Abstract
The human ankyrin-1 gene (ANK1) contains 3 tissue-specific alternative promoters. We have shown previously that the erythroid-specific ankyrin 1 (ANK1E) core promoter contains a 5′ DNase I hypersensitive site (HS) with barrier insulator function that prevents gene silencing in vitro and in vivo. Mutations in the ANK1E barrier region lead to decreased ANK1 mRNA levels and hereditary spherocytosis. In this report, we demonstrate a second ANK1E regulatory element located in an adjacent pair of DNase I HS located 5.6 kb 3′ of the ANK1E promoter at the 3′ boundary of an erythroid-specific DNase I–sensitive chromatin domain. The 3′ regulatory element exhibits enhancer activity in vitro and in transgenic mice, and it has the histone modifications associated with an enhancer element. One of the ANK1E 3′HS contains an NF-E2 binding site that is required for enhancer function. We show that a chromatin loop brings the 3′ enhancer and NF-E2 into proximity with the 5′ barrier region including the ANK1E core promoter. These observations demonstrate a model for the tissue-specific activation of alternative promoters that may be applicable to the ∼ 30% of mammalian genes with alternative promoters that exhibit distinct expression patterns.
Introduction
Hereditary spherocytosis (OMIM 182900) is an inherited hemolytic anemia that affects approximately 1 in 2000 people.1-3 Hereditary spherocytosis takes its name from the spherical-shaped erythrocytes observed on the peripheral blood smear of affected patients.1 Clinically, hereditary spherocytosis patients present with anemia that can vary from nearly asymptomatic to severe and transfusion-dependent.1 Other hematologic findings include reticulocytosis, elevated mean corpuscular hemoglobin concentration, and increased erythrocyte osmotic fragility after incubation.1 The most commonly mutated gene in hereditary spherocytosis is ANK1; this gene encodes a critical component of the erythrocyte membrane skeleton, the ankyrin 1 protein.1 Less frequent mutations associated with hereditary spherocytosis have been described in the SLC4A1 (band 3), SPTB (β-spectrin), EPB4.2 (band 4.2), and SPTA (α-spectrin) genes.1 Most of these mutations occur in the coding sequence of these genes and lead to functional protein deficiencies. Despite advances in diagnostic DNA sequencing, roughly 50% of hereditary spherocytosis patients do not have a molecular diagnosis.1
Roughly 30% of mammalian genes have multiple, distinct alternative promoters and first exons,4 and genes expressed in erythroid cells contain an even higher frequency of alternative promoters and first exons (35%).5 The human ANK1 gene contains 3 distinct, tissue-specific alternative promoters and first exons that are spliced to a common exon 2 in different cell types. The ANK1B promoter and first exon is expressed exclusively in brain and muscle cells and is located 138 kb upstream of exon 2. The erythroid-specific ANK1E promoter and first exon is located 39 kb upstream of exon 2. The ANK1A promoter and first exon is expressed in many cell types and is located 9 kb upstream of exon 2.6
We have studied the core ANK1E promoter extensively. The ANK1E core promoter activity is contained in a 271-bp region between −15 and −286 upstream of the ATG start codon.7 The promoter activity of this fragment is dependent on GATA1 binding and addition of 2.7 kb of upstream sequence did not increase the basal level of expression in either reporter assays in K562 cells or in transgenic mice.6,8 The identification of mutations in noncoding sequences upstream of the ANK1E coding sequence in hereditary spherocytosis patients has led to additional insights. One such mutation is a dinucleotide deletion in the 5′ untranslated (but transcribed) region at positions −72/−73 relative to the ATG start codon.6,9 This mutation disrupts core promoter function by decreasing the binding of the transcription complex TFIID, leading to reduced transcription from the ANK1E promoter.6 Other patients with hereditary spherocytosis have linked −108 T-to-C and −153 G-to-A substitution variants10 that we showed resided within a DNase I hypersensitive site (HS) located between −100 bp and −300 bp upstream of the ATG start codon. We identified this region as a barrier insulator that prevents gene silencing in vitro and permits position-independent, copy number–dependent, uniform expression in transgenic mice.8,11,12 The −108/−153 variant in the ANK1E barrier insulator was associated with loss of barrier insulator function and reduced transcription from the ANK1E promoter.8,11
Enhancer elements have been shown to be important regulatory elements for many different genes and are associated with the recruitment of transcription factors and the transcriptional machinery.13 Although many enhancer elements are located within or adjacent to core promoters, recent work has demonstrated that enhancers also may be located far from the promoters they activate.13 The classic example of long-range enhancer effects is the vertebrate β-globin gene clusters where enhancer elements in the locus control region (LCR) interact with individual globin gene promoters located ≥ 40 kb downstream.14 In vertebrates, the interaction of the β-like globin promoters and the LCR enhancers is associated with the presence of the CCCTC-binding protein CTCF.15 Recent studies of global CTCF binding have found that CTCF binds to barrier elements that organize the genome, to enhancer blockers that prevent enhancer-promoter interactions, as well as to individual enhancers and promoters.16 These latter interactions are proposed to be the result of the formation of a chromatin loop that brings barrier, promoter, and/or enhancer elements and their associated trans-acting proteins into proximity.17 In addition to the β-globin gene clusters, CTCF-mediated enhancer/promoter loops have been described in the APO,18 HERC2,10 Kit,19 and INFG10 loci, all of which have only a single promoter region.
The regulation of genes with distinct, tissue-restricted alternative promoters and first exons is less well understood. We hypothesized that the ANK1E barrier insulator works in conjunction with a distant erythroid-specific enhancer element to induce erythroid-specific expression of ANK1E. To test this hypothesis, we defined the structural and functional characteristics of the chromatin surrounding the ANK1E promoter. We identified a DNase I HS located 5.6 kb 3′ of the ANK1E promoter that marks a 3′ boundary of an erythroid-specific active chromatin domain. The 3′ HS was shown to be a tissue-specific enhancer element that was dependent on the binding of the transcription factor NF-E2. Using a ChIP-loop assay, we show that the tissue-specific enhancer element interacts with the ANK1E barrier element/promoter via a chromatin loop in erythroid K562 cells but not in neuronal SH-SY5Y cells. We conclude that the interaction of the 5′ ANK1E barrier and 3′ enhancer element is responsible for the tissue-specific expression of the ANK1E promoter.
Methods
DNase I HS mapping
Southern blot analysis was performed to fine map 3′HS1 and 2 as described previously.8 DNA from DNase I–treated nuclei was digested with NsiI (New England Biolabs) and compared with digests of high-molecular-weight DNA with NsiI and 1 of 8 restriction enzymes that cut within the region.
Luciferase reporter assay
Overlapping fragments of the ANK1E region from 5′HS to 500 bp downstream of 3′HS2 were inserted 5′ of an ANK1E promoter/luciferase or a thymidine kinase (TK)/luciferase reporter gene.7 Selected fragments also were cloned located 3′ to the luciferase gene. We coelectroporated 20 μg of test plasmid and 0.5 μg of the Renilla luciferase control plasmid into K562 or SH-SY5Y cells using a Gene Pulser Xcell (Bio-Rad Laboratories) at 200 V at 960 μF. After 48 hours, luciferase and Renilla luciferase activity were measured using the Dual-Luciferase Reporter Assay System (Promega) on a Fluoroskan Ascent FL microplate fluorometer and luminometer (Thermo Fisher Scientific).8
ChIP
K562 and SH-SY5Y cells were cross-linked as described by Steiner et al.20 Sonication was performed with a Sonicator 3000 (Misonix; K562 cells: power level 5 for 30 seconds total, with 1 second on and 0.5 second off; SH-SY5Y cells: power level 0.5 for 10 seconds total, 1 second on and 0.5 second off). ChIP was performed using the Magna ChIP A assay kit (17-610; Millipore),20 with anti-GATA1 (sc-265), –NF-E2 (sc-22 827), -USF2 (sc-862), -USF1 (sc-8983), -PRMT1, and -RNA polymerase II (sc-130851; all from Santa Cruz Biotechnology), -CTCF (07-729; Millipore), or -CARM1 (PRMT4, ab84370; Abcam) antibodies. The sequences of the PCR primers are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In many cases, these are not the same primers we used in our previous study.8 The custom genomic tiling array including the ANK1 locus and 100 kb of 5′ and 3′ flanking DNA20 was analyzed using the Tamalpais peak-calling algorithm.21 PCR reactions were performed on a 7500 Real Time PCR System with SYBR Green PCR master mix (Applied Biosystems).22
Barrier insulator assays
Barrier insulator assays were performed as described in Pikaart et al, but with the enhanced green fluorescent protein (EGFP) reporter gene instead of the IL-2 receptor.23 HS4 (HS4) from the chicken β-globin locus (+ control), 3′HS1, 3′HS2 were used to flank a mouse HS2-human β-globin promoter-EGFP reporter gene construct. Linearized constructs (200 ng) and pRSVneo (20 ng) were electroporated into K562 cells using a Gene Pulsar Xcell at 200 V, 960 μF. Stable clones were picked, expanded in 750 μg/mL G418, and analyzed for GFP expression on an FACS Calibur (BD Biosciences) using FlowJo software (TreeStar). Clones were grown in the absence of G418 for 2 to 22 weeks with periodic analysis of GFP expression. The presence of an intact GFP gene was confirmed by southern blot analysis.12
A human β-globin gene alone or flanked by 3′HS1, 3′HS2, or 3′HS1 and 2 was injected into fertilized mouse eggs.24 Transgene copy number was determined by southern blot analysis of DNA isolated from tail-snips. The level of human β-globin mRNA was determined by RNase protection, as described previously,24 in RBCs obtained by retro-orbital bleeding of adult anemic mice. Human β-globin protein was analyzed in fixed RBCs obtained from adult retro-orbital bleeding of adult anemic mice using an FITC-conjugated anti–human β-globin antibody (sc-21757 FITC; Santa Cruz Biotechnology) as described previously.25
ChIP-loop assay
ChIP-loop was performed as described previously,26 with the following modifications. Chromatin was cross-linked as described under “ChIP,” resuspended in 1 X DpnII buffer (New England Biolabs), incubated with 0.1% SDS at 65°C for 10 minutes, and then Triton X-100 added to a final concentration of 10% before digestion with 400 U of DpnII (New England Biolabs) overnight at 37°C. Digestion was estimated to be 95% to 100% complete. DpnII was heat inactivated for 20 minutes at 65°C, and chromatin was immunoprecipitated as described above using 25 μg of anti–NF-E2, -GATA1, -CTCF, -polymerase (Pol) II, or control antibodies overnight at 4°C with shaking. Chromatin fragments were ligated using 400 units of T4 ligase at 16°C for 3 hours, digested with 100 μg of proteinase K (Sigma) and 250 μg of RNase I (Sigma) at 68°C for 6 hours.27 DNA was purified using a PCR Purification Kit (28104; QIAGEN). Ligation products were amplified using touchdown PCR28 1 hold at 95°C for 3 minutes; 10 down cycles of 3 stages: 95°C for 30 seconds, 70°C for 45 seconds (with the temperature decreased 1°C each cycle to final temperature of 60°C), followed by 1 minute at 72°C; 25 cycles of 95°C for 30 seconds, 60°C for 45 seconds, 72°C for 1 minute, and a final hold at 72°C for 5 minutes.28 PCR products were visualized on a 2% agarose gel. The sequences of the PCR primers are listed in supplemental Table 1.
Results
Analysis of a downstream DNase I hypersensitive site in the ANK1 promoter region
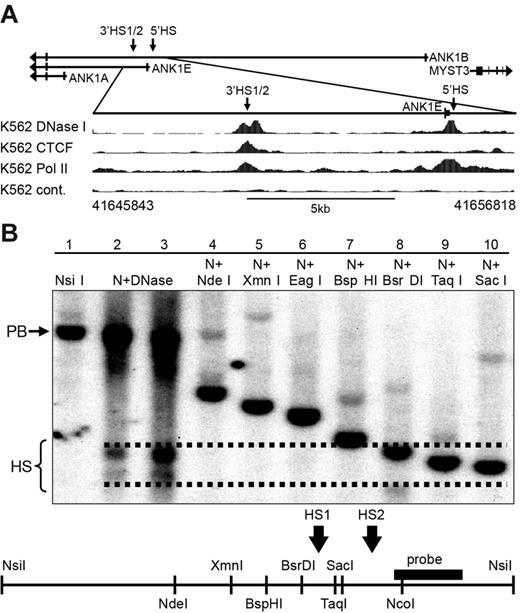
We have shown previously that a DNase I–sensitive domain extending approximately 6 kb downstream of the ANK1E promoter was present in K562 cells (supplemental Figure 1) but not in SH-SY5Y (supplemental Figure 2) or Jurkat cells.8 Consistent with this observation, the ENCODE UW DNase data shows a DNase I HS approximately 6 kb downstream from the ANK1E 5′HS (Figure 1A); we have previously characterized this HS. Southern blot fine mapping of the 3′HS revealed 2 adjacent HSs: a 211-bp BsrDI/SacI (3′HS1; hg19 chr8: 41650224-41649986) and a 620-bp SacI/NcoI (3′HS2; hg19 chr8: 41649986-41649367; Figure 1B).
DNA binding protein occupancy and DNase I HS analysis in the ANK1 promoter region in K562 cells. (A top) Schematic representation of the ANK1 promoter region showing all 3 alternative promoters. Arrows indicate the location of DNase I HSs. (Bottom) ENCODE data for DNase I HSs (DNase I), CTCF, and RNA Pol II binding. The hg19 coordinates of the expanded 11-kb region are shown below the panel. (B) Southern blot analysis of the 3′ANK1E 3′DNase I HS. Lane 1 contains high-molecular-weight K562 DNA digested with NsiI to reveal a 5.3-kb parent band (PB). Lanes 2 and 3 contain K562 DNA extracted from chromatin treated with a low and a high concentration of DNase I, respectively, and digested with NsiI (N + DNase), which revealed 2 DNase I HSs (HS). Lanes 4 to 10 contain high-molecular-weight DNA digested with NsiI and the indicated second enzyme to identify the boundaries of ANK1E 3′HS1 and 3′HS2. A map of the region showing the restriction sites and the probe used for southern blotting is shown below the data. The dashed lines indicate our best estimate of the boundaries of the HSs shown in lanes 2 and 3.
DNA binding protein occupancy and DNase I HS analysis in the ANK1 promoter region in K562 cells. (A top) Schematic representation of the ANK1 promoter region showing all 3 alternative promoters. Arrows indicate the location of DNase I HSs. (Bottom) ENCODE data for DNase I HSs (DNase I), CTCF, and RNA Pol II binding. The hg19 coordinates of the expanded 11-kb region are shown below the panel. (B) Southern blot analysis of the 3′ANK1E 3′DNase I HS. Lane 1 contains high-molecular-weight K562 DNA digested with NsiI to reveal a 5.3-kb parent band (PB). Lanes 2 and 3 contain K562 DNA extracted from chromatin treated with a low and a high concentration of DNase I, respectively, and digested with NsiI (N + DNase), which revealed 2 DNase I HSs (HS). Lanes 4 to 10 contain high-molecular-weight DNA digested with NsiI and the indicated second enzyme to identify the boundaries of ANK1E 3′HS1 and 3′HS2. A map of the region showing the restriction sites and the probe used for southern blotting is shown below the data. The dashed lines indicate our best estimate of the boundaries of the HSs shown in lanes 2 and 3.
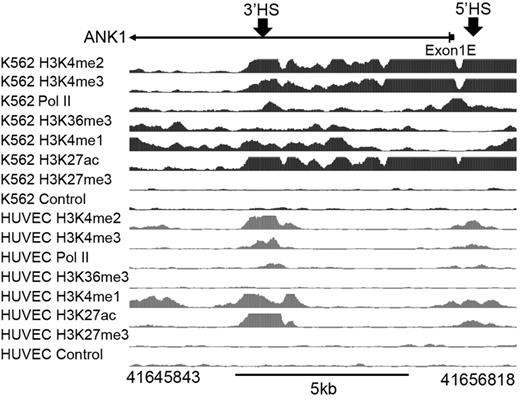
The ENCODE ChIPSeq data29 for K562 cells demonstrate the presence of active histone marks over the promoter region and ANK1E 3′HS (H3K4me2 and H3K4me3) and in the transcribed region (H3K36me3). Enhancer-associated histone modifications (H3K4me1 and H3K27ac) are present in the ANK1E 3′HS region as well as the region 5′ of the ANK1E 5′HS, even though there is no functional enhancer activity in the 5′HS region.8 These modifications terminate downstream of ANK1E 3′HS (Figure 2), although they are not replaced by histone modifications associated with repression. Human vascular endothelial cells (in which the ANK1E promoter is inactive) have lower levels of the active histone marks between ANK1E 5′HS and 3′HS. We conclude that the ANK1E 3′HS is located at the boundary of an erythroid-specific open chromatin domain.
Analysis of histone modifications across the ∼ 11-kb ANK1E promoter region in K562 and HUVECs. (Top) Location of the ANK1E promoter (exon 1E) and ANK1E 5′HS and ANK1E 3′HS1/2 (indicated by arrows). (Middle) Broad ChIPSeq profiles for histone modifications in K562 cells. (Bottom) ENCODE ChIPSeq profiles for histone modifications in HUVECs. The hg19 coordinates of the 11-kb region are shown below the panel.
Analysis of histone modifications across the ∼ 11-kb ANK1E promoter region in K562 and HUVECs. (Top) Location of the ANK1E promoter (exon 1E) and ANK1E 5′HS and ANK1E 3′HS1/2 (indicated by arrows). (Middle) Broad ChIPSeq profiles for histone modifications in K562 cells. (Bottom) ENCODE ChIPSeq profiles for histone modifications in HUVECs. The hg19 coordinates of the 11-kb region are shown below the panel.
DNA-binding proteins in the ANK1E promoter region
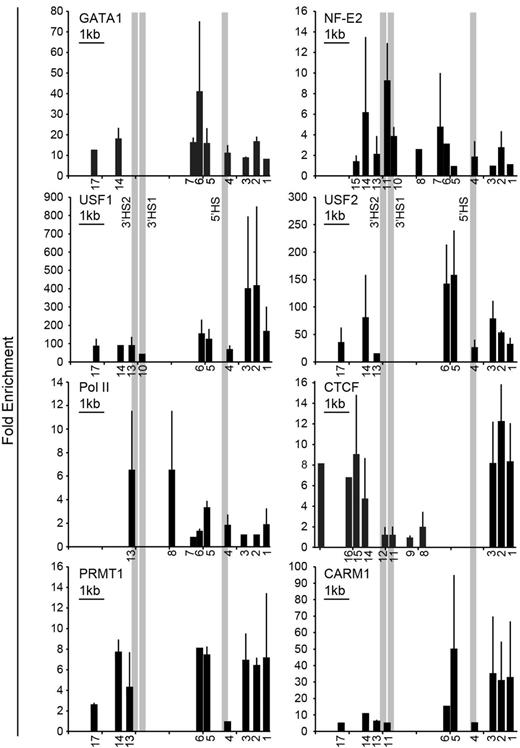
We have shown previously that a consensus GATA1 binding sequence binds GATA1 in vitro and is required for ANK1E promoter activity.7 To map DNA-binding proteins in this region, we performed ChIP analysis over the DNase I–sensitive domain in the ANK1E promoter region. GATA1 binding was detected in the ANK1E 5′HS region, and NF-E2 binding was detected in the ANK1E 3′HS1/2 region as well as in the ANK1E 5′HS region20 (also see Figure 3). ANK1E 3′HS1 contains an NF-E2 consensus sequence that binds NF-E2 in vitro (supplemental Figure 3). No GATA1 consensus sequences and no GATA1 binding were detected in this region. The barrier-associated proteins USF1 and USF2 were found to occupy sites upstream of ANK1E 5′HS and downstream of ANK1E 3′HS2 (Figure 3). Consistent with the ENCODE ChIPSeq data (Figures 1–2), we showed that that RNA polymerase II is concentrated over ANK1E 5′HS and 3′HS1/2 (Figure 3). Similarly, we found that the chromatin architectural protein CTCF occupies sites upstream of ANK1E 5′HS and downstream of ANK1E 3′HS2 (Figure 3). PCR-based ChIP assays also demonstrated the occupancy of the barrier-associated proteins PRMT1 and CARM1 in the ANK1E 5′ HS and 3′HS regions (Figure 3).
Validation of DNA binding protein occupancy. ChIP analysis of K562 chromatin for occupancy by the erythroid-specific transcription factors GATA1 and NF-E2; RNA Pol II; the chromatin architectural protein CTCF; and the barrier-associated proteins USF1, USF2, PRMT1, and CARM1. The numbers below the x-axes indicate the locations of the primers used (see supplemental Table 1 for coordinates) and are distributed to scale. A 1-kb scale bar is shown in the top left corner of each graph. The absence of a number below the x-axis indicates that the region either was not tested or the result was not significant. The y-axis shows the fold enrichment over precipitation with immunoglobulin G. The bars represent the results of multiple ChIP experiments. Each ChIP experiment was done in triplicate, and the standard deviation is displayed. Each bar shown is significant at P < .05 or lower compared with control. The CTCF results are the combination of several ChIP experiments and the relative enrichment cannot be compared.
Validation of DNA binding protein occupancy. ChIP analysis of K562 chromatin for occupancy by the erythroid-specific transcription factors GATA1 and NF-E2; RNA Pol II; the chromatin architectural protein CTCF; and the barrier-associated proteins USF1, USF2, PRMT1, and CARM1. The numbers below the x-axes indicate the locations of the primers used (see supplemental Table 1 for coordinates) and are distributed to scale. A 1-kb scale bar is shown in the top left corner of each graph. The absence of a number below the x-axis indicates that the region either was not tested or the result was not significant. The y-axis shows the fold enrichment over precipitation with immunoglobulin G. The bars represent the results of multiple ChIP experiments. Each ChIP experiment was done in triplicate, and the standard deviation is displayed. Each bar shown is significant at P < .05 or lower compared with control. The CTCF results are the combination of several ChIP experiments and the relative enrichment cannot be compared.
3′HS1 is an NF-E2–dependent, cis-acting enhancer element
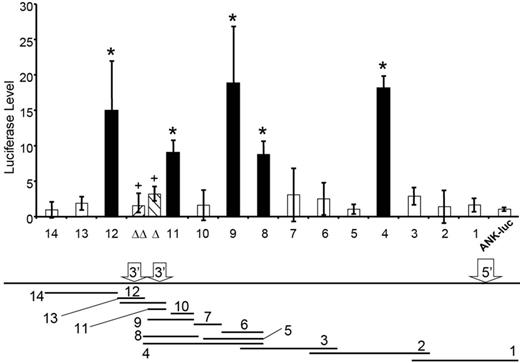
To determine whether the ANK1E chromatin domain contained regulatory elements, we cloned 14 overlapping fragments 5′ to an ANK1E promoter/luciferase reporter gene (ANK1E/luc) and transfected them into K562 cells. Only those fragments containing ANK1E 3′HS1 (fragments 4, 8, 9, 11, and 12; Figure 4) significantly increased ANK1E/luc expression (P < .01). ANK1E 3′HS1 increased luciferase expression (in either orientation) when adjacent to the ANK1E promoter but not when located 3′ to the luciferase reporter gene (supplemental Figure 4). We compared the activity of 3′HS1 fragments containing a wild-type (CTGACTCATATCT) NF-E2 binding sequence, a mutated (CTGACTCATATAT) NF-E2 binding sequence, and a mutation (CCAACTCATAT) that disrupts the binding of both NF-E2 and AP1. Both mutations decreased luciferase levels to background levels in K562 cells (Figure 4). Substitution of the heterologous TK promoter for the ANK1E promoter produced similar results. In neuronal SH-SY5Y cells, there was no effect of 3′HS1 on either ANK1E or TK promoter activity (supplemental Figure 4). These data suggest that 3′HS1 and its NF-E2 binding site must be juxtaposed near the ANK1E promoter to achieve its full activity in erythroid cells.
An enhancer element in the ANK1E DNase I–sensitive chromatin domain. Overlapping fragments of the ∼ 11 kb region (shown below with the locations of 5′HS, 3′HS1, and 3′HS2) were cloned into an ANK1/luciferase vector and transfected into K562 cells. The bar graph shows the relative luciferase expression of each fragment compared with the ANK1E promoter alone. The black bars indicate fragments with significantly higher levels of luciferase expression (*P < .01). Mutation of the NF-E2 (Δ) or NF-E2/AP1 (ΔΔ) binding sites abolishes the activity of 3′HS1 (+, P < .01; compared with wild-type fragment 11).
An enhancer element in the ANK1E DNase I–sensitive chromatin domain. Overlapping fragments of the ∼ 11 kb region (shown below with the locations of 5′HS, 3′HS1, and 3′HS2) were cloned into an ANK1/luciferase vector and transfected into K562 cells. The bar graph shows the relative luciferase expression of each fragment compared with the ANK1E promoter alone. The black bars indicate fragments with significantly higher levels of luciferase expression (*P < .01). Mutation of the NF-E2 (Δ) or NF-E2/AP1 (ΔΔ) binding sites abolishes the activity of 3′HS1 (+, P < .01; compared with wild-type fragment 11).
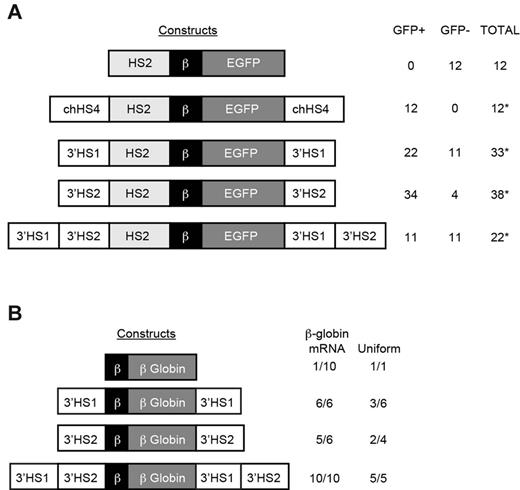
Analysis of barrier activity of 3′HS1 and 3′HS2 in vitro and in vivo
Because the ANK1E 3′HS region marks the boundary between DNase I–sensitive and -resistant chromatin and is occupied by barrier-associated proteins, we hypothesized that ANK1E 3′HS1 and 2 are barrier insulators. We tested this hypothesis using the in vitro K562 gene silencing assay.23 Fragments containing ANK1E 3′HS1 alone, 3′HS2 alone, or 3′HS1 and 2 together were used to flank a reporter construct consisting of the mouse HS2 enhancer, the human β-globin (huβ-globin) promoter, and the EGFP gene (HS2β-EGFP; Figure 5A). After transfection into K562 cells, clonal selection, and growth in nonselective medium, GFP was silenced in 12 of 12 HS2β-EGFP clones within 4 weeks. When the HS2β-EGFP was flanked with the well-described cHS4, 0 of 12 clones were silenced at 22 weeks. We observed a significant reduction in gene silencing at 22 weeks when HS2β-EGFP was flanked by 3′HS1 (11 of 33 silenced), 3′HS2 (4 of 38 silenced), or 3′HS1 and 2 (11 of 22 silenced). However, when these results are compared with the results with the ANK1E 5′ HS barrier or cHS4, in which no silent clones were observed,8 we conclude that 3′HS1 and 2 do not contain barrier activity. The partial resistance to silencing is consistent with the observation that 3′HS1 contains an enhancer element.
In vitro and in vivo analysis of barrier activity. (A) Constructs on the left were cotransfected into K562 cells with a pRSVneo plasmid, and individual clones were isolated in G418. After transfer to nonselective medium, the clones were assayed for GFP expression. The table at the right shows the number of GFP-expressing (GFP+) and -silenced (GFP−) clones. Significant resistance to silencing is indicated by asterisks (*P < .05). (B) Constructs on the left were used to generate transgenic mice. The table on the right shows the number of lines expressing huβ-globin mRNA (isolated from red blood cells obtained by bleeding of anemic adult mice) and the number of lines with uniform expression of huβ-globin protein (isolated from red blood cells obtained by bleeding of adult mice) for each line with sufficient levels of huβ-globin protein for analysis.
In vitro and in vivo analysis of barrier activity. (A) Constructs on the left were cotransfected into K562 cells with a pRSVneo plasmid, and individual clones were isolated in G418. After transfer to nonselective medium, the clones were assayed for GFP expression. The table at the right shows the number of GFP-expressing (GFP+) and -silenced (GFP−) clones. Significant resistance to silencing is indicated by asterisks (*P < .05). (B) Constructs on the left were used to generate transgenic mice. The table on the right shows the number of lines expressing huβ-globin mRNA (isolated from red blood cells obtained by bleeding of anemic adult mice) and the number of lines with uniform expression of huβ-globin protein (isolated from red blood cells obtained by bleeding of adult mice) for each line with sufficient levels of huβ-globin protein for analysis.
We generated transgenic mice with the huβ-globin gene alone or flanked by ANK1E 3′HS1, 2, or both to test barrier function in vivo. F1 mice containing 1 to 10 copies of the intact transgenes (as determined by southern blot analysis of DNA extracted from tail snips) were analyzed for huβ-globin mRNA and protein expression. As expected, huβ-globin mRNA was detected in only 1 of 10 lines containing the unflanked huβ-globin gene. In transgenic lines with the huβ-globin gene flanked by either ANK1E 3′HS1 or 3′HS2, 6 of 6 and 5 of 6 lines expressed huβ-globin mRNA, respectively. There was no correlation between transgene copy number and mRNA levels in these lines. Among 3′HS1 and 3′HS2 lines demonstrating detectable levels of human β-globin protein, variegated expression was observed in 3 of 6 and 2 of 4 lines, respectively (Figure 5B; Table 1). When both 3′HS1 and 2 flanked the huβ-globin gene, 10 of 10 lines expressed huβ-globin mRNA, with a significant correlation between huβ-globin mRNA level and transgene copy number, and uniform expression of huβ-globin protein in 5 of 5 lines with detectable levels of human β-globin protein (r2 = 0.59, P < .01; Table 1; Figure 5B). We conclude that neither 3′HS1 nor 3′HS2 has barrier activity.
Analysis of 3′HS1 and 3′HS2 function in transgenic mice
| Construct . | Line . | Copy no. . | huβ-mRNA/mα-mRNA . | huβ-mRNA/copy* . | Uniform, variegated . | ||||
|---|---|---|---|---|---|---|---|---|---|
| β-globin | A | 1 | 0.001 + 0.0005† | 0.001 | ND | ||||
| B | 2 | 0.0025 + 0.002† | 0.001 | ND | |||||
| C | 1 | 0.002 + 0.001† | 0.002 | ND | |||||
| D | 1 | 0.001 + 0.0001† | 0.001 | ND | |||||
| E | 3 | 0.000 + 0.0003† | 0.000 | ND | |||||
| F | 1 | 0.066 + 0.012 | 0.066 | U | |||||
| G | 1 | 0.001 + 0.0001† | 0.001 | ND | |||||
| H | 2 | 0.002 + 0.001† | 0.001 | ND | |||||
| I | 1 | 0.001 + 0.0003† | 0.001 | ND | |||||
| J | 1 | 0.002 + 0.0005† | 0.002 | ND | |||||
| Mean: 0.008 + 0.02 | |||||||||
| 3′HS1-β-globin-3′HS1 | A | 10 | 2.47 + 0.46 | 0.247 | U | ||||
| B | 1 | 0.17 + 0.04 | 0.17 | U | |||||
| C | 4 | 0.22 + 0.024 | 0.06 | V | |||||
| D | 3 | 0.23 + 0.012 | 0.078 | V | |||||
| E | 3 | 0.37 + 0.02 | 0.12 | U | |||||
| F | 4 | 2.26 + 0.17 | 0.565 | V | |||||
| Mean: 0.21 + 0.19 P = 0.009‡ | |||||||||
| 3′HS2-β-globin-3′HS2 | A | 1 | 0.033 + 0.005 | 0.033 | U | ||||
| B | 1 | 0.011 + 0.002 | 0.011 | V | |||||
| C | 1 | 0.002 + 0.001† | 0.002 | ND | |||||
| D | 2 | 0.028 + 0.01 | 0.014 | U | |||||
| E | 2 | 0.018 + 0.002 | 0.009 | ND | |||||
| F | 1 | 0.047 + 0.012 | 0.047 | V | |||||
| Mean: 0.02 + 0.02 P = 0.186‡ | |||||||||
| 3′HS1 + 2-β-globin-3′HS1 + 2 | A | 4 | 0.127 + 0.034 | 0.032 | U | ||||
| B | 5 | 0.05 + 0.008 | 0.01 | ND | |||||
| C | 2 | 0.083 + 0.012 | 0.042 | U | |||||
| D | 3 | 0.012 + 0.006 | 0.004 | ND | |||||
| E | 6 | 0.192 + 0.055 | 0.032 | U | |||||
| F | 1 | 0.053 + 0.013 | 0.053 | ND | |||||
| G | 1 | 0.037 + 0.013 | 0.037 | ND | |||||
| H | 3 | 0.147 + 0.062 | 0.049 | U | |||||
| I | 9 | 0.22 + 0.037 | 0.025 | U | |||||
| J | 5 | 0.13 + 0.016 | 0.026 | U | |||||
| Mean: 0.031 + 0.016 | |||||||||
| Construct . | Line . | Copy no. . | huβ-mRNA/mα-mRNA . | huβ-mRNA/copy* . | Uniform, variegated . | ||||
|---|---|---|---|---|---|---|---|---|---|
| β-globin | A | 1 | 0.001 + 0.0005† | 0.001 | ND | ||||
| B | 2 | 0.0025 + 0.002† | 0.001 | ND | |||||
| C | 1 | 0.002 + 0.001† | 0.002 | ND | |||||
| D | 1 | 0.001 + 0.0001† | 0.001 | ND | |||||
| E | 3 | 0.000 + 0.0003† | 0.000 | ND | |||||
| F | 1 | 0.066 + 0.012 | 0.066 | U | |||||
| G | 1 | 0.001 + 0.0001† | 0.001 | ND | |||||
| H | 2 | 0.002 + 0.001† | 0.001 | ND | |||||
| I | 1 | 0.001 + 0.0003† | 0.001 | ND | |||||
| J | 1 | 0.002 + 0.0005† | 0.002 | ND | |||||
| Mean: 0.008 + 0.02 | |||||||||
| 3′HS1-β-globin-3′HS1 | A | 10 | 2.47 + 0.46 | 0.247 | U | ||||
| B | 1 | 0.17 + 0.04 | 0.17 | U | |||||
| C | 4 | 0.22 + 0.024 | 0.06 | V | |||||
| D | 3 | 0.23 + 0.012 | 0.078 | V | |||||
| E | 3 | 0.37 + 0.02 | 0.12 | U | |||||
| F | 4 | 2.26 + 0.17 | 0.565 | V | |||||
| Mean: 0.21 + 0.19 P = 0.009‡ | |||||||||
| 3′HS2-β-globin-3′HS2 | A | 1 | 0.033 + 0.005 | 0.033 | U | ||||
| B | 1 | 0.011 + 0.002 | 0.011 | V | |||||
| C | 1 | 0.002 + 0.001† | 0.002 | ND | |||||
| D | 2 | 0.028 + 0.01 | 0.014 | U | |||||
| E | 2 | 0.018 + 0.002 | 0.009 | ND | |||||
| F | 1 | 0.047 + 0.012 | 0.047 | V | |||||
| Mean: 0.02 + 0.02 P = 0.186‡ | |||||||||
| 3′HS1 + 2-β-globin-3′HS1 + 2 | A | 4 | 0.127 + 0.034 | 0.032 | U | ||||
| B | 5 | 0.05 + 0.008 | 0.01 | ND | |||||
| C | 2 | 0.083 + 0.012 | 0.042 | U | |||||
| D | 3 | 0.012 + 0.006 | 0.004 | ND | |||||
| E | 6 | 0.192 + 0.055 | 0.032 | U | |||||
| F | 1 | 0.053 + 0.013 | 0.053 | ND | |||||
| G | 1 | 0.037 + 0.013 | 0.037 | ND | |||||
| H | 3 | 0.147 + 0.062 | 0.049 | U | |||||
| I | 9 | 0.22 + 0.037 | 0.025 | U | |||||
| J | 5 | 0.13 + 0.016 | 0.026 | U | |||||
| Mean: 0.031 + 0.016 | |||||||||
U indicates uniform expression (100% of cells express human β-globin); V, variegated expression (< 100% of cells express human β-globin); and ND, level of human β-globin protein was insufficient for analysis.
huβ-mRNA/copy = huβ-mRNA/mα-mRNA divided by copy number for each line.
mRNA levels below 0.005 are indistinguishable from background and are scored as negative.
Compared with 3′HS1 + 2-β-globin-3′HS1 + 2.
Consistent with the observation that ANK1E 3′HS1 increases expression in transient transfection assays when adjacent to a promoter, the level of huβ-globin mRNA per transgene copy number was significantly higher when ANK1E 3′HS1 was adjacent to the huβ-globin transgene compared with the level in the ANK1E 3′HS2 transgenic lines (P < .01; Table 1). When ANK1E 3′HS1 was positioned further from the huβ-globin promoter by the addition of 3′HS2, the level of expression was similar to that seen in animals flanked by ANK1E 3′HS2 alone (P < .01; Table 1). These observations confirm that ANK1E 3′HS1 must be adjacent to the promoter to exert its positive regulatory function in vivo as well as in vitro.
The ANK1E 5′HS and 3′HS regions interact in K562 cells
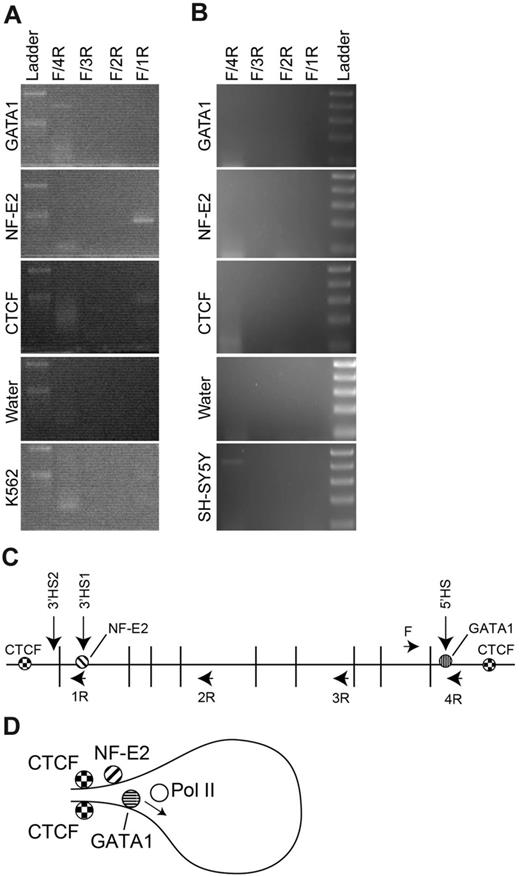
We hypothesized that NF-E2 and the chromatin architectural protein CTCF were involved in the formation of an erythroid-specific chromatin loop. To test this hypothesis, we used the ChIP-loop assay.26 Cross-linked K562 chromatin was digested with DpnII and immunoprecipitated with anti-GATA1, –NF-E2, or -CTCF antibodies. After enrichment, the DpnII ends were ligated, and PCR analysis was performed using a forward primer near the ANK1E promoter and 4 reverse primers in the region between ANK1E 5′HS and 3′HS1 and 2 (Figure 6). In GATA1-enriched chromatin, only ligation products from adjacent ends in ANK1E promoter region could be amplified. Ligation products representing the interaction of the ANK1E 5′HS and 3′HS1 and 2 regions were amplified from both NF-E2– and CTCF-enriched chromatin (Figure 6). Sequence analysis of the PCR products confirmed the predicted ligation reaction. Other than linearly adjacent fragments, no PCR products representing ligation between ANK1E 5′HS and other regions were identified in neuronal SH-SY5Y chromatin (Figure 6). We conclude that the 5′HS/ANK1E and 3′HS1 and 2 regions interact through the cooperation of NF-E2 and CTCF in K562 cells.
ChIP-loop analysis of the ANK1E promoter region. K562 or SH-SY5Y chromatin was cross-linked, digested with DpnII, immunoprecipitated, and ligated. (A) PCR analysis of ligated K562 chromatin. Each panel represents an analysis of chromatin precipitated with the indicated antibody and amplified with the 4 sets of primers. The top molecular weight marker is 500 bp and the lower band is 300 bp (Ladder). (B) PCR analysis of ligated SH-SY5Y chromatin. Each panel represents an analysis of chromatin precipitated with the indicated antibody and amplified with the 4 sets of primers. The top molecular weight marker is 500 bp followed by 400 bp, 300 bp, 200 bp, and 100 bp (Ladder). (C) Schematic representation of the ANK1E promoter region showing the 5′ and 3′ HS, the location of the DpnII sites, and the location of the PCR primers used to detect interactions (see supplemental Table 1 for coordinates). (D) Model for the chromatin loop in K562 cells that is revealed by precipitation with anti–NF-E2 and -CTCF.
ChIP-loop analysis of the ANK1E promoter region. K562 or SH-SY5Y chromatin was cross-linked, digested with DpnII, immunoprecipitated, and ligated. (A) PCR analysis of ligated K562 chromatin. Each panel represents an analysis of chromatin precipitated with the indicated antibody and amplified with the 4 sets of primers. The top molecular weight marker is 500 bp and the lower band is 300 bp (Ladder). (B) PCR analysis of ligated SH-SY5Y chromatin. Each panel represents an analysis of chromatin precipitated with the indicated antibody and amplified with the 4 sets of primers. The top molecular weight marker is 500 bp followed by 400 bp, 300 bp, 200 bp, and 100 bp (Ladder). (C) Schematic representation of the ANK1E promoter region showing the 5′ and 3′ HS, the location of the DpnII sites, and the location of the PCR primers used to detect interactions (see supplemental Table 1 for coordinates). (D) Model for the chromatin loop in K562 cells that is revealed by precipitation with anti–NF-E2 and -CTCF.
Discussion
In this study, we explored the ANK1E promoter region to identify regulatory elements that control the tissue-specific expression of ANK1E. In addition to providing a mechanism for the erythroid-specific expression of the ANK1E alternative promoter, our studies offer an opportunity to test the predictions of the histone code in the ANK1E promoter region. The H3K4me1 and H3K27ac histone modifications described in the ANK1E 3′HS region are consistent with the enhancer activity that we observed in this region. These modifications are not seen in HUVECs and other nonerythroid cells, consistent with our observation that the enhancer activity is erythroid-specific. We have shown that the region between the ANK1E core promoter and 3′HS is DNase I–sensitive only in erythroid cells. This observation is consistent with the presence of histone modifications including H3K4me2, and H3K4me3, both of which are associated with active transcription in this region in erythroid K562 cells, where the ANK1E promoter is active, but not in HUVECs where the ANK1E and ANK1B promoters are not active.6
However, our functional analysis of the ANK1E regulatory region also identified discrepancies from the predicted histone code. The region 2 kb upstream of the ANK1E 5′HS region also bears the enhancer-associated H3K4me1 and H3K27ac histone modifications in K562 chromatin, despite that we have found no evidence for the presence of positive regulatory elements in this region in vitro7 or in vivo.8 The chromatin loop between ANK1E 5′HS and 3′HS reconciles these discrepancies. We propose that the chromatin loop that brings the NF-E2–bound ANKIE 3′HS enhancer into juxtaposition to the ANK1E core promoter, allowing NF-E2 and enhancer-associated histone modifications to be mapped to both regions.
Most chromatin loops are associated with occupancy by the architectural DNA binding protein CTCF that maintains a tissue-specific active chromatin domain within a locus. In erythroid cells, CTCF has been shown to contribute to the formation of chromatin loops in the β-globin locus in association with cohesin.30,31 Tissue-specific chromatin loops are achieved, at least in part, through the association of architectural proteins and tissue-specific transcription factors. In the murine β-globin locus, GATA1 binds HS2 of the LCR and LDB1; this binding stabilizes the CTCF-mediated LCR and β-globin promoter interaction. In the absence of GATA1, LDB1 does not bind, and the loop is not stabilized.32 In the human erythroid cells, the human LDB1 homolog NL1 forms an activation loop between the GATA1-bound LCR and the γ-globin gene.33
Erythroid gene expression is critically dependent on the expression of erythroid transcription factors.34 The ANK1E core promoter is fully dependent on GATA1 for basal activity.7 Although this region has no consensus NF-E2 binding sites, we observed NF-E2 occupancy in K562 chromatin upstream of 5′HS. Interactions between GATA1 and NF-E2 are important components of erythroid gene expression. For example, deficiency of either GATA1 or NF-E2 in K562 cells reduces LCR-promoter loop interactions and transcription in the β-globin locus.35 Furthermore, NF-E2 has been shown to cooperate with USF1 to recruit RNA Pol II and increase expression of globin gene promoters.36
These findings suggest a mechanism by which the ANK1E core promoter becomes activated in erythroid cells, but silent in other cell types where alternative tissue-specific ANK1 core promoters are active. We propose a model in which the expression of NF-E2 in erythroid cells allows interactions with CTCF that bring the ANK1E 3′HS1 enhancer and NF-E2 into juxtaposition with the GATA1-dependent ANK1E promoter. With the establishment of the chromatin loop, interactions of NF-E2 with USF1 recruit RNA Pol II to specifically activate transcription of the ANK1E promoter and first exon. In the absence of NF-E2 and GATA1, the ANK1E promoter does not become active. This model predicts that an analogous neuronal-specific chromatin loop involving the ANK1B core promoter is responsible for its activation and highlights the critical role of higher order chromatin configurations in gene regulation. Finally, our model predicts that mutations in 3′HS enhancer region will be found among the ∼ 50% of ankyrin-deficient hereditary spherocytosis patients where a mutation in the coding sequence or the core promoter region of the ANK1 gene has not been identified.1
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by intramural funds from National Human Genome Research Institute (D.M.B.) and grants HL65448 and DK62039 from the National Institutes of Health (P.G.G.).
National Institutes of Health
Authorship
Contribution: A.O.Y., L.A.S., N.E.S, A.P.C., E.D.R., J.Y.L, C.W., and L.J.G. performed the research; and A.O.Y., P.G.G., and D.M.B. analyzed the data, designed the research, prepared the figures, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David M. Bodine, Genetics and Molecular Biology Branch, National Human Genome Research Institute, National Institutes of Health, Bldg 49, Rm 4A04, 49 Convent Dr, MSC 4442, Bethesda, MD 20892; e-mail: tedyaz@mail.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal