Abstract
Approximately 10% of B-cell acute lymphoblastic leukemias (B-ALLs) overexpress the cytokine receptor subunit CRLF2, which may confer a poor prognosis. CRLF2 binds its ligand thymic stromal lymphopoietin (TSLP) as a heterodimer with IL7R. Subsets of CRLF2-overexpressing B-ALLs also have a gain-of-function CRLF2 F232C mutation or activating mutations in JAK2. Whether these mutant alleles confer differences in signaling has not been addressed. Through a domain mutation analysis, we demonstrate a distinct dependence on the CRLF2 intracellular tyrosine Y368 in signaling by CRLF2 F232C, but not signaling induced by TSLP or through CRLF2/mutant JAK2. In contrast, CRLF2 signaling in each context is strictly dependent on both the CRLF2 box1 domain and the intracellular tryptophan W286. Using a global quantitative analysis of tyrosine phosphorylation induced by TSLP, we previously identified TSLP-induced phosphorylation of multiple kinases implicated in B-cell receptor signaling, including Lyn, Btk, Hck, Syk, MAPK8, MAPK9, and MAPK10. We now demonstrate that cells dependent on CRLF2/mutant JAK2 have reduced phosphorylation at these targets, suggesting that the kinases promote TSLP-mediated proliferation but serve as negative regulators of CRLF2/mutant JAK2 signaling. Thus, targetable nodes downstream of CRLF2 differ based on the presence or absence of additional mutations in CRLF2 signaling components.
Introduction
The cytokine thymic stromal lymphopoietin (TSLP) was first identified as a growth factor that could support murine B-cell development in the absence of IL7.1 Multiple groups subsequently cloned the mouse receptor for TSLP,2-5 which was termed cytokine receptor-like factor 2 (CRLF2), and then the human TSLP6 and CRLF2.7,8 Both mouse and human CRLF2 signal as heterodimers with the IL7R subunit, but the homology across species is limited for both TSLP and CRLF2. Thus, the mouse ligand does not signal through the human receptor complex and vice versa.
Several differences in signaling exist between the murine and human TSLP receptors. Homodimerization of the intracellular portion of murine Crlf2 by fusion to an alternate transmembrane domain does not promote signaling and proliferation2,3 but homodimerization of the human CRLF2 does.9,10 Human TSLP robustly stimulates proliferation and phosphorylation of STAT transcription factors in myeloid dendritic cells and CD4+ T cells that are dependent on JAK1 and JAK2.11,12 In contrast, murine Tslp weakly stimulates phosphorylation of Stat1 and Stat5 in mouse CD4+ T cells13 and Jak1 and Jak2 are dispensable for murine Tslp-dependent proliferation.14
We and others recently identified rearrangements of the CRLF2 locus in 5%-10% of adult and pediatric B-lineage acute lymphoblastic leukemias (B-ALLs).15-18 Rearrangement links the full-length coding sequence of CRLF2 to alternate transcriptional control, either through translocation with the immunoglobulin heavy chain locus or through an intrachromosomal deletion involving P2RY8. Additional cases of B-ALL that lack CRLF2 locus rearrangements highly express CRLF2 through unknown mechanisms.19 The majority of CRLF2 overexpressing B-ALLs also harbor gain-of-function mutations in a component of the TSLP signaling complex; these include: (1) mutations in JAK2 and less commonly JAK1, (2) cysteine substitutions in IL7R that promote constitutive homodimerization or heterodimerization with CRLF2,20 and (3) rarely an F232C substitution in the juxtamembrane region of CRLF2, which results in constitutive homodimerization through a disulfide bridge.18,21
Although 30%-50% of CRLF2-rearranged B-ALL harbor activating JAK2 mutations, multiple aspects of this association remain unexplained. The JAK2 mutations in B-ALL primarily affect exon 16, whereas the JAK2 V617F mutation observed in more than 50% of myeloproliferative neoplasms22,23 has never been reported in B-ALL.24 This is somewhat in contrast with the JAK1 mutations observed in CRLF2-rearranged B-ALL. Among the previously reported JAK1 mutations are JAK1 V658F, which is the corresponding mutation to JAK2 V617F.24 Of note, the overwhelming majority of B-ALL cases with JAK2 mutations (and most cases with JAK1 mutations) also overexpress CRLF2.15-18,25 Cytokine receptor association is required for JAK enzymatic activity,26 but JAK2 can signal through many different cytokine receptors. It remains unclear why CRLF2 appears to be an essentially obligate scaffold for mutant JAK2 signaling in B-ALL. The close association between CRLF2 and JAK2 exon 16 mutants suggests a requirement for cooperative signaling, yet a large fraction of CRLF2-rearranged B-ALL cases lack JAK2 mutations. The differences in signaling downstream of CRLF2/mutant JAK2 or CRLF2 in the absence of a JAK2 mutation have not been explored.
Several previous studies have used the IL3-dependent Ba/F3 cell line to interrogate aspects of human CRLF2 signaling. Exposure of Ba/F3 cells coexpressing human CRLF2 and IL7R to TSLP drives IL3-independent proliferation (see Figure 1A) as well as activation of Stat1, Stat3, and Stat5 that is dependent on Jak1 and Jak2,7,27 which recapitulates signaling in human cells. Although TSLP signaling requires CRLF2, IL7R, JAK2, and JAK1, the mutant alleles of CRLF2 and JAK2 can signal without the full complex. Most notably, IL7R and TSLP are dispensable for signaling through either CRLF2 F232C alone or CRLF2 with JAK2 mutants in BaF3 cells (Figure 1A), although signaling through these alleles also promotes the phosphorylation of Stat5.16,18,20
High expression of CRLF2 is associated with a poor prognosis in adult and high-risk pediatric cases of B-ALL.18,19 For example, Chen et al recently reported that in a cohort of 499 high-risk pediatric B-ALL cases, CRLF2 overexpression and minimal residual diseases were the only factors statistically associated with a reduced relapse-free survival.19 Aberrant TSLP signaling also plays a role in Th2 responses that drive atopic disorders,28 as well as tumor-associated stroma in breast29,30 and pancreatic cancers.31 Thus, there is a clear therapeutic need for agents that target CRLF2 signaling in both malignant and nonmalignant diseases.32
We took 2 approaches to explore the differences between signaling in response to TSLP and signaling involving mutant alleles of CRLF2 and JAK2. First, we performed a domain mutation analysis to define the essential components of CRLF2 necessary for proliferation and JAK/STAT activation in response to TSLP, through CRLF2 F232C or through CRLF2 with mutant JAK2. Second, we built on our previous study of TSLP signaling33 to carry out a global quantitative phosphoproteomic analysis of CRLF2/mutant JAK2-induced tyrosine phosphorylation.
Methods
Cell culture and reagents
Ba/F3 (ATCC) cells were maintained in RPMI 1640 medium (Invitrogen) with 10% FCS (Invitrogen) and 500 pg/mL IL3 (Millipore), or 1 ng/mL TSLP (R&D Systems). Ba/F3 were stably transduced with CRLF2 (MSCVpuro), IL7R (MSCV-IRES-GFP), JAK2 (MSCVneo), and mouse Jak2 (MSCV-IRES-GFP) with or without activating mutations in the pseudokinase domain (R683G/S/T or V617F) as indicated. The B-ALL cell lines MUTZ-5 and MHH-CALL4 were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen, and grown in RPMI 1640 with 20% FBS. Construction of plasmids by PCR cloning and retroviral infections were performed as previously described.18,34
Proliferation assays
Proliferation was measured manually by cell counting every 2 days, starting with a concentration of 105 cells/mL. Cells were washed twice before plating to remove residual IL3.
Fluorescence assorted cell sorting
To test the cell surface expression of CRLF2 and IL7R, approximately 1 × 106 Ba/F3 cells were stained for 30 minutes with phycoerythrin (PE)–labeled anti–human TSLP receptor antibody (eBioscience; 1:20 dilution) and/or AF647-labeled hIL7R/CD127 (BD Bioscience; 1:100 dilution) at room temperature. Next, cells were washed, followed by fluorescence detection using a BD FACSCanto II flow cytometer (BD Bioscience). Ba/F3 cells transduced with both CRLF2 and IL7R constructs were (after selection for puromycin resistance), sorted for GFP positivity to obtain a homogenous CRLF2/IL7R Ba/F3 population.
siRNA and nucleofection
For each siRNA experiment, 2 × 106 Ba/F3 cells were resuspended in 100 μL cell line nucleofector solution V (Lonza) containing 300mM of either siRNA against mouse Jak2 (Thermo Scientific) or nontargeting siRNA (Thermo Scientific). Cells were nucleofected using the Nucleofector II device (Amaxa Biosystems) according to the manufacturer's instructions and cultured in 2.5 mL of media.
Additional methods can be found in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results
CRLF2 and mutant JAK2 directly interact
To assay CRLF2 signaling, we used the well-established Ba/F3 cell line, which is absolutely dependent on IL3 for growth. We ectopically expressed wild-type components of the TSLP signaling complex or mutant alleles identified in patients with B-ALL (Figure 1A). We previously demonstrated that expression in Ba/F3 cells of: (1) CRLF2 with IL7R and exposure to TSLP, (2) CRLF2 with JAK2 R683G, or (3) CRLF2 F232C alone promotes proliferation in the absence of IL3.18
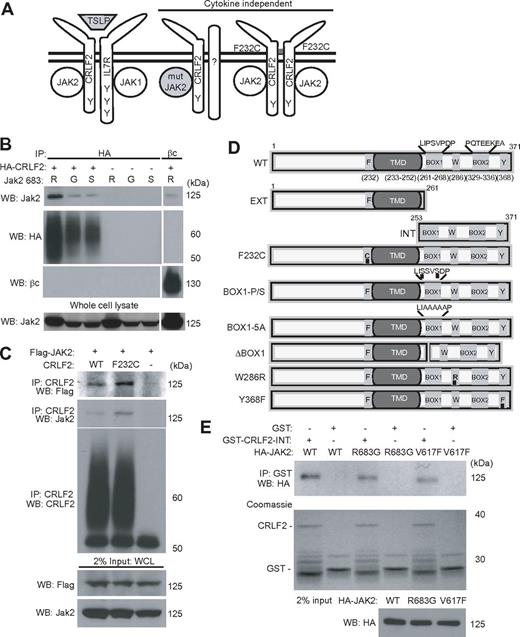
Signaling through human CRLF2 involves JAK2. (A) CRLF2 signaling can be activated by either ligand or gain-of-function mutations within CRLF2, IL7R, JAK1, or JAK2. On the left, CRLF2 can heterodimerize with IL7R and respond to TSLP through JAK1 and JAK2. In the middle, CRLF2 can signal through a gain-of-function mutant JAK2, in which case involvement of a dimerization partner is unknown. On the right, CRLF2 F232C can homodimerize through a disulfide bond.18 C-terminal tyrosines (Y) within CRLF2 and IL7R are indicated. (B) Coimmunoprecipitation of HA-CRLF2 with mouse Jak2 R683 (wild-type), R683G, and R683S in Ba/F3 lysates. Coimmunoprecipitation of wild-type Jak2 and βc is included as a control. (C) Coimmunoprecipitation of FLAG-tagged human JAK2 with CRLF2 or CRLF2 F232C in Ba/F3 lysates. (D) A series of mutation and deletion constructs were generated in CRLF2 to define the role of specific residues (F232, W286, Y368) or domains (Box 1, cytoplasmic, extracellular). Modified amino acids are underlined. Numbers above or below constructs indicate codons. (E) Coimmunoprecipitation of GST or GST-CRLF2-INT generated in E coli with HA-JAK2 (wild-type, WT; R683G or V617F) generated by in vitro translation.
Signaling through human CRLF2 involves JAK2. (A) CRLF2 signaling can be activated by either ligand or gain-of-function mutations within CRLF2, IL7R, JAK1, or JAK2. On the left, CRLF2 can heterodimerize with IL7R and respond to TSLP through JAK1 and JAK2. In the middle, CRLF2 can signal through a gain-of-function mutant JAK2, in which case involvement of a dimerization partner is unknown. On the right, CRLF2 F232C can homodimerize through a disulfide bond.18 C-terminal tyrosines (Y) within CRLF2 and IL7R are indicated. (B) Coimmunoprecipitation of HA-CRLF2 with mouse Jak2 R683 (wild-type), R683G, and R683S in Ba/F3 lysates. Coimmunoprecipitation of wild-type Jak2 and βc is included as a control. (C) Coimmunoprecipitation of FLAG-tagged human JAK2 with CRLF2 or CRLF2 F232C in Ba/F3 lysates. (D) A series of mutation and deletion constructs were generated in CRLF2 to define the role of specific residues (F232, W286, Y368) or domains (Box 1, cytoplasmic, extracellular). Modified amino acids are underlined. Numbers above or below constructs indicate codons. (E) Coimmunoprecipitation of GST or GST-CRLF2-INT generated in E coli with HA-JAK2 (wild-type, WT; R683G or V617F) generated by in vitro translation.
Both mouse Crlf2 and human CRLF2 interact with JAK2,12 whereas IL7R interacts with JAK1. To determine whether human CRLF2 interacts with gain-of-function mutants of mouse Jak2, we coexpressed CRLF2 with an N-terminal hemagglutinin (HA) tag along with Jak2 R683G or Jak2 R683S in Ba/F3 cells and performed immunoprecipitation with antibody against HA followed by immunoblotting for Jak2. CRLF2 interacts with both wild-type mouse Jak2 and the R683 mutants (Figure 1B). As a negative control, we did not detect mouse βc in the CRLF2 protein complex although we did recover Jak2 after immunoprecipitation for mouse βc, which is known to interact with Jak2 as a component of IL3 signaling.35 To determine whether wild-type CRLF2 or CRLF2 F232C specifically coimmunoprecipitate with human JAK2, we expressed triple FLAG-tagged human JAK2. As shown in Figure 1C, both CRLF2 and CRLF2 F232C interact with the tagged JAK2. Except where noted, the subsequent experiments in Ba/F3 cells all used human JAK2.
To confirm that CRLF2 and JAK2 interact within human cells, we introduced CRLF2 F232C and wild-type CRLF2 into human UT7 leukemia cells either alone or with HA-tagged JAK2. Both CRLF2 alleles coimmunoprecipitated with JAK2 (supplemental Figure 1). Wild-type CRLF2 also coimmunoprecipitated with HA-JAK2 R683G (supplemental Figure 1). In addition, JAK2 coimmunoprecipitated with CRLF2 in the human B-ALL cell lines MHH-CALL4 and MUTZ-5 (supplemental Figure 1), which both harbor IGH@-CRLF2 translocations and activating JAK2 exon 16 mutations.17
To clarify whether the CRLF2-JAK2 interaction is direct, we generated a GST-tagged cytoplasmic fragment of CRLF2 (GST-CRLF2-INT; Figure 1D) in Escherichia coli and expressed HA-tagged JAK2, JAK2 R683G, and JAK2 V617F using in vitro translation. The HA-tagged JAK2 proteins coimmunoprecipitated with GST-CRLF2-INT but not with GST alone (Figure 1E), indicating that human CRLF2 directly interacts with both wild-type and mutant JAK2.
CRLF2 box1 and W286 are essential for TSLP-induced signaling but Y368 is not
Mutation in the box1 domain or the homologous tryptophan and tyrosine residues within mouse Crlf2 is known to block Tslp-induced proliferation.36 To delineate the contributions of these cytoplasmic CRLF2 regions to signaling induced by TSLP, through CRLF2 F232C or involving the CRLF2 with JAK2 R683G, we generated a series of CRLF2 mutants within the box1 domain as well as the conserved W286 and Y368 residues (Figure 1D).
As expected, Ba/F3 cells expressing human CRLF2 or IL7R alone failed to proliferate in response to TSLP 1 ng/mL, but cells expressing both cytokine receptor subunits readily proliferated in response to TSLP (Figure 2A). Cells expressing CRLF2 W286R and IL7R failed to proliferate in response to TSLP. Similarly, mutation of the first 2 conserved proline residues in the CRLF2 box1 domain to serines (box1P/S) blocked TSLP-dependent proliferation (Figure 2A). As we previously showed,37 mutation of the single human CRLF2 intracellular tyrosine Y368 to phenylalanine had only a minor effect on proliferation (Figure 2A). In contrast, mutation of the C-terminal tyrosine (Y358) of murine Crlf2 was previously shown to block proliferation of NAG8/7 cells.36 The same findings were observed in cells harboring IL7R with wild-type or mutated CRLF2 on exposure to a higher concentration (30 ng/mL) of TSLP (supplemental Figure 2). Flow cytometric analyses using antibodies against extracellular domains of both CRLF2 and IL7R revealed that cell surface expression of both CRLF2 and IL7R is not affected by the box1, W286R, or Y368F mutation in Ba/F3 cells (supplemental Figure 4).
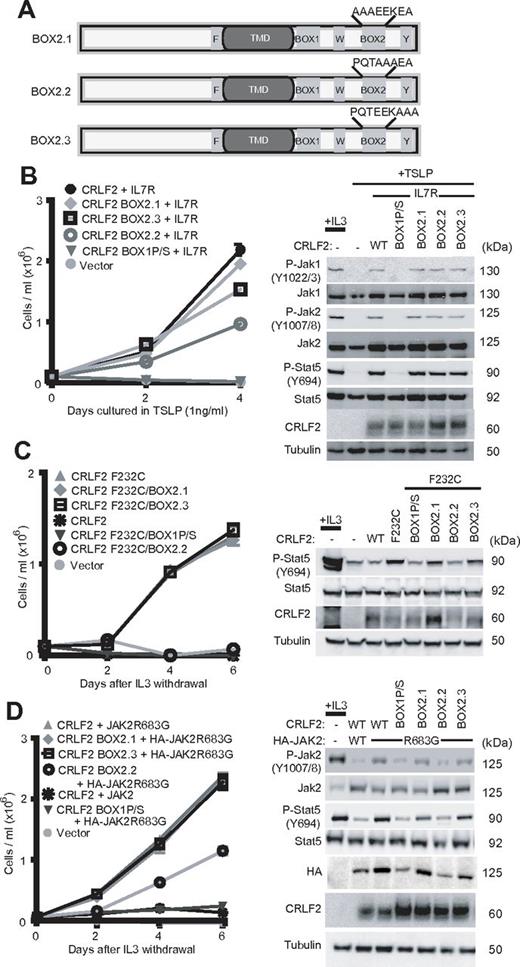
CRLF2 box1, W286R and Y368F mutations in TSLP and CRLF2 F232C signaling. (A) Ba/F3 cells expressing the indicated alleles were grown in TSLP beginning day 0 and cells were counted every other day. Inset shows immunoblotting for the indicated proteins. (B) Immunoblotting was performed with antibodies against indicated total or phospho (P−) proteins. Tyrosine phospho-sites are included. Abbreviations: WT, wild-type; P/S, box1-P/S; WR, W286R; and YF, Y368F. (C) Identical to panel A except cells were grown in the absence of cytokines. (D) Immunoblotting as in (B) P-STAT5/STAT5 indicates the ratio of band intensities quantified for each lane. (E) Ba/F3 cells expressing WT CRLF2 or CRLF2 F232C alone or with HA-tagged JAK2 R683G were subjected to immunoprecipitation (IP) using an antibody against total JAK2 followed by immunoblotting (Western blot [WB]) for the indicated proteins. WCL indicates whole cell lysates.
CRLF2 box1, W286R and Y368F mutations in TSLP and CRLF2 F232C signaling. (A) Ba/F3 cells expressing the indicated alleles were grown in TSLP beginning day 0 and cells were counted every other day. Inset shows immunoblotting for the indicated proteins. (B) Immunoblotting was performed with antibodies against indicated total or phospho (P−) proteins. Tyrosine phospho-sites are included. Abbreviations: WT, wild-type; P/S, box1-P/S; WR, W286R; and YF, Y368F. (C) Identical to panel A except cells were grown in the absence of cytokines. (D) Immunoblotting as in (B) P-STAT5/STAT5 indicates the ratio of band intensities quantified for each lane. (E) Ba/F3 cells expressing WT CRLF2 or CRLF2 F232C alone or with HA-tagged JAK2 R683G were subjected to immunoprecipitation (IP) using an antibody against total JAK2 followed by immunoblotting (Western blot [WB]) for the indicated proteins. WCL indicates whole cell lysates.
We performed immunoblotting to determine the effects of the box1, W286R, and Y368F mutations in CRLF2 on TSLP-induced phosphorylation of Jak1, Jak2, and Stat5. TSLP treatment of Ba/F3 cells coexpressing CRLF2 with IL7R resulted in increased phosphorylation (P−) of Jak1, Jak2, and Stat5, which was absent in cells expressing either the box1 or W286R mutants of CRLF2 (Figure 2B). Consistent with the data on proliferation (Figure 2A), the Y368F mutation had little or no effect on P-Jak1, P-Jak2, or P-Stat5 (Figure 2B).
We next examined the effects of these mutations on CRLF2 F232C-driven signaling. Expression of CRLF2 F232C alone promotes cytokine-independent proliferation in Ba/F3 cells, although the cells proliferate less rapidly than Ba/F3-CRLF2/IL7R cells exposed to TSLP (Figure 2C). Both the W286R and box1P/S mutations blocked CRLF2 F232C-dependent proliferation in Ba/F3 cells (Figure 2C). In contrast with the results from Ba/F3 cells dependent on TSLP, CRLF2 Y368F also completely blocked proliferation in Ba/F3-CRLF2 F232C cells (Figure 2C). Coexpression of IL7R had no effect on proliferation of either Ba/F3-CRLF2 F232C or Ba/F3-CRLF2 F232C/Y368F cells (supplemental Figure 5). Immunoblotting demonstrated a slight but consistent increase in P-STAT5 but no increase in P-Jak1 or P-Jak2 in cells dependent on CRLF2 F232C (Figures 2D and 4C). To more sensitively assay JAK2 phosphorylation, we performed immunoprecipitation of Jak2 followed by immunoblotting for P-Jak2, which showed a small increase in P-Jak2 in Ba/F3-CRLF2 F232C cells compared with Ba/F3 cells expressing wild-type CRLF2 (Figure 2E). Cells expressing CRLF2 F232C with box1, W286R, or Y368F mutation had reduced phosphorylation of Stat5, consistent with the effects on proliferation (Figure 2D). Neither the box1, W286R, nor the Y368F mutations significantly affected surface expression of either CRLF2 F232C or IL7R (supplemental Figure 4).
Next, we examined the effects of CRLF2 mutations on signaling by leukemia-associated JAK2 mutants in Ba/F3 cells. As we previously reported,18 expression of wild-type CRLF2 in combination with HA-JAK2 failed to stimulate the growth of Ba/F3 cells (Figure 3A-B). However, coexpression of CRLF2 with either JAK2 R683G or JAK2 V617F readily stimulated proliferation (Figure 3C-D). Coexpression of IL7R had no effect on proliferation of Ba/F3 cells expressing CRLF2 with wild-type JAK2, JAK2 R683G or JAK2 R683S (supplemental Figure 5). In addition, exposure to TSLP did not further promote the growth of Ba/F3 cells that expressed CRLF2/IL7R with JAK2 R683G or JAK2 R683S (supplemental Figure 5). Like signaling induced by TSLP (Figure 2A) and signaling through CRLF2 F232C (Figure 2B), both the W286R and box1P/S mutations in CRLF2 completely blocked proliferation in combination with either JAK2 R683G or JAK2 V617F (Figure 3C-D). The CRLF2 Y368F mutation had only a minor effect on proliferation in Ba/F3 cells expressing CRLF2 with JAK2 R683G or JAK2 V617F (Figure 3C-D), which mirrors human TSLP signaling (Figure 2A) but differs from cells dependent on CRLF2 F232C (Figure 2C).
CRLF2 box1, W286R and Y368F mutations in CRLF2/mutant JAK2 signaling. (A-D) Ba/F3 cells expressing the indicated alleles were grown in the absence of cytokines beginning day 0 and cells were counted every other day. Insets show immunoblotting for the indicated proteins. (E) Immunoblotting was performed with antibodies against indicated total or phospho (P−) proteins either in the presence of IL3 or 24 hours after IL3 withdrawal. Tyrosine phospho-sites are included. (F) Coimmunoprecipitation of FLAG-tagged JAK2 with CRLF2 constructs in Ba/F3 lysates. (G) Coimmunoprecipitation of GST or GST-CRLF2int (WT or carrying the indicated mutations) generated in E coli with HA-tagged JAK2 generated by in vitro translation. P/S indicates box1-P/S; WR, W286R; YF, Y368F; RG, R683G; and VF, V617F.
CRLF2 box1, W286R and Y368F mutations in CRLF2/mutant JAK2 signaling. (A-D) Ba/F3 cells expressing the indicated alleles were grown in the absence of cytokines beginning day 0 and cells were counted every other day. Insets show immunoblotting for the indicated proteins. (E) Immunoblotting was performed with antibodies against indicated total or phospho (P−) proteins either in the presence of IL3 or 24 hours after IL3 withdrawal. Tyrosine phospho-sites are included. (F) Coimmunoprecipitation of FLAG-tagged JAK2 with CRLF2 constructs in Ba/F3 lysates. (G) Coimmunoprecipitation of GST or GST-CRLF2int (WT or carrying the indicated mutations) generated in E coli with HA-tagged JAK2 generated by in vitro translation. P/S indicates box1-P/S; WR, W286R; YF, Y368F; RG, R683G; and VF, V617F.
Consistent with the data on proliferation and similar to TSLP-stimulated cells (Figure 2C), the box1 and W286R mutations in CRLF2 but not the Y368F mutation blocked the phosphorylation of Jak2 and Stat5 in Ba/F3-CRLF2/JAK2 R683G cells (Figure 3E). Of note, the level of HA-JAK2 protein was consistently higher in cell lines with functional CRLF2 signaling (Figure 3C-E), suggesting that ongoing signaling may stabilize HA-JAK2.
Mutation of the box1 domain of cytokine receptors like EPOR and GP130 can block association with JAK2.38 To determine whether the CRLF2 box1 domain is essential for interaction with human JAK2, we generated additional CRLF2 mutants with multiple alanine substitutions in the box1 domain (box1-5A) or complete deletion of the domain (Δbox1; Figure 1D). We performed coimmunoprecipitation of lysates from Ba/F3 cells expressing combinations of CRLF2 with triple FLAG-tagged human JAK2. None of the CRLF2 box1 mutants lost the ability to interact with JAK2 (Figure 3F). As expected, the CRLF2-EXT fragment that does not include the intracellular domain (Figure 1D) was unable to coimmunoprecipitate with FLAG-tagged JAK2 (Figure 3F). To clarify whether the mutant CRLF2 proteins directly interact with JAK2, we immunoprecipitated GST-CRLF2-INT polypeptides that harbor the box1P/S, W286R, or Y368F mutations after mixture with HA-JAK2. None of the mutations abrogated the direct interaction (Figure 3G), indicating that loss of CRLF2 signaling from these mutations does not involve differences in the interaction with JAK2.
The box2 domain is essential for signaling by CRLF2 F232C
In addition to the box1 domain, the cytoplasmic portion of CRLF2 contains a putative and less well-characterized box2 domain defined by (V/L)E(V/L)L.8 The box2 motif is present in several type I cytokine receptors, and is required in some cases for full activation of Jak2.39,40
To address the functional significance of the CRLF2 box2 domain, we generated multiple CRLF2 and CRLF2 F232C alleles with triple alanine substitutions (Figure 4A). We coexpressed wild-type CRLF2 or its mutants with IL7R in Ba/F3 cells and exposed the cells to TSLP 1 ng/mL or 30 ng/mL. The CRLF2-box2.2 allele had the greatest effect on proliferation, although none of the mutants completely lost the ability to proliferate in the absence of IL3 (Figure 4B and supplemental Figure 2). TSLP-induced phosphorylation of Jak1, Jak2, and Stat5 was unaffected by any of the box2 mutations (Figure 4B). In contrast, CRLF2 F232C containing the box2.2 mutation (but not box2.1 or box2.3) completely lost the ability to proliferate in the absence of IL3, which was associated with a reduction in P-Stat5 (Figure 4C). Similar to TSLP-stimulated cells, the box2.2 mutation had limited effects on proliferation and signaling in cells expressing CRLF2 with JAK2 R683G (Figure 4D). Thus, the box2 domain, similar to Y368, was essential for signaling by CRLF2 F232C but was dispensable for signaling through wild-type CRLF2, either involving mutant JAK2 or with IL7R in response to TSLP. Flow cytometry demonstrated similar cell surface expression of CRLF2 and IL7R in cells expressing wild-type CRLF2 and any of the box2 mutants (supplemental Figure 3).
Mutation of the box2 domain of CRLF2 partially block signaling. (A) Triple alanine mutations were introduced across the CRLF2 box2 domain. (B) Ba/F3 cells expressing the indicated alleles were grown in TSLP beginning day 0 and cells were counted every other day. Inset shows immunoblotting for the indicated proteins. (C-D) Identical to panel B except cells were grown in the absence of cytokines.
Mutation of the box2 domain of CRLF2 partially block signaling. (A) Triple alanine mutations were introduced across the CRLF2 box2 domain. (B) Ba/F3 cells expressing the indicated alleles were grown in TSLP beginning day 0 and cells were counted every other day. Inset shows immunoblotting for the indicated proteins. (C-D) Identical to panel B except cells were grown in the absence of cytokines.
CRLF2 F232C signaling requires Jak2
Despite the interaction of CRLF2 F232C with Jak2 (Figure 1C), immunoprecipitation followed by immunoblotting detected only a slight increase in phosphorylation of Jak2 at the well-characterized activation site Y1007/Y1008 site in Ba/F3-CRLF2 F232C cells (Figure 2E). To clarify whether this phosphorylation is associated with dependence on JAK2, we treated Ba/F3-CRLF2 F232C cells with siRNA targeting mouse Jak2 or with a control siRNA. Although the control siRNA had no effect, the siRNA targeting Jak2 reduced proliferation of Ba/F3-CRLF2 F232C cells, but not Ba/F3 cells expressing the oncogenic fusion protein ATIC-ALK, which does not signal through Jak2 (Figure 5A).
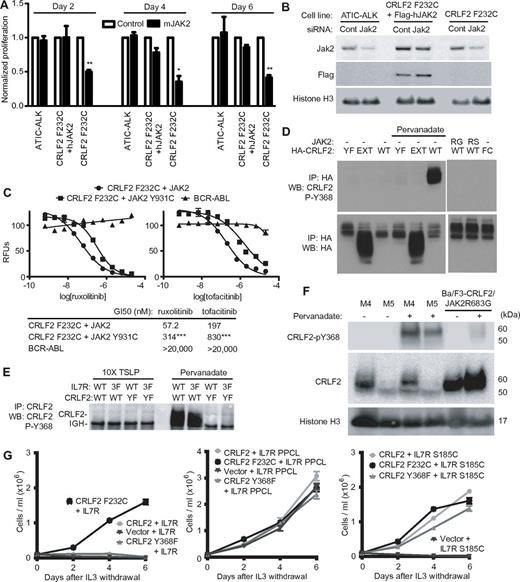
Signaling through CRLF2 F232C requires JAK2 but not phosphorylation of CRLF2 Y368. (A) siRNA against mouse Jak2 was transfected into Ba/F3 cells expressing the indicated alleles and proliferation was assayed after 48 hours (*P < .05, **P < .01 compared with control siRNA in the same line by paired, Student t test). (B) Western blotting for indicated proteins after siRNA transfection. (C) Ba/F3 cells expressing the indicated alleles were grown for 48 hours in the absence of cytokines and in the presence of increasing concentrations of compounds. RFU indicates relative fluorescence units. All assays were performed in quadruplicate. Error bars indicate the SEM (***P < .001 compared with Ba/F3-CRLF2 F232C/JAK2). (D) IP of HA-tagged CRLF2 constructs from Ba/F3 cells expressing the indicated alleles followed by WB for CRLF2 phospho-Y368 or HA. Where indicated, cells were pretreated for 15 minutes with pervanadate. FC indicates F232C. (E) IP using anti-CRLF2 antibody in Ba/F3 cells expressing WT CRLF2 or CRLF2 Y368F (YF) with WT IL7R or IL7R-3F (3F) followed by immunoblotting with a specific antibody against CRLF2 P-Y368. IGH indicates mouse immunoglobulin heavy chain. (F) MHH-CALL4 (M4) and MUTZ-5 (M5) human B-ALL cells and Ba/F3-CRLF2/JAK2 R683G cells were treated with vehicle or pervanadate and then lysates were collected and immunoblotted for the indicated proteins. (G) Ba/F3 cells expressing the indicated combinations of CRLF2 and IL7R alleles were cultured in the absence of IL3 and cells were counted every other day. Error bars indicate SD.
Signaling through CRLF2 F232C requires JAK2 but not phosphorylation of CRLF2 Y368. (A) siRNA against mouse Jak2 was transfected into Ba/F3 cells expressing the indicated alleles and proliferation was assayed after 48 hours (*P < .05, **P < .01 compared with control siRNA in the same line by paired, Student t test). (B) Western blotting for indicated proteins after siRNA transfection. (C) Ba/F3 cells expressing the indicated alleles were grown for 48 hours in the absence of cytokines and in the presence of increasing concentrations of compounds. RFU indicates relative fluorescence units. All assays were performed in quadruplicate. Error bars indicate the SEM (***P < .001 compared with Ba/F3-CRLF2 F232C/JAK2). (D) IP of HA-tagged CRLF2 constructs from Ba/F3 cells expressing the indicated alleles followed by WB for CRLF2 phospho-Y368 or HA. Where indicated, cells were pretreated for 15 minutes with pervanadate. FC indicates F232C. (E) IP using anti-CRLF2 antibody in Ba/F3 cells expressing WT CRLF2 or CRLF2 Y368F (YF) with WT IL7R or IL7R-3F (3F) followed by immunoblotting with a specific antibody against CRLF2 P-Y368. IGH indicates mouse immunoglobulin heavy chain. (F) MHH-CALL4 (M4) and MUTZ-5 (M5) human B-ALL cells and Ba/F3-CRLF2/JAK2 R683G cells were treated with vehicle or pervanadate and then lysates were collected and immunoblotted for the indicated proteins. (G) Ba/F3 cells expressing the indicated combinations of CRLF2 and IL7R alleles were cultured in the absence of IL3 and cells were counted every other day. Error bars indicate SD.
Immunoblotting of Ba/F3 cells 48 hours after transfection of the Jak2-specific siRNA demonstrated approximately 80% loss of Jak2 protein compared with cells transfected with the control siRNA (Figure 5B). Coexpression of a human JAK2 cDNA that is resistant to the mouse Jak2 siRNA reversed the effect on proliferation in CRLF2 F232C cells (Figure 5A), confirming that the Jak2 siRNA effect is on-target.
We recently identified mutations at JAK2 Y931 that confer resistance to a panel of enzymatic Jak2 inhibitors in cells dependent on CRLF2/JAK2 signaling.32 To confirm that Jak2 is essential for CRLF2 F232C-mediated proliferation, we generated Ba/F3 lines that coexpress CRLF2 F232C with either human JAK2 or JAK2 Y931C, and then treated the cells with the JAK2 inhibitors ruxolitinib or tofacitinib. The Y931C mutation in JAK2 resulted in 4- to 6-fold resistance to both agents, consistent with dependence of CRLF2 F232C signaling on JAK2 (Figure 5C). In contrast, the Y931C mutation had no effect on sensitivity to the HSP90 inhibitor 17-AAG (GI50, 39.5nM vs 35.6nM for wild-type JAK2; supplemental Figure 6), which we previously demonstrated promotes the degradation of JAK2.32 Together, these results indicate that CRLF2 F232C signaling requires JAK2 enzymatic activity.
CRLF2 Y368 is not phosphorylated in response to TSLP
The phosphorylation of intracellular tyrosines on cytokine receptors can mediate the recruitment of Stat5. To determine whether Y368, the sole cytoplasmic tyrosine residue in CRLF2, is phosphorylated in response to TSLP, we generated a specific monoclonal antibody against P-Y368 by peptide injection in rabbits. Pervanadate stimulation of Ba/F3 cells expressing CRLF2 resulted in potent phosphorylation at Y368 that was not observed in cells expressing CRLF2 Y368F or the extracellular portion of CRLF2, demonstrating the specificity of the antibody (Figure 5D). Cells expressing HA-CRLF2 F232C or HA-CRLF2 with JAK2 R683G or JAK2 R683S had no detectable P-Y368 in the absence of pervanadate (Figure 5D). We also performed immunoprecipitation of lysates from Ba/F3-CRLF2/IL7R cells stimulated with TSLP using an antibody against CRLF2. Mass spectrometry of the immunoprecipitated proteins recovered only unphosphorylated CRLF2 C-terminal heptapeptide (codons 365-371; data not shown).
In addition to the single Y368 in CRLF2, human IL7R contains 3 intracellular tyrosine residues. Mutation of any 3 of the 4 tyrosines in the CRLF2/IL7R complex does not block proliferation and phosphorylation of Stat5 in response to TSLP, but mutation of all 4 tyrosines completely blocks proliferation.37 Thus, Ba/F3 cells expressing CRLF2 with an IL7R allele that contains all 3 intracellular tyrosine residues mutated to phenylalanine are dependent on CRLF2 Y368.37 We hypothesized that phosphorylation of Y368 could be amplified in the absence of tyrosine residues in IL7R. Ba/F3-CRLF2/IL7R and Ba/F3-CRLF2/IL7R-3F cells had similar levels of expression of both CRLF2 and IL7R proteins (supplemental Figure 7). However, stimulation with TSLP did not induce detectable phosphorylation of Y368 in Ba/F3 cells expressing CRLF2 with either IL7R or IL7R-3F (Figure 5E). The human CRLF2-rearranged B-ALL cell lines MUTZ-5 and MHH-CALL4 also had no detectable phosphorylation of CRLF2 Y368 (Figure 5F).
Gain-of-function mutations in IL7R were recently reported in cases of T-lineage ALL and CRLF2-rearranged B-ALL.20,41 The IL7R insPPCL allele is capable of homodimerization and signaling in the absence of CRLF2, whereas the IL7R S185C allele requires heterodimerization with CRLF2 (Figure 5F).20 As expected, both alleles conferred IL3 independent growth in Ba/F3 cells expressing CRLF2 Y368F (Figure 5F). Thus, Y368 is only essential for signaling in the setting of CRLF2 F232C, but even in this context does not undergo detectable phosphorylation.
Differences in the phosphoproteome of cells dependent on TSLP or CRLF2/JAK2 R683G
The analysis above failed to identify differences in STAT5 activation or domain requirements between TSLP-induced signaling and signaling through CRLF2 with mutant JAK2. To more broadly screen for differences in kinase targets between the 2 pathways, we built on our previously published global quantitative phosphoproteomic analysis of the TSLP signaling network.33 Using stable isotope labeling by amino acids in cell culture (SILAC; Figure 6A), we previously identified 226 proteins whose phosphorylation status was modulated by TSLP stimulation, including several members of the Src and Tec family of kinases.33
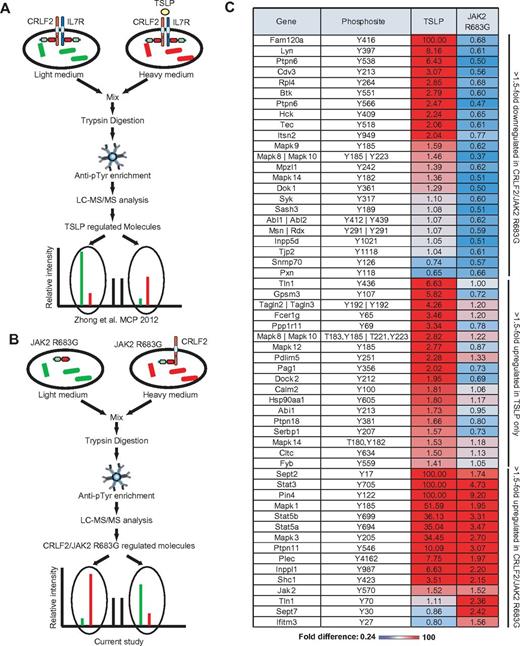
SILAC-based phosphoproteomic analysis of CRLF2-dependent signaling. (A) Schematic illustration of our previous SILAC study (Zhong et al33 ) which identified tyrosine phosphorylation sites induced by TSLP.33 (B) The same SILAC approach was used to identify tyrosine phosphorylation sites regulated by CRLF2 in the presence of JAK2 R683G in Ba/F3 cells. IL3 was withdrawn for 6 hours and lysates were mixed, subjected to phosphotyrosine enrichment and the enriched phosphopeptides were analyzed by LC-MS/MS. (C) The heatmap shows differences between tyrosine phosphorylation regulated by TSLP and by CRLF2/JAK2 R683G.
SILAC-based phosphoproteomic analysis of CRLF2-dependent signaling. (A) Schematic illustration of our previous SILAC study (Zhong et al33 ) which identified tyrosine phosphorylation sites induced by TSLP.33 (B) The same SILAC approach was used to identify tyrosine phosphorylation sites regulated by CRLF2 in the presence of JAK2 R683G in Ba/F3 cells. IL3 was withdrawn for 6 hours and lysates were mixed, subjected to phosphotyrosine enrichment and the enriched phosphopeptides were analyzed by LC-MS/MS. (C) The heatmap shows differences between tyrosine phosphorylation regulated by TSLP and by CRLF2/JAK2 R683G.
We used the same SILAC-based quantitative phosphoproteomic approach to identify changes in tyrosine phosphorylation induced by CRLF2/JAK2 R683G (Figure 6B). “Heavy” Ba/F3-CRLF2/JAK2 R683G cells and “light” Ba/F3-JAK2 R683G cells were starved in media lacking IL3 for 6 hours, lysed, mixed, and subjected to tryptic digestion. The tyrosine phosphorylated-peptides enriched by anti-phosphotyrosine antibodies were analyzed on an LTQ-Orbitrap Velos mass spectrometer. The acquired mass spectral data from 3 biologic replicate experiments were processed and analyzed using the Proteome Discoverer platform (supplemental Table 1). We identified and quantified a total of 372 tyrosine-phosphorylated peptides, derived from 303 phosphoproteins (supplemental Table 2).
We chose a 1.5-fold cutoff for increased phosphorylation and a 0.67-fold cutoff for decreased phosphorylation, as in our previous study.33 We identified 46 tyrosine phosphopeptides as up-regulated and 48 as down-regulated in CRLF2/JAK2 R683G cells compared with JAK2 R683G cells (supplemental Table 2). Consistent with the data from immunoblotting (Figure 5D-E), we failed to identify phosphorylation of CRLF2 Y368 in either Ba/F3-CRLF2/IL7R cells stimulated with TSLP33 or Ba/F3-CRLF2/JAK2 R683G cells (supplemental Table 2). In contrast, we did identify phosphorylation of JAK1 Y1033/4 in Ba/F3-CRLF2/JAK2 R683G cells (supplemental Table 2) that was not detected by immunoblotting (Figure 3E), suggesting that this approach is more sensitive at some phospho-sites.
We compared the change of phosphorylation level on tyrosine residues induced by CRLF2/JAK2 R683G with that induced by TSLP (supplemental Table 3, Figure 6C). As expected, both TSLP and CRLF2/JAK2 R683G promoted the phosphorylation of Stat5b, Stat5a, Stat3, and Jak2 (Figure 6C), all of which were previously demonstrated to be activated by both TSLP and CRLF2/JAK2 R683G signaling.18
The most notable difference between the cell lines was that several members of the Tec, Src, and MAPK family kinases that were phosphorylated in cell stimulated with TSLP had reduced phosphorylation in CRLF2/JAK2 R683G cells. These kinases include Lyn, Btk, Hck, Tec, Syk, MAPK8 (JNK), MAPK9 (JNK2), and MAPK10 (JNK3). In addition, phosphorylation of the phosphatases Ptpn6 (SHP1) and Inppd5 (SHIP-1), which regulate multiple pathways including JAK/STAT signaling in hematopoietic cells, as well as the kinase interacting partner Dok1 were increased by TSLP but not by CRLF2/JAK2 R683G (Figure 6C).
Discussion
Using both domain analysis and phosphoproteomics, we identified commonalities and differences in signaling between cells dependent on TSLP (through CRLF2/IL7R), CRLF2 F232C, or CRLF2 with mutant JAK2. First, we demonstrated that human CRLF2 directly interacts with both wild-type and gain-of-function human JAK2 proteins. Signaling in any of the 3 contexts requires both the box1 domain and W286 within CRLF2. Mutations in the box1 motifs of the growth hormone receptor, erythropoietin receptors and gp130 prevent physical association of JAK2 with the receptor.38,42 However, mutation or deletion of the human CRLF2 box1 domain did not abrogate the interaction with JAK2, either within Ba/F3 cells or using purified proteins in vitro. A previous study of the interferon receptor 2 demonstrated greater dependence on the box1 domain for signal activation and on the box2 domain for interaction with JAK1.39 Similarly, cells dependent on TSLP or CRLF2 with mutant JAK2 required a functional box1 domain for signaling and proliferation but mutation of the box2 domain had a much lesser effect.
The sole intracellular tyrosine in CRLF2 (Y368) was dispensable for signaling either in response to TSLP or through CRLF2/mutant JAK2. In contrast, Y368 was essential for proliferation and signaling in cells dependent on CRLF2 F232C. The homodimer of CRLF2 F232C/Y368F lacks any intracellular tyrosines, further supporting the importance of at least 1 tyrosine to mediate signaling. Unexpectedly, we failed to detect phosphorylation of Y368 under any conditions, either using a specific antibody against phospho-Y368 or by immunoprecipitation followed by mass spectrometry. Although we cannot rule out the possibility that low-level or very transient phosphorylation occurs and plays a role in signaling, our data suggest that phosphorylation of Y368 remains primarily unphosphorylated, whether in cells responding to TSLP, on signaling through F232C or CRLF2/mutant JAK2, or even when partnered with an IL7R mutant that lacks cytoplasmic tyrosine residues.
The CRLF2 F232C mutation occurs in 5%-10% of B-ALL cases that harbor a CRLF2 rearrangement.15,18,21 Mice transplanted with bone marrow transduced with CRLF2 F232C develop a myeloproliferative disorder through lineage-unrestricted oncogene activation of STAT5.21 CRLF2 F232C alone is also adequate to confer STAT5 phosphorylation and IL3-independent growth in Ba/F3 cells, but does not promote phosphorylation of JAK2 Y1007/1008 in these cells. Although JAK2 Y1007/1008 phosphorylation is apparent in cells responding to TSLP or expressing CRLF2/mutant JAK2, it remains possible that phosphorylation of JAK2 in cells dependent on CRLF2 F232C involves other sites. CRLF2 F232C cells are dependent on JAK2, based on the siRNA experiments in Figure 5A. In fact, the enzymatic activity of JAK2 is required for CRLF2 F232C-induced proliferation as these cells become resistant to JAK2 enzymatic inhibitors on coexpression of CRLF2 F232C with the JAK2 Y931C allele (Figure 5C).
The identification of therapeutic targets in CRLF2-rearranged B-ALL remains a high priority, as these leukemias are associated with high rates of relapse.18,25 Our data indicates that, independent of additional mutations in CRLF2 complex components, signaling through CRLF2 is dependent on JAK2. A consistent finding across our data were that mutant JAK2 levels were higher in cells with functional CRLF2 signaling (Figures 3C-E and 4D), suggesting that CRLF2 signaling both promotes mutant JAK2 phosphorylation and stabilizes the protein. A clinical trial of the JAK1/JAK2 enzymatic inhibitor ruxolitinib is currently enrolling through the Children's Oncology Group study ADVL1011. We recently demonstrated that treatment of MUTZ-5 and MHH-CALL4 with JAK2 enzymatic inhibitors results in paradoxical hyperphosphorylation of JAK2 and continued phosphorylation of STAT5.34 These lines are resistant to JAK2 inhibitors currently in trials (GI50 for ruxolitinib and tofacitinib > 15-30μM in both lines) but highly sensitive to inhibitors of HSP90, which promote the destabilization and degradation of JAK2 (GI50 for AUY922 < 0.05μM in both lines).34 AUY922 was more effective than the JAK2 inhibitor BVB808 in mice xenografted with primary human CRLF2-rearranged B-ALL, independent of whether the leukemia harbored a JAK2 mutation. Thus, JAK2 can be targeted in CRLF2-rearranged B-ALL cells by HSP90 inhibition, but these cells are capable of maintaining JAK2 phosphorylation and signaling in the presence of enzymatic JAK2 inhibitors.34
To search for additional therapeutic targets downstream of CRLF2, we performed a global assessment of tyrosine phosphorylation that identified several differences downstream of TSLP and CRLF2/JAK2 signaling. First and foremost among these, Tec, Src, and MAPK family kinases that were phosphorylated in response to TSLP (Figure 6C) were down-regulated in cells dependent on CRLF2/JAK2 R683G. This suggests that these kinases (Lyn, Btk, Hck, Tec, Syk, JNK, JNK2, and JNK3) drive proliferation downstream of TSLP but serve as negative regulators of CRLF2/mutant JAK2 signaling. In contrast, the phosphatases Ptpn6 and SHIP-1, which are phosphorylated in the setting of TSLP stimulation but lose phosphorylation downstream of CRLF2/mutant JAK2 serve as negative regulators only downstream of TSLP. Strikingly, all of these factors are components of B-cell receptor signaling,43 suggesting that TSLP can drive the activation of this pathway.
Two important aspects of CRLF2 signaling remain poorly defined. First, JAK1 partners with IL7R and not CRLF2 (Figure 1), so signaling may differ between complexes that harbor a JAK1 versus a JAK2 activating mutation. Of note, JAK1 was phosphorylated in Ba/F3-CRLF2/IL7R cells exposed to TSLP (Figure 2B) but not in Ba/F3-CRLF2/JAK2 R683G cells (Figure 3E). Thus, signaling through JAK1 does not appear to be required in the presence of JAK2 R683G.
The second important aspect of CRLF2 signaling that remains unclear is the role of endogenous TSLP in stimulating human CRLF2-rearranged B-ALL cells in situ. Tasian et al recently reported that in vitro stimulation of CRLF2-rearranged B-ALL specimens with 25 ng/mL TSLP (after serum starvation) promoted increases in the phosphorylation of STAT5 and S6, regardless of whether the specimens did or did not harbor JAK2 mutations.44 It is unclear how well these experiments mimic in vivo conditions, but the findings raise the intriguing possibility that all CRLF2-rearranged B-ALLs receive mitogenic stimulation within the bone marrow and/or peripheral blood from endogenous TSLP. Addressing this question will be challenging, as mouse TSLP does not activate the human CRLF2/IL7R complex. Thus, either the development of humanized mice with congenic B-ALL or a human TSLP knock-in approach may be necessary to faithfully address the effects of endogenous TSLP on human B-ALL in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Andrew Lane, and Scott Rodig for helpful discussion and assistance with experiments.
This work was supported by National Institutes of Health (NIH) R01-CA151898-01 (D.M.W.), a contract HHSN268201000032C from the NHLBI (A.P.), grant S10RR023025 from the High End Instrumentation Program of the NIH (A.P.), and NIH Roadmap grant “Technology Center for Networks and Pathways” U54 RR 020839 (A.P.).
National Institutes of Health
Authorship
Contribution: D.V.B. designed and performed research, analyzed data, and wrote the paper; J.Z. designed and performed research, analyzed data, and wrote the paper; C.D. designed and performed research; M.-S.K. and N.K. designed and performed research; L.B. designed and performed research; O.W. performed research and contributed vital new reagents; J.T. performed research and analyzed data; A.P. designed research, contributed vital new reagents, and analyzed data; A.Y. designed research, analyzed data, and wrote the paper; and D.M.W. designed research, contributed vital new reagents, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David Weinstock, Dana-Farber Cancer Institute, 450 Brookline Ave, Dana 510B, Boston, MA 02215; e-mail: dweinstock@partners.org.
References
Author notes
A.Y. and D.M.W. contributed equally to this work.

![Figure 2. CRLF2 box1, W286R and Y368F mutations in TSLP and CRLF2 F232C signaling. (A) Ba/F3 cells expressing the indicated alleles were grown in TSLP beginning day 0 and cells were counted every other day. Inset shows immunoblotting for the indicated proteins. (B) Immunoblotting was performed with antibodies against indicated total or phospho (P−) proteins. Tyrosine phospho-sites are included. Abbreviations: WT, wild-type; P/S, box1-P/S; WR, W286R; and YF, Y368F. (C) Identical to panel A except cells were grown in the absence of cytokines. (D) Immunoblotting as in (B) P-STAT5/STAT5 indicates the ratio of band intensities quantified for each lane. (E) Ba/F3 cells expressing WT CRLF2 or CRLF2 F232C alone or with HA-tagged JAK2 R683G were subjected to immunoprecipitation (IP) using an antibody against total JAK2 followed by immunoblotting (Western blot [WB]) for the indicated proteins. WCL indicates whole cell lysates.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/14/10.1182_blood-2012-02-413252/4/m_zh89991297170002.jpeg?Expires=1767704612&Signature=mpHHEwH2cm5N3TpPVyW0rOb7oe7zlzmofMGTbYeY~oOaVfdoJOIsgNCtI8nwElIL7EfNUA67q5hHvWdQCVH47S~GoLaQOyichsU8cncs1zs3C4nIBEwjiohfsILP8sK9uTF14~MRQYVND6JtwcBQQ2e~uThcD-rrgg0fs25eunKM3~1t2vp4erWlkHeRhdu5WX8c-mmQW3mRJ4loKUETkDttX80PRk2a68AhLr1DOhgm3VqkpowijxlU~TfWxHtI4qwNbejoyZce2y4Mp19WCNUhdL7xc5hcExlVUBefrvikxjsmsOhTl-g9jryE~WPZXvKkWl9FPbOIZNxFcrAl~Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal