Abstract
The outcome of allogeneic hematopoietic cell transplantation is influenced by donor/recipient genetic disparity at loci both inside and outside the MHC on chromosome 6p. Although disparity at loci within the MHC is the most important risk factor for the development of severe GVHD, disparity at loci outside the MHC that encode minor histocompatibility (H) antigens can elicit GVHD and GVL activity in donor/recipient pairs who are otherwise genetically identical across the MHC. Minor H antigens are created by sequence and structural variations within the genome. The enormous variation that characterizes the human genome suggests that the total number of minor H loci is probably large and ensures that all donor/recipient pairs, despite selection for identity at the MHC, will be mismatched for many minor H antigens. In addition to mismatch at minor H loci, unrelated donor/recipient pairs exhibit genetic disparity at numerous loci within the MHC, particularly HLA-DP, despite selection for identity at HLA-A, -B, -C, and -DRB1. Disparity at HLA-DP exists in 80% of unrelated pairs and clearly influences the outcome of unrelated hematopoietic cell transplantation; the magnitude of this effect probably exceeds that associated with disparity at any locus outside the MHC.
Introduction
Genetic nonidentity between donor and recipient is the key to the therapeutic efficacy of allogeneic hematopoietic cell transplantation (HCT) for malignant disease, but it is also the root of GVHD, its primary limitation. Pioneering studies in the late 1960s and early 1970s led to the critical discovery that donor/recipient genetic nonidentity at the MHC on chromosome 6p is the single most important risk factor for the development of severe GVHD.1 The subsequent implementation of donor selection procedures according to donor/recipient MHC matching established with serologic assays and mixed lymphocyte culture revolutionized the nascent field of allogeneic marrow transplantation and enabled rapid growth during the subsequent decade in the annual number of transplantations performed, as well as considerable improvement in the likelihood of a successful transplantation outcome. Nonetheless, clinically significant GVHD still developed in a sizeable fraction of recipients of bone marrow grafts from sibling donors with whom they were genotypically identical throughout the MHC region on both copies of chromosome 6p. This observation suggested that genetic loci outside the MHC, encoding putative histocompatibility determinants that were collectively referred to as minor histocompatibility (H) antigens, could also influence a recipient's likelihood of developing GVHD or benefitting from GVL activity.1,2 The intervening years have witnessed steady progress in elucidating the nature of minor H antigens and the genes that encode them and have seen the identification and characterization of other complex genetic loci that also influence histocompatibility in the allogeneic HCT setting. Here, we review the manner in which genetic identity or nonidentity between donor and recipient at loci both inside and outside the MHC can influence the outcome of allogeneic HCT, emphasizing wherever possible the underlying immunobiologic principles, and suggest an agenda for future research. This review focuses primarily on the contribution of donor/recipient genetic disparity to histocompatibility in the allogeneic HCT setting, but does not discuss immunotherapeutic strategies for exploiting disparity at specific loci, which has been covered in a recent review.3
The MHC: establishing immunologic identity
The MHC spans > 3.3 megabases of genomic sequence on the short arm of chromosome 6 and includes several hundred protein-coding genes. Improved understanding of the MHC has driven steady evolution of the laboratory procedures used to evaluate potential HCT donors and recipients to establish that they are “MHC matched.” As recently as 10-15 years ago, histocompatibility typing involved serologic assays to evaluate compatibility for products of the class I MHC loci such as HLA-A and HLA-B, combined with mixed lymphocyte culture to assess compatibility for products of class II MHC loci such as HLA-DR. Current histocompatibility typing entails targeted sequencing of a limited number of exons in specific class I and class II MHC genes. For identification of potential unrelated donor candidates, the National Marrow Donor Program currently recommends sequencing exons 2 and 3 of the HLA-A, -B, and -C genes as well as exon 2 of HLA-DRB1 (Figure 1).4 Donor and recipient sequences in exon 2, and occasionally exon 3, of the HLA-DQB1 gene are also typically assessed during the unrelated donor search process with the use of medium resolution molecular techniques, primarily to ensure an optimal match at HLA-DRB1, but these HLA-DQB1 exons are not routinely evaluated with direct sequencing. Because the 7 exons in the HLA-A, -B, -C, and -DRB1 genes of the intended recipient and the prospective donor(s) that are sequenced during the process of unrelated donor selection are, on average, 275 nucleotides in length, routine histocompatibility typing for unrelated donor HCT involves determining the DNA sequence in a cumulative interval of only 1925 nucleotides per haploid genome. This represents less than one thousandth of the genetic sequence within the MHC and less than one millionth of the 3.3 × 109 bases in the haploid human genome. How is it that such a small fraction of the sequence within the MHC, and a miniscule fraction of the sequence in the entire genome, can serve as the master regulator of histocompatibility?
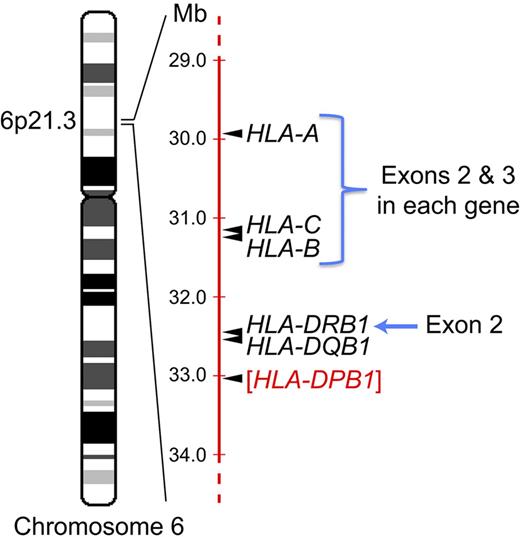
Selection of unrelated donors for allogeneic HCT involves sequencing specific exons in 4 different genes on chromosome 6p. The relative position of the HLA-A, HLA-B, HLA-C, and HLA-DRB1 genes in the MHC on the short arm of chromosome 6 at band 6p21.3 is indicated, as are the specific exons within each gene that are routinely sequenced in the process of unrelated donor selection for allogeneic HCT. Most transplantation centers do not as yet perform routine sequencing of exon 2 in the HLA-DQB1 gene, but donor/recipient matching for sequences in this exon is typically performed with medium resolution molecular techniques. The relative position of the HLA-DPB1 gene (in red), which plays a role in histocompatibility in the allogeneic HCT setting but at this time is not routinely sequenced during the donor selection process, is also indicated.
Selection of unrelated donors for allogeneic HCT involves sequencing specific exons in 4 different genes on chromosome 6p. The relative position of the HLA-A, HLA-B, HLA-C, and HLA-DRB1 genes in the MHC on the short arm of chromosome 6 at band 6p21.3 is indicated, as are the specific exons within each gene that are routinely sequenced in the process of unrelated donor selection for allogeneic HCT. Most transplantation centers do not as yet perform routine sequencing of exon 2 in the HLA-DQB1 gene, but donor/recipient matching for sequences in this exon is typically performed with medium resolution molecular techniques. The relative position of the HLA-DPB1 gene (in red), which plays a role in histocompatibility in the allogeneic HCT setting but at this time is not routinely sequenced during the donor selection process, is also indicated.
The products of HLA-A, -B, and -C, and the heterodimer formed by the products of HLA-DRA and HLA-DRB1, are transmembrane proteins whose extracellular domains form a characteristic “groove” comprising 2 conserved α helices and a conserved β-pleated sheet region. Solution of the structure of the HLA-A2 molecule in 19875 serendipitously provided the critical insight that the extracellular grooves in class I MHC molecules noncovalently bind small peptides, thereby illustrating how MHC molecules literally “present” antigenic peptides to T lymphocytes. Subsequent studies have reported an analogous function for the grooves formed by the extracellular domains of the heterodimeric class II MHC molecules.6 The groove in class I MHC molecules binds a wide variety of short peptides (typically 8-11 residues in length) derived primarily from the degradation of endogenous cellular proteins, and the corresponding groove formed by the α and β subunits of class II MHC molecules binds slightly longer peptides thought to be derived primarily, but not exclusively, from extracellular sources. The peptide-binding groove, also referred to as the antigen-recognition site, of HLA-A, -B, and -C molecules is encoded by exons 2 and 3 of the corresponding genes (Figure 2A-B), whereas the peptide-binding groove of HLA-DR molecules is formed by the product of exon 2 of HLA-DRB1, in conjunction with the product of exon 2 of the minimally polymorphic HLA-DRA (Figure 2C-D). Thus, the 1925 nucleotides sequenced in each haploid genome encode the antigen-recognition sites of HLA-A, -B, -C, and -DR in their entirety.
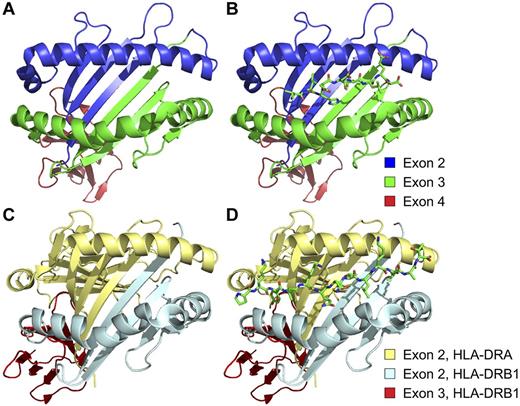
The exons sequenced during the process of donor selection for allogeneic HCT encode critical portions of the peptide-binding grooves of class I and class II MHC molecules. (A) Structure of the extracellular portion of the HLA-A*02:01:01 molecule, indicating the regions encoded by exons 2 (blue), 3 (green), and 4 (red) of the A*02:01:01 allele of the HLA-A gene. (B) Structure of HLA-A*02:01:01 with the peptide VLHDDLLEA, the minor histocompatibility antigen HA-1, bound in its peptide-binding groove. (C) Structure of the extracellular portion of the heterodimeric HLA-DR4 molecule, showing the moieties encoded by exons 2 (light blue) and 3 (red) of the HLA-DRB1 gene, as well as the moiety encoded by exon 2 of the minimally polymorphic HLA-DRA gene (pale yellow). (D) Structure of HLA-DR4 with a peptide derived from influenza hemagglutinin, PKYVKQNTLKLAT, bound in its peptide-binding groove. The HLA-A*02:01:01 and HLA-DR4 structures were derived from Protein Data Bank (PDB) accession nos. 3D25107 and 1J8H,108 and rendered with Pymol.109
The exons sequenced during the process of donor selection for allogeneic HCT encode critical portions of the peptide-binding grooves of class I and class II MHC molecules. (A) Structure of the extracellular portion of the HLA-A*02:01:01 molecule, indicating the regions encoded by exons 2 (blue), 3 (green), and 4 (red) of the A*02:01:01 allele of the HLA-A gene. (B) Structure of HLA-A*02:01:01 with the peptide VLHDDLLEA, the minor histocompatibility antigen HA-1, bound in its peptide-binding groove. (C) Structure of the extracellular portion of the heterodimeric HLA-DR4 molecule, showing the moieties encoded by exons 2 (light blue) and 3 (red) of the HLA-DRB1 gene, as well as the moiety encoded by exon 2 of the minimally polymorphic HLA-DRA gene (pale yellow). (D) Structure of HLA-DR4 with a peptide derived from influenza hemagglutinin, PKYVKQNTLKLAT, bound in its peptide-binding groove. The HLA-A*02:01:01 and HLA-DR4 structures were derived from Protein Data Bank (PDB) accession nos. 3D25107 and 1J8H,108 and rendered with Pymol.109
The ensemble of peptides presented by the complement of class I and class II MHC molecules on the surface of a given cell constitute the MHC-associated “immunopeptidome”7 of that cell and establish its immunologic identity. The HLA-A, -B, -C, and -DRB1 loci are the most polymorphic genes in the human genome (Figure 3), and virtually all sequence polymorphism in these genes that has been shown to influence histocompatibility affects residues located in the α helices or the β-pleated sheet region that collectively form the peptide-binding grooves. The molecules encoded by the A*02:01:01 and A*03:01:01 alleles of HLA-A, for example, are not identical at 22 of 300 residues, 14 of which are located in the peptide-binding domain (Figure 4). This polymorphism has an enormous influence on the repertoire of peptides bound by a given MHC molecule and, therefore, profoundly shapes the immunologic identity of each cell. Polymorphism in MHC molecules also influences the conformation of the complexes formed by peptides and MHC molecules, as well as the interaction of T-cell antigen receptors with peptide/MHC complexes.
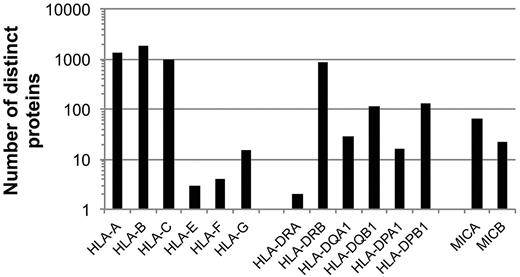
The HLA-A, -B, -C, and -DRB1 loci are the most polymorphic genes in the entire human genome. The number of distinct HLA-A, -B, -C, -E, -F, -G, -DRA, -DRB1, -DQA1, -DQB1, -DPA1, -DPB1, MICA, and MICB proteins collectively encoded by all known alleles at the corresponding MHC loci is indicated. Data were taken from http://www.ebi.ac.uk/imgt/hla/stats.html; accessed February 13, 2012.
The HLA-A, -B, -C, and -DRB1 loci are the most polymorphic genes in the entire human genome. The number of distinct HLA-A, -B, -C, -E, -F, -G, -DRA, -DRB1, -DQA1, -DQB1, -DPA1, -DPB1, MICA, and MICB proteins collectively encoded by all known alleles at the corresponding MHC loci is indicated. Data were taken from http://www.ebi.ac.uk/imgt/hla/stats.html; accessed February 13, 2012.
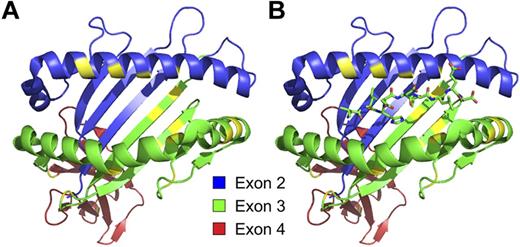
The majority of polymorphic residues that distinguish different alleles of class I and class II molecules are located in positions that influence peptide binding or interaction with T-cell receptors. Structure of HLA-A*02:01:01 without (A) and with (B) the HA-1 minor H antigen bound in its peptide-binding groove, with the regions encoded by exons 2, 3, and 4 of the A*02:01:01 allele of HLA-A colored as in Figure 2. The specific residues in HLA-A*02:01:01 that are nonidentical with those in the HLA-A*03:01:01 molecule are indicated in yellow.
The majority of polymorphic residues that distinguish different alleles of class I and class II molecules are located in positions that influence peptide binding or interaction with T-cell receptors. Structure of HLA-A*02:01:01 without (A) and with (B) the HA-1 minor H antigen bound in its peptide-binding groove, with the regions encoded by exons 2, 3, and 4 of the A*02:01:01 allele of HLA-A colored as in Figure 2. The specific residues in HLA-A*02:01:01 that are nonidentical with those in the HLA-A*03:01:01 molecule are indicated in yellow.
The MHC-associated immunopeptidome and the origin of minor H antigens
GVHD and GVL occur in recipients of allogeneic hematopoietic cell grafts from donors with whom they are “MHC matched” because the repertoire of MHC-bound peptides presented on the surface of recipient cells is not perfectly identical to the repertoire of peptides presented on donor cells due to donor/recipient genetic disparity outside the MHC. Although peptides derived from the degradation of MHC-encoded proteins have been shown to be presented on the surface of cells in association with class I or class II MHC molecules,8-12 most MHC-associated peptides are derived from proteins encoded by genes located outside the MHC. The peptides associated with MHC molecules are derived from a broad spectrum of nuclear, cytoplasmic, transmembrane, and even cryptic proteins that are encoded by genes located throughout the genome, with representatives from every chromosome.7,13,14 Disparity between MHC-matched donor/recipient pairs for specific peptides, that is, the presence of a peptide in the immunopeptidome of the recipient and its absence from that of the donor, or the converse, can elicit T-cell responses that, in turn, can contribute to GVHD/GVL or, alternatively, to graft rejection. Peptides encoded by polymorphic loci outside the MHC and presented by an MHC molecule are functionally defined as minor H antigens.
As of April 2012, ≥ 49 different genes that encode minor H antigens recognized by either CD8+ or CD4+ T cells have been identified (Figure 5). Although most of these genes are located on autosomes, with virtually every chromosome represented, ≥ 6 different genes on the Y chromosome encode male-specific minor H (H-Y) antigens. Both sequence variation and structural variation in the human genome can give rise to minor H antigens. Most autosomally encoded minor H antigens discovered to date are created by nonsynonymous single nucleotide polymorphisms (nsSNPs) in protein coding sequences that give rise to single amino acid polymorphisms. The minor H antigen HA-1,15 arguably the most intensively studied human minor H antigen, exemplifies this phenomenon. A common nsSNP in the KIAA0223 gene creates a His ↔ Arg amino acid polymorphism at residue 139 in the corresponding protein. A peptide spanning this polymorphism, VLHDDLLEA, binds to HLA-A*02:0115 and can stimulate peptide-specific, HLA-A*02:01-restricted CD8+ T-lymphocyte responses in HLA-A*02:01+ donor/recipient pairs who are discordant for the KIAA0223 allele that encodes HA-1 (present in 1 member of the pair and absent in the other). Other autosomal minor H antigens are created by SNPs that alter mRNA slice sites16,17 or create stop codons,18 and another is created by a common small insertion polymorphism that creates a shift in reading frame.19 Several autosomal minor H antigens are created by SNPs in cryptic, noncanonical open reading frames in “genes” such as HMHB120 and C19orf4821 that are otherwise not known to generate a protein product, or in the nominal 3′ “untranslated” region of protein-coding genes.22 Some autosomal SNP loci can encode antigenic peptides that are naturally presented by ≥ 2 MHC alleles.23
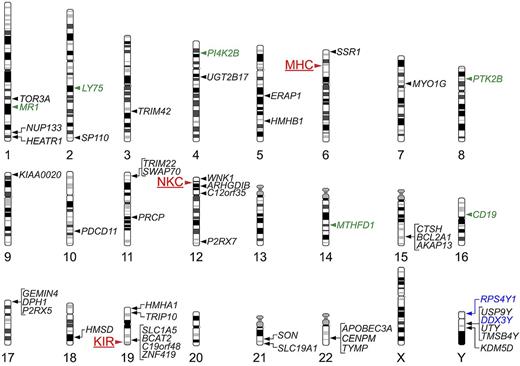
Map of genetic loci that can influence histocompatibility in the allogeneic HCT setting. The chromosomal location of the MHC, and of 2 other multigene clusters, the NKC and the KIR locus, are indicated by red labels and arrowheads to the left of the corresponding chromosomes. The chromosomal locations of genes that have been shown to encode T lymphocyte–defined minor H antigens are indicated by labels and arrowheads to the right of the corresponding chromosomes; genes that encode class I MHC-restricted minor H antigens recognized by CD8+ T cells are indicated by black labels, those that encode class II MHC-restricted minor H antigens recognized by CD4+ T cells are indicated by green labels, and those that encode both class I and class II MHC-restricted minor H antigens are indicated by blue labels.
Map of genetic loci that can influence histocompatibility in the allogeneic HCT setting. The chromosomal location of the MHC, and of 2 other multigene clusters, the NKC and the KIR locus, are indicated by red labels and arrowheads to the left of the corresponding chromosomes. The chromosomal locations of genes that have been shown to encode T lymphocyte–defined minor H antigens are indicated by labels and arrowheads to the right of the corresponding chromosomes; genes that encode class I MHC-restricted minor H antigens recognized by CD8+ T cells are indicated by black labels, those that encode class II MHC-restricted minor H antigens recognized by CD4+ T cells are indicated by green labels, and those that encode both class I and class II MHC-restricted minor H antigens are indicated by blue labels.
Minor H antigens presented by class I and class II MHC molecules probably play distinct roles in GVHD, GVL, rejection, and other transplantation phenomena. Class I MHC molecules are constitutively expressed on all nucleated cells save neurons and germ cells. Constitutive expression of class II molecules, in contrast, is generally limited to professional antigen-presenting cells and other subsets of both normal and malignant hematopoietic cells, but their expression can be readily induced on a wide variety of nonhematopoietic cells by inflammatory stimuli. It is therefore possible that, under noninflammatory conditions, spontaneous or induced T-cell responses against class II–restricted minor H antigens could contribute selectively to GVL activity in patients with class II–positive malignancies.
Clinical significance of donor/recipient mismatching at autosomal minor H loci
Retrospective analyses of donor/recipient mismatching at autosomal loci encoding class I MHC-restricted minor H antigens recognized by CD8+ T cells have evaluated whether mismatching is significantly associated with transplantation outcome. For most of those studies, mismatching has been defined as the presence of an allele that encodes a minor H antigen in 1 member of a transplant donor/recipient pair, but not the other, and in which both donor and recipient express the class I MHC molecule that presents the minor H antigen to T cells. In general, those studies did not evaluate whether T-cell responses against the relevant minor H antigen could be detected, but they simply assessed whether the donor/recipient genotypes were permissive for a T-cell response. Moreover, they did not consider the potential effects of immunodominance, a phenomenon that reflects intrinsic differences in the ability of T-cell epitopes to elicit a detectable immune response. Several small studies have suggested that donor/recipient disparity for minor H antigens created by nsSNPs in autosomal genes is significantly associated with important transplantation outcomes such as the incidence of GVHD, relapse, mortality, or survival, but few of these associations have been validated in large, multicenter transplant cohorts. The clinical significance of donor/recipient disparity for the HLA-A*0201–restricted minor H antigen HA-1 in HLA-A*02:01+ transplant pairs, for example, has been studied more intensively than any other minor H antigen,24-34 in part because of the high allele frequency of HLA-A*02:01 in European and North American transplant cohorts. Disparity for HA-1 in MHC-matched, HLA-A*02:01+ donor/recipient pairs has not consistently been associated with any transplantation outcome in these studies, and by far the largest and most comprehensive studies of HA-1 disparity in related28 and unrelated33 transplant pairs have shown no significant association with any of the several outcomes examined.
The UGT2B17 gene on chromosome 4q is homozygously deleted in a fraction of every human population that has been studied to date,35 and peptides derived from the UGT2B17 protein can act as minor H antigens in donor/recipient pairs in which 1 member of the pair expresses the gene but the other is homozygously deleted.36 UGT2B17 encodes minor H antigens presented by ≥ 3 MHC class I molecules: HLA-A*29:02, -B*44:03, and -A*02:06,36-38 and it is very probable that additional UGT2B17-derived peptides can bind to other MHC alleles and also function as minor antigens. UGT2B17 is a cell surface protein and can also serve as the target for antibody responses that develop in UGT2B17+ recipients of hematopoietic cell grafts from UGT2B17− donors.39 These observations suggest that the UGT2B17 protein can elicit alloreactive immune responses in a large fraction of the donor/recipient pairs in which disparity at the UGT2B17 locus exists, regardless of the specific MHC class I and class II molecules expressed by each pair. The potential clinical significance of UGT2B17-specific immune responses has been suggested by a large retrospective study of donor/recipient mismatching at UGT2B17 in 3 separate transplant cohorts comprising 1345 MHC-identical sibling transplant pairs, which suggested a statistically significant association between donor/recipient disparity at this locus and risk of developing acute GVHD.39 Testing this association in other large transplant cohorts is an important research priority.
H-Y–specific immune responses in sex-mismatched HCT
Sex-mismatched allogeneic HCT represents a unique situation in which the donor and recipient genomes are discordant for a large, complex genetic element: the Y chromosome. Although most of the genes on the human Y chromosome are unique to the Y and not found in a female genome, most of these are expressed only within the testis, which is an immunologic sanctuary.40,41 A small subset of ∼ 15 Y chromosome–specific genes, however, are expressed both within the testis and in a broad range of other tissues,42 including both normal hematopoietic progenitor cells,43 mature blood cells, and malignant blood cells. When expressed outside the immunologic sanctuary of the testis, the products of several of these genes are potently immunogenic and can elicit both CD8+ and CD4+ male-specific T-cell responses in MHC-matched but sex-mismatched donor/recipient pairs. The male-specific minor H antigens encoded by these Y chromosome genes are known as H-Y antigens.
Human H-Y–specific T cells were first identified in a heavily transfused female patient with aplastic anemia who had rejected a bone marrow graft from her MHC-identical brother.44,45 H-Y–specific T cells can also be readily isolated from male recipients of MHC-matched female grafts.46 At least 6 Y chromosome genes, including KDM5D (formerly known as SMCY), UTY, DDX3Y, USP9Y, TMSB4Y, and RPS4Y, encode H-Y antigens recognized by either CD8+ or CD4+ T cells,47-52 and 4 of these genes encode multiple H-Y antigens that are presented by multiple MHC alleles. A recent study reported that KDM5D and UTY each encode ≥ 6 distinct H-Y epitopes that are presented by HLA-A*02:01 alone,53 suggesting that the total number of H-Y epitopes encoded by these genes is large. The capacity to encode multiple minor H antigens distinguishes the H-Y genes from most of the autosomal minor antigen-encoding genes highlighted in Figure 5, which typically encode only a single minor H antigen presented by a single MHC allele.
The products of the H-Y genes are also the targets of antibody responses that commonly develop in male recipients of female hematopoietic cell grafts.54,55 Simultaneous CD4+ T-cell and antibody responses to specific H-Y gene products such as DDX3Y suggest a coordinated T- and B-cell response.56,57 Similar to the H-Y–specific T-cell response, the main targets of the H-Y–specific antibody response appear to be KDM5D, UTY, and DDX3Y.55,58
Clinical observations suggest that, in the aggregate, female immune responses against H-Y antigens have a measurable effect on the outcome of sex-mismatched allogeneic HCT. The highest rate of graft rejection after MHC-matched HCT for aplastic anemia is observed in female recipients of MHC-matched male grafts,59,60 suggesting that recipient T cells recognizing H-Y antigens on donor hematopoietic progenitor cells may contribute to rejection. H-Y–specific T-cell responses are also probably important in transplantations with sex mismatch in the opposite direction. Male recipients of grafts from MHC-matched female donors have the highest rates of both acute and chronic GVHD of any donor/recipient sex combination,61-65 and the duration of immune suppressive therapy required to treat chronic GVHD in male recipients of female grafts is longer than for any other donor/recipient sex combination.66 These observations suggest that H-Y–specific T-cell responses make a measurable contribution to GVHD in male recipients with female donors. The rate of relapse after transplantation is also lower in these patients than in any other donor/recipient sex combination,61,63,65 suggesting that H-Y–specific T-cell responses contribute to GVL activity as well as to GVHD. Antibody responses to H-Y proteins are also associated with both chronic GVHD and maintenance of remission,55 but whether these antibody responses contribute meaningfully to GVHD, or simply serve as markers for it, remains unclear. The decrease in relapse rate observed in male recipients with female donors is more than negated by the increased morbidity and mortality that they experience because of GVHD, however, and, consequently, these patients do not experience improved survival.65
Unlike the autosomal loci that typically encode a single minor H antigen presented by a single MHC allele, Y chromosome genes such as KDM5D encode multiple H-Y antigens that are presented by multiple MHC alleles. H-Y–specific responses thus probably develop in a larger proportion of any given transplant cohort. The H-Y genes also encode both class I– and class II–restricted minor H antigens that stimulate CD8+ and CD4+ T cells, respectively, which is usually not the case for autosomal minor H loci. H-Y genes are encoded on a single genetic element, the Y chromosome, and are therefore in complete linkage disequilibrium. Finally, H-Y proteins also elicit B-cell responses in sex-mismatched graft recipients, which, with the single exception of UGT2B17, have not yet been observed with autosomally encoded minor H antigens. Consequently, simultaneous and potentially synergistic T- and B-cell immune responses to multiple H-Y antigens are possible, and in fact probable, in male recipients with female donors.
How many distinct minor H loci probably exist?
The exploration of human genetic variation has been one of the most exciting and productive scientific efforts of the past decade. Global collaborative initiatives such as the HapMap Project67-69 and the 1000 Genomes Project70 have found that genetic diversity both within and between human populations is far greater than was appreciated when the first reference genome was completed in 2000. The fruits of these initiatives have not only profoundly accelerated the identification of autosomal loci that encode minor H antigens,38,71-73 but they have also suggested that the total number of human minor H antigens is probably quite large, far larger, in fact, than the number identified to date. On the basis of data generated by the pilot phase of the 1000 Genomes Project, it has been estimated that the genome of any person will differ from the reference human genome sequence at 10 000-11 000 nsSNPs.70 By extension, therefore, the genomes of any 2 unrelated persons probably differ at a similar number of nsSNPs. The recent rapid accumulation of whole exome and genome sequence data has also enabled the identification of a steadily increasing number of exons in specific genes and entire genes that, like UGT2B17, are deleted in otherwise healthy persons.74 Peptides derived from these exons and genes could also function as minor H antigens in appropriately discordant (absent/present) donor/recipient pairs. These observations imply that the total number of minor H antigen loci that might elicit a T-cell response in the allogeneic HCT setting is extremely large, and, therefore, that all allogeneic donor/recipient transplant pairs, regardless of their biologic relation, will be mismatched for a considerable number of minor H antigens.
Can we find autosomal minor H antigens that are particularly important in HCT?
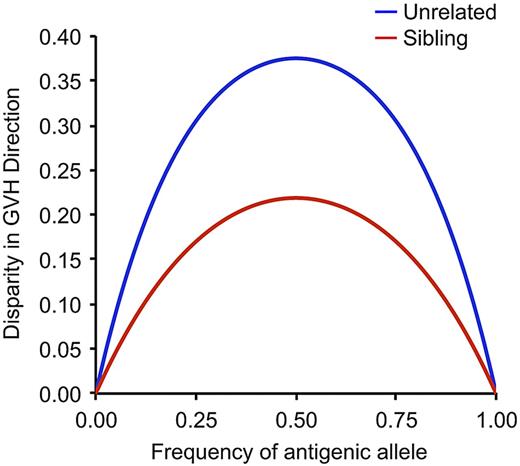
Given the large number of autosomal minor H loci that probably exist, are there any at which donor/recipient disparity is clinically significant, and, if so, how might we identify them? How large a transplant cohort, for example, would be required to determine whether disparity at a specific minor H locus is significantly associated with the risk of developing acute GVHD? The requisite cohort size depends on both the magnitude of the effect associated with donor/recipient disparity at that locus and the frequencies of the antigenic and nonantigenic alleles at that locus in the transplant population. The expected donor/recipient disparity at a hypothetical minor H locus will be maximal when the frequency of the antigenic allele in the population from which the donor and recipient are drawn is 0.5 and declines to 0 as this allele frequency approaches the 2 extremes of 0 and 1 (Figure 6).75 In addition, at any given frequency of the antigenic allele, the expected disparity will be greater for unrelated pairs than for full sibling donor/recipient pairs. With the use of the distribution of expected disparity as a function of antigenic allele frequency, one can calculate the requisite size of a transplant cohort that would need to be studied to determine with 90% power whether donor/recipient disparity for a specific minor H antigen is associated with a 10% or 20% relative increase in the risk of acute GVHD (Table 1), assuming that all donor/recipient pairs in the cohort express the appropriate MHC-restricting allele for the minor H antigen in question. This analysis does not take immunodominance into account. The requisite cohort size is always smaller for unrelated donor/recipient pairs than for full siblings. Nonetheless, almost 4500 unrelated donor/recipient pairs are needed to provide 90% power to detect a 10% increase in the risk of acute GVHD from 50% to 55% if the frequency of the antigenic allele is 0.5, and donor/recipient disparity for the minor H antigen is therefore maximal (Table 1). This cohort size is far larger than the number of donor/recipient pairs available for analysis at any transplantation center in North America. Definitive identification of clinically significant autosomal minor H antigen loci will require large, coordinated multicenter studies with many thousands of participating donor/recipient pairs.
Expected donor/recipient disparity at a hypothetical locus encoding a minor histocompatibility antigen is a function of the frequency of the antigenic allele and, for any given frequency of the antigenic allele, is greater for unrelated than for sibling donor/recipient pairs. Adapted from Martin.75
Expected donor/recipient disparity at a hypothetical locus encoding a minor histocompatibility antigen is a function of the frequency of the antigenic allele and, for any given frequency of the antigenic allele, is greater for unrelated than for sibling donor/recipient pairs. Adapted from Martin.75
Statistical power analysis
| Frequency of antigenic allele . | GVHD . | |
|---|---|---|
| 50% vs 55% . | 50% vs 60% . | |
| 0.1 | ||
| Unrelated donor | 7642 | 1891 |
| Sibling donor | 13 330 | 3296 |
| 0.2 | ||
| Unrelated donor | 5326 | 1319 |
| Sibling donor | 8362 | 2069 |
| 0.3 | ||
| Unrelated donor | 4723 | 1169 |
| Sibling donor | 7490 | 1853 |
| 0.4 | ||
| Unrelated donor | 4519 | 1119 |
| Sibling donor | 6296 | 1558 |
| 0.5 | ||
| Unrelated donor | 4469 | 1106 |
| Sibling donor | 6128 | 1517 |
| Frequency of antigenic allele . | GVHD . | |
|---|---|---|
| 50% vs 55% . | 50% vs 60% . | |
| 0.1 | ||
| Unrelated donor | 7642 | 1891 |
| Sibling donor | 13 330 | 3296 |
| 0.2 | ||
| Unrelated donor | 5326 | 1319 |
| Sibling donor | 8362 | 2069 |
| 0.3 | ||
| Unrelated donor | 4723 | 1169 |
| Sibling donor | 7490 | 1853 |
| 0.4 | ||
| Unrelated donor | 4519 | 1119 |
| Sibling donor | 6296 | 1558 |
| 0.5 | ||
| Unrelated donor | 4469 | 1106 |
| Sibling donor | 6128 | 1517 |
Statistical power analysis to estimate the number of transplant donor/recipient pairs that would need to be studied to provide 90% power to detect a hypothetical 10% (50%-55%) or 20% (50%-60%) increase in the risk of developing acute GVHD attributable to donor/recipient disparity at a specific minor H locus, as a function of (1) the frequency of the antigenic allele in the population from which the donor and recipient are derived, and (2) the relationship of the donor to the recipient (unrelated vs full sibling). It is assumed for this analysis that the donor/recipient pairs are matched for HLA-A, -B, -C, and -DRB1, and that they all express the MHC-restricting allele for the minor H antigen. The requisite number of pairs when the frequency, f, of the antigenic allele is > 0.5 is the same as the requisite number when the frequency of the antigenic allele is 1 − f.
Genome-wide characterization of donor/recipient genetic identity in related and unrelated pairs
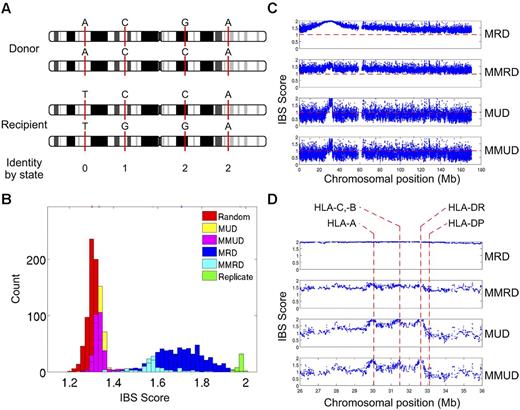
Analysis of donor and recipient genotypes at ∼ 500,000 SNP loci distributed across the genome with the use of high-density SNP arrays confirms the long-held assumption that donor/recipient genetic identity in related and unrelated pairs,76 both of whom are selected solely for identity across specific exons in HLA-A, -B, -C, and -DRB1, differs substantially both within and outside the MHC (Figure 7). Across the entire genome, related donor/recipient pairs, as expected, show greater average genetic identity than unrelated pairs (Figure 7B-C). Over the 4-megabase interval that contains the MHC, both related and unrelated pairs show peaks of near-perfect genetic identity, reflecting the selection for identity across specific exons in HLA-A, -B, -C, and -DRB1. The fine structure of donor/recipient genetic identity within the MHC in related and unrelated pairs, however, is profoundly different. Whereas related MHC-matched pairs show a single broad peak of perfect genetic identity that encompasses the entire MHC, reflecting the inheritance of 2 identical chromosome segments spanning this region (identity by descent), unrelated MHC-matched pairs show incomplete genetic identity in the intervals between HLA-A, -B/C, and -DRB1, reflecting the fact that these pairs are selected solely for identity by state at HLA-A, -B, -C, and -DRB1 (Figure 7C-D). Donor/recipient genetic identity centromeric of HLA-DRB1 decays quite rapidly in unrelated donor/recipient pairs, because this region contains multiple hot spots of recombination (Figure 8).
SNP genotyping with the use of high-density DNA arrays shows the fine structure of donor-recipient genetic identity both within and outside the MHC. (A) Definition of the concept of donor/recipient genetic identity by state (IBS). The genotypes of a hypothetical HCT donor/recipient pair at 4 SNPs located in different intervals on chromosome 1 are indicated by the letters above the corresponding positions on the donor and recipient chromosomes; the IBS score between donor and recipient at each of the 4 SNPs (either 0, 1, or 2 alleles shared) is indicated at the bottom of the panel immediately below the position of each SNP. (B) Distribution of the average IBS across 14 098 SNPs, all with minor allele frequency ≥ 0.2, on chromosome 6 for 1378 HCT donor/recipient pairs who received a transplant at the Fred Hutchinson Cancer Research Center between 1992 and 2004; genotypes were determined with the Affymetrix Human SNP 5.0 chip.76 The transplant pairs were classified as matched related donor (MRD; n = 595), mismatched related donor (MMRD; n = 122), matched unrelated donor (MUD; n = 347), or mismatched unrelated (MMUD; n = 302) pairs based on their relationship to one another and on their degree of HLA matching, as determined by sequencing of their HLA-A, -B, -C, -DRB1, and -DQB1 alleles (matched, 10 of 10 alleles; mismatched, < 10 of 10 alleles). The distribution of average IBS across the same 14 098 SNPs between 661 randomly selected pairs of persons in the cohort and between replicate genotypes for 45 persons is also shown. (C) The haplotype-based IBS in a sliding window of 7 adjacent SNPs calculated for all 26 814 SNPs on chromosome 6 with < 10% missing genotypes is plotted for the MRD, MMRD, MUD, and MMUD donor/recipient pairs. The haplotype-based IBS in a window of SNPs is the statistical expectation of the number of haplotypes shared by the donor and recipient, given their unphased SNP genotypes within that interval. (D) Magnified view of the haplotype-based IBS data from panel C for all of the SNPs in a 10-Mb interval on chromosome 6p that spans the entire MHC. The location of the HLA-A, -C, -B, -DRB1, and -DQB1 genes is indicated by the dashed vertical red lines.
SNP genotyping with the use of high-density DNA arrays shows the fine structure of donor-recipient genetic identity both within and outside the MHC. (A) Definition of the concept of donor/recipient genetic identity by state (IBS). The genotypes of a hypothetical HCT donor/recipient pair at 4 SNPs located in different intervals on chromosome 1 are indicated by the letters above the corresponding positions on the donor and recipient chromosomes; the IBS score between donor and recipient at each of the 4 SNPs (either 0, 1, or 2 alleles shared) is indicated at the bottom of the panel immediately below the position of each SNP. (B) Distribution of the average IBS across 14 098 SNPs, all with minor allele frequency ≥ 0.2, on chromosome 6 for 1378 HCT donor/recipient pairs who received a transplant at the Fred Hutchinson Cancer Research Center between 1992 and 2004; genotypes were determined with the Affymetrix Human SNP 5.0 chip.76 The transplant pairs were classified as matched related donor (MRD; n = 595), mismatched related donor (MMRD; n = 122), matched unrelated donor (MUD; n = 347), or mismatched unrelated (MMUD; n = 302) pairs based on their relationship to one another and on their degree of HLA matching, as determined by sequencing of their HLA-A, -B, -C, -DRB1, and -DQB1 alleles (matched, 10 of 10 alleles; mismatched, < 10 of 10 alleles). The distribution of average IBS across the same 14 098 SNPs between 661 randomly selected pairs of persons in the cohort and between replicate genotypes for 45 persons is also shown. (C) The haplotype-based IBS in a sliding window of 7 adjacent SNPs calculated for all 26 814 SNPs on chromosome 6 with < 10% missing genotypes is plotted for the MRD, MMRD, MUD, and MMUD donor/recipient pairs. The haplotype-based IBS in a window of SNPs is the statistical expectation of the number of haplotypes shared by the donor and recipient, given their unphased SNP genotypes within that interval. (D) Magnified view of the haplotype-based IBS data from panel C for all of the SNPs in a 10-Mb interval on chromosome 6p that spans the entire MHC. The location of the HLA-A, -C, -B, -DRB1, and -DQB1 genes is indicated by the dashed vertical red lines.
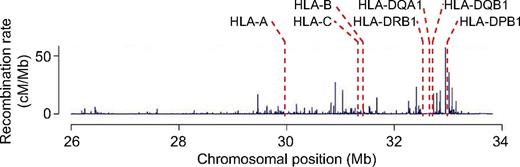
Weak linkage between the HLA-DP loci and the class I and telomeric portion of the class II regions of the MHC is primarily attributable to hot spots of recombination that lie just telomeric of HLA-DP. Map of observed recombination rate within an 8-Mb interval of chromosome 6p that spans the classic MHC. Adapted from de Bakker et al with permission.110
Weak linkage between the HLA-DP loci and the class I and telomeric portion of the class II regions of the MHC is primarily attributable to hot spots of recombination that lie just telomeric of HLA-DP. Map of observed recombination rate within an 8-Mb interval of chromosome 6p that spans the classic MHC. Adapted from de Bakker et al with permission.110
This genome-wide analysis of donor/recipient genetic identity indicates that the benefit of matching donors and recipients for HLA-A, -B, -C, and -DRB1 differs in related and unrelated pairs. Full siblings who have inherited from their parents the same 2 segments of chromosome 6p spanning the MHC will, with high probability, also be identical at HLA-DQ and -DP, as well as at all other loci within the MHC. This, in general, is not true for unrelated donor/recipient pairs selected solely for identity across specific exons within HLA-A, -B, -C, and -DRB1. Despite identity at these 4 critical loci, a large proportion of unrelated pairs have genetic disparity at other MHC loci such as MICA, MICB, HLA-DQ, HLA-DP, and many loci in the class III region, among many others. The probability of disparity in unrelated pairs at HLA-DP is particularly high, 80% in large studies,77 because of the recombination hot spots located between HLA-DR and HLA-DP (Figure 8) and the resulting weak linkage that exists between these 2 loci.
Dissecting the effect of genetic disparity within the MHC in unrelated HCT
The MHC exhibits unique patterns of extended haplotype structure,78,79 and an analysis of haplotype mismatching within the MHC in unrelated donor/recipient pairs that were allele-matched at HLA-A, -B, -C, -DRB1, and -DQB1 found that MHC haplotype mismatching was associated with a significantly increased risk of acute GVHD.80 Subsequent analysis of highly conserved MHC haplotypes in a cohort of HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 matched Japanese unrelated pairs confirmed that information encoded within the haplotype influences the risk of developing acute GVHD.79 The identity of additional MHC loci at which donor/recipient mismatching may influence transplantation outcomes such as GVHD in unrelated HCT remains the focus of considerable current investigation and has prompted numerous analyses of donor/recipient matching at novel candidate loci such as MICA81,82 as well as at more traditional candidates such as HLA-DQ and HLA-DP.
The clinical significance of donor/recipient identity and nonidentity at HLA-DP in unrelated HCT has received far more scrutiny than any other candidate MHC locus. This reflects the fact that the products of HLA-DPA1 and HLA-DPB1 form heterodimers that are highly homologous to HLA-DR and HLA-DQ molecules and are constitutively expressed on the surface of many types of both normal and malignant hematopoietic cells, where they present a diverse repertoire of peptides, as yet poorly characterized, to CD4+ T cells. The expression of HLA-DP molecules on both normal and malignant hematopoietic cells implies a potential role for HLA-DP–restricted T cells in both GVHD and GVL. Laboratory analyses of peripheral blood T cells from recipients of HLA-DP–mismatched grafts have shown prominent HLA-DP–restricted T-cell responses associated with graft rejection,83,84 acute GVHD,85,86 and GVL activity after donor leukocyte infusion.87 A large genome-wide association study in Japanese patients identified a SNP in the HLA-DP region at which donor/recipient mismatch was significantly associated with GVHD.88 In addition, 3 large retrospective analyses of HLA-DP matching in unrelated donor HCT have found significant associations between matching status and the incidence of relapse after transplantation.77,89,90 A number of studies have suggested that not all HLA-DP mismatches are equivalent and have led to a model of “permissive” and “nonpermissive” mismatches,84,91-94 but this model conflicts with results of several recent laboratory studies of HLA-DP–specific T-cell alloreactivity.95-97 Nonetheless, a recent international study of 8539 unrelated donor transplantations classified according to this model as having permissive, nonpermissive, or no HLA-DP mismatch found a significant association between the presence of nonpermissive mismatches and decreased overall survival.77
Clinical significance of genetic variation in genes encoding NK and γδ T-cell receptors and their ligands
Natural killer (NK) cells and T lymphocytes bearing γδ T-cell receptors comprise a small fraction of the lymphocytes in peripheral blood but nonetheless make important contributions to immune interactions that mediate GVHD and GVL after allogeneic HCT. NK cells express a complex array of activating and inhibitory receptors, many of which are encoded in the killer cell immunoglobulin-like receptor (KIR) locus within the leukocyte receptor complex on chromosome 19q13.4, or in the natural killer complex (NKC) on chromosome 12p13 (Figure 5). The KIR genes are both highly polymorphic and highly homologous to one another, and the gene content of the KIR locus is itself polymorphic, with a variety of KIR haplotypes found in the population that contain a core set of framework genes (KIR3DL3, KIR3DP1, KIR2DL4, and KIR3DL2) found in all virtually haplotypes, along with a variable number of additional KIR genes. The natural ligands for several of the inhibitory KIR receptors are epitopes present on subsets of HLA-B and -C molecules; the natural ligands for most of the activating KIR receptors remain poorly defined. T cells bearing γδ receptors exhibit features of both innate and adaptive immune cells. Approximately one-half of all γδ T cells in blood express a highly conserved Vγ9Vδ2 TCR that recognizes a range of endogenous and microbial phosphoantigens.98,99 γδ T cells comprise a majority of the intraepithelial T cells in the gut, and intestinal γδ T cells primarily express Vδ1 TCRs that bind CD1c molecules,100 as well as MICA and MICB, polymorphic MHC class I–like molecules that are encoded in the MHC.101 Most γδ T cells also express NKG2D, the product of the KLRK1 gene in the NKC; NKG2D is also a receptor for MICA, MICB, and other stress-induced proteins such as the UL16-binding proteins.
The functional significance of polymorphism in the genes encoding NK and γδ TCRs and their ligands, particularly as it pertains to allogeneic HCT, is poorly defined. It is unknown, for example, whether the products of the allelic variants of the different KIR genes have different affinities for their specific epitopes on HLA-B and -C molecules, or perhaps recognize different epitopes altogether. Although the polymorphism in MICA and MICB map preferentially to the contact region with NKG2D,102 the extent to which this polymorphism affects the interaction of MICA and MICB with NKG2D has not been comprehensively defined. Because many of the genes encoding NK and γδ TCRs and their ligands are located not in the MHC, but rather in the KIR locus or the NKC, transplant donor/recipient pairs selected only for MHC identity will exhibit genetic identity for the KIR locus or the NKC in ∼ 25% of full sibling pairs but rarely, if ever, in unrelated pairs. Much current research, however, is focused on dissecting how donor and recipient KIR and NKC genotypes influence transplantation outcome, and in particular how the products of donor KIR genes expressed by donor NK and T cells may interact with the class I MHC molecules expressed by recipient cells.103-106 Recent studies have reported that specific donor KIR haplotypes or genes are associated with a significantly reduced rate of relapse in patients with acute myelogenous leukemia undergoing T-replete unrelated donor103,104 or T-depleted sibling donor105 HCT, and with reduced risk of acute GVHD after unrelated HCT, regardless of recipient diagnosis.106
Summary and conclusions
Matching the donor and recipient for the short DNA sequences that encode the peptide-binding regions of HLA-A, -B, -C, and -DRB1 does not also match them for the diverse set of peptides presented by those molecules on the surface of donor and recipient cells. The enormous sequence and structural variations that characterize the human genome ensures that the MHC-associated immunopeptidomes of no 2 persons will be the same, unless they are identical twins, and therefore also ensures that all allogeneic donor/recipient transplant pairs, regardless of MHC identity, will be mismatched for scores if not hundreds of minor H antigens. There are as yet no consistently validated data to show that donor/recipient mismatch for any single minor H antigen created by an autosomal SNP is associated with a measurable effect on transplantation outcome, although measurable effects may indeed exist. The effects of donor/recipient disparity for UGT2B17, or of donor/recipient sex mismatch in allogeneic HCT, which are probably mediated by donor immune responses against the UGT2B17 or Y chromosome–encoded proteins, respectively, are clinically measurable because donor/recipient disparity for UGT2B17 or for the Y chromosome enables simultaneous coordinated CD8+ and CD4+ T-cell as well as B-cell responses against multiple epitopes. The identification of autosomal SNP-encoded minor H antigens at which donor/recipient disparity is clinically significant will probably require multicenter studies with large numbers of donor/recipient pairs, because the effect of disparity at any single autosomal SNP is probably small and any associated signal will be difficult to identify in the background because of responses against the hundreds or thousands of other such disparities. The search for clinically significant autosomal SNPs will also more readily succeed if the study population comprises MHC-identical sibling pairs. Although the expected donor/recipient disparity at any autosomal locus will be lower in sibling compared with unrelated pairs, making it necessary to study larger cohorts, the potent effect of donor/recipient disparity at loci within the MHC, particularly disparity at HLA-DP, which exists in 80% of unrelated pairs, is likely to obscure the effect of coexisting disparity at autosomal loci. Transplant donor/recipient pairs, related and unrelated alike, selected for identity at the MHC will rarely, if ever, be matched at the highly polymorphic KIR locus on chromosome 19 and the NKC on chromosome 12. Determining whether donor/recipient genetic identity at KIR or the NKC is associated with any clinical benefit, or any clinical cost, will therefore be extremely challenging. It seems probable that the specific pairing of donor KIR and NKC genotypes with recipient MHC genotype will be a more important determinant of transplantation outcome than the identity or nonidentity between donor and recipient KIR and NKC genotypes.
Acknowledgments
The authors thank Kathleen Kroeger of the Cytogenetics Laboratory at the Seattle Cancer Care Alliance for her assistance with the preparation of Figure 5 and Helen E. Heslop of Baylor College of Medicine for her helpful comments on the manuscript.
This work was supported by the National Institutes of Health (grants HL094260, HL105914, and AI033484).
National Institutes of Health
Authorship
Contribution: E.H.W. proposed this review and wrote the manuscript; and X.C.Z., S.L., W.F., B.E.S., and P.J.M. generated and analyzed data presented in Table 1 and Figures 6 and 7. All authors read and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Edus H. Warren, Program in Immunology, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D3-100, PO Box 19024, Seattle WA 98109-1024; e-mail: ehwarren@u.washington.edu.