Hence, increased secretion of TGFβ by patient-derived MSC25 rather directly contributes to the functional impairment of HSPC. Still, the functional impairment of HSPCs in MM patients might be associated with the altered functional properties of MSC.
In addition, we observed a comprehensive transcriptional deregulation of pathways and single genes related to the organization of the cell cytoskeleton, cell migration and cell adhesion in HSPC subsets of MM patients. Consistently, we found an altered actin structure with abnormal aggregates as well as a reduced expression of CD44 in HSPCs of MM patients. CD44 plays a pivotal role in the adhesion of HSPCs to their principal ligand hyaluronan (HA) and SDF-1–induced cell polarization and motility.42 Cellular response to these chemotactic stimuli requires the association of CD44 with actin followed by activation of several signal transduction pathways to induce the dynamic reorganization of the actin cytoskeleton.43 Thus, both SDF-1–induced directional migration and adhesion to HA were significantly decreased in the patients' HSPC. We also observed, albeit to a lesser extent, decreased adhesion to FN which depends on β-integrin binding followed by signal transduction via an integrin-cytoskeletal association and, eventually, reorganization of the actin cytoskeleton.44 The adhesion to FN may thus be hampered by disturbances in the integrin signaling because of a defective association of the integrin with the cytoskeleton. Because the CD44- and β-integrin–mediated migration as well as adhesion are essential for HSPC homing and repopulation of the BM,42,45 the latter one also because of induction of stem cell quiescence on adhesion,46 these deficits might contribute to the depletion of HSPCs and hematopoietic impairment in MM patients. In combination with the alterations of MM MSCs that provide a less hospitable environment for HSPCs and a more supportive one for malignant PC,9 this may create an advantage for clonal PCs over HSPCs in the competition for potentially overlapping BM niches and lead to the replacement of HSPCs as recently suggested.16 Activation of TGFβ signaling along with decreased CD44 expression in normal HSPCs of MM patients contrasts with the findings in cancer stem cells of several solid tumor entities where activation of TGFβ signaling leads to up-regulation of CD44 expression which promotes self-renewal, invasion, and metastasis.43 This discrepancy, despite striking similarities of malignant and nonmalignant stem cells,34 might be because of the nonmalignant phenotype of HSPCs of MM patients. In favor of this assumption, TGFβ is able to promote context-dependent opposed transcriptional responses in nonmalignant and malignant cells through the same promoter region.47,48
Transplantation into the MM-free BM microenvironment of NOG mice revealed normal short-term and even enhanced long-term engraftment along with normal differentiation capacities of MM HSPCs indicating a transient functional impairment that depends on MM-related microenvironmental influences. This is in accordance with the clinical experience as rapid and sustained engraftment is achieved in the majority of MM patients after elimination of malignant PCs from the BM by high-dose cytotoxic therapy.49 Increased engraftment might be due to a compensatory rebound after discontinuation of the inhibitory effects which requires a recovery period exceeding the time provided by in vitro assays. In support of this view, increased numbers of HSPCs have recently been reported in MM patients after efficacious high-dose therapy and autologous HSPC transplantation.16
In conclusion, our data show that symptomatic MM leads to functional impairment and diminution of all HSPC subsets with an emphasis on early erythroid precursors. These effects are apparently transient and largely dependent on the MM-related BM microenvironment. Among those, we found a particular role of activated TGFβ signaling for the functional impairment of HSPCs.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Annemarie Koch for excellent technical assistance.
This work was supported by grants from Leukämie Liga e.V., Düsseldorf, Germany and the Forschungskommission of the Heinrich-Heine-University, Düsseldorf, Germany.
I. Bruns is the recipient of an American Society of Hematology–European Hematology Association (ASH-EHA) research exchange award. A.C. has been awarded an EHA research fellowship. U.S. is the recipient of a Howard Temin Award of the National Cancer Institute, a New Investigator Award of the Leukemia Research Foundation, and the Diane and Arthur B. Belfer Faculty Scholar in Cancer Research of the Albert Einstein College of Medicine.
National Institutes of Health
Authorship
Contribution: I. Bruns designed the study, supervised the experiments, analyzed data, and wrote the manuscript; R.-P.C. performed most of the experiments, analyzed data, and reviewed the manuscript; I. Brueckmann performed all in vivo experiments and analyzed data; J.F., S.G., and S.B. performed experiments, analyzed data, and reviewed the manuscript; J.C.F, F.R., C.M.W., F.A.S., F.Z., B.B., and T.S. analyzed data and reviewed the manuscript; A.-N.H., C.Z., and M.J. provided patient samples; and U.S., R.F., G.K., A.C., A.T., and R.H. designed the study and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ingmar Bruns, Department of Hematology, Oncology, and Clinical Immunology, Heinrich-Heine-University, Moorenstrasse 5, 40225 Düsseldorf, Germany; e-mail: brunsin@med.uni-duesseldorf.de.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal