Abstract
The intracellular location of nucleic acid sensors prevents recognition of extracellular self-DNA released by dying cells. However, on forming a complex with the endogenous antimicrobial peptide LL37, extracellular DNA is transported into endosomal compartments of plasmacytoid dendritic cells, leading to activation of Toll-like receptor-9 and induction of type I IFNs. Whether LL37 also transports self-DNA into nonplasmacytoid dendritic cells, leading to type I IFN production via other intracellular DNA receptors is unknown. Here we found that LL37 very efficiently transports self-DNA into monocytes, leading the production of type I IFNs in a Toll-like receptor-independent manner. This type I IFN induction was mediated by double-stranded B form DNA, regardless of its sequence, CpG content, or methylation status, and required signaling through the adaptor protein STING and TBK1 kinase, indicating the involvement of cytosolic DNA sensors. Thus, our study identifies a novel link between the antimicrobial peptides and type I IFN responses involving DNA-dependent activation of cytosolic sensors in monocytes.
Introduction
Type I IFNs, such as IFN-α and IFN-β, are key cytokines in the antiviral host defense because of their ability to limit viral replication in infected cells.1,2 In addition, type I IFNs shape antiviral immune responses by promoting maturation of conventional DCs (cDCs), survival and proliferation of T cells, activation of NK cells, and differentiation of B cells into antibody-secreting plasma cells.1-4 Type I IFN induction is mediated by innate immune recognition of viral nucleic acids in endosomal compartments (during viral entry), or in the cytosol (during viral replication).5 In the endosome, viral DNA is recognized by Toll-like receptor-9 (TLR9), which is primarily expressed in plasmacytoid dendritic cells (pDCs).6 TLR9 recognizes hypomethylated CpG islets and signals via the adaptor molecule MyD88, leading to the activation of IRF7 in pDCs.6 In the cytosol, several DNA receptors have been identified and shown to be responsible for TLR9-independent sensing of viral DNA and type I IFN production by non-pDCs. Cytosolic DNA receptors include DNA-dependent activator of IFN-regulatory factors,7,8 RNA polymerase III,9 IFI16, a protein that belongs to the PYHIN protein family,10 and the recently described helicase DDX41 expressed by dendritic cells.11 All these receptors recognize B form double-stranded DNA regardless of its sequence5,12,13 and signal via the adaptor protein STING (stimulator of interferon genes) and Tank-binding kinase 1 (TBK1), which activates and phosphorylates IRF3.5,14,15 Another cytosolic DNA receptor, which induces type III IFNs and signals via IRF7 and not IRF3, is Ku70, a protein also involved in DNA repair.16
DNA is abundantly released into the extracellular environment by dying cells in the context of tissue injury. This self-DNA is normally nonimmunogenic because of its rapid extracellular degradation and because of the intracellular seclusion of DNA recognizing receptors.17 However, we recently found that an endogenous antimicrobial peptide called LL37 derived from epithelial cells and neutrophils can break the innate immune tolerance to self-DNA by protecting extracellular DNA from degradation and by efficiently transporting it into endosomal compartments of pDCs.18 This process leads to the activation of TLR9 and to the production of type I IFNs,18 indicating that the immunogenicity of extracellular self-DNA is under control of endogenous antimicrobial peptides, such as LL37. This innate immune activation pathway was found to be relevant in the context of skin injury19 but also in IFN-driven autoimmunity in psoriasis and systemic lupus erythematosus (SLE).18,20
IFN-driven autoimmunity has also been linked to TLR9-independent sensing of self-DNA. Indeed, DNase II deficiency leads to accumulation of self-DNA, uncontrolled type I IFN induction, and SLE-like autoimmunity that cannot be reversed in the absence of TLR9 or MyD88.21 Furthermore, circulating immune complexes containing self-DNA trigger DC activation via TLR9-independent pathways in SLE.22,23 These studies suggest that innate sensing of self-DNA is not confined to endosomes but may also occur via cytosolic receptors. However, the mechanisms that regulate the transport of extracellular self-DNA into the cytosol for TLR9-independent type I IFN induction are unclear.
Here we investigated the possibility that endogenous antimicrobial peptides also transport self-DNA into cytosolic compartments leading to type I IFN expression by non-pDCs. LL37 was indeed found to efficiently transport extracellular self-DNA into the cytosol of monocytes and trigger induction of type I IFNs. This process was mediated by cytosolic DNA sensors as it was mediated via the adaptor protein STING and TBK1 kinase signaling and required recognition of B form double-stranded DNA regardless of its sequence. Thus, we uncover an endogenous mechanism for the transport of extracellular self-DNA into cytosolic compartments for type I IFN induction in non-pDCs, controlled by endogenous antimicrobial peptides.
Methods
Reagents
The synthetic peptide LL37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES) and mutated LL37 peptides Mut+1 (LLGDFFAVSKEKIGAEFVRIVQAIKDFLRNLVPRTES) and Mut+8 (KGGDFFRKSKEKDGKEFKRERQRGKDFKRNKVPRTES) were purchased from Innovagen. The following synthetic oligos of a phosphodiester backbone were produced by TriLink BioTechnologies: CpG DNA (sense 5′ XTC GTC GTT TTG TCG TTT TGT CGT TG 3′; antisense 5′ XAA CGA CAA AAC GAC AAA ACG ACG AG 3′), GpC DNA (sense 5′ XTG CTG CTT TTG TGC TTT TGT GCT TG 3′; antisense 5′ XAA GCA CAA AAG CAC AAA AGC AGC AG 3′), poly A (5′ AAA AAA AAA AAA AAA AAA 3′), poly T (5′ TTT TTT TTT TTT TTT TTT 3′). Synthetic polydeoxynucleotides poly (dA:dT) ̇ poly (dT:dA) and poly (dC:dG) ̇ poly (dG:dC) were purchased from Sigma-Aldrich. CpG-containing plasmid (pBR322) was purchased from Invitrogen. CpG-free plasmid (pCpG-mcs G2) was obtained from Invivogen. All DNA sequences were used at a final concentration of 10 μg/mL for stimulation of human cells unless otherwise stated. Human genomic DNA (2 mg/mL) was purchased from BioChain or extracted from U937 cells following standard procedures. Highly purified vaccinia virus (VV; lister strain) and human herpes simplex virus (HSV) virus were purchased from Advanced Biotechnologies. Purified genomic DNA from VV and HSV virus was extracted with the use of QIAamp DNA Mini Kit (QIAGEN) according to the manufacturer's instruction for Blood and Body Fluid Spin Protocol. Bafilomycin was obtained from Sigma-Aldrich. DNase I (800 U/mL) was from Roche Diagnostics.
Generation of DNA-LL37 complexes
DNA complexes were generated by mixing 2 μg of human genomic DNA (self-DNA) or synthetic DNA sequences (final concentration, 10 μg/mL) with 10 μg LL37 (final concentration, 50 μg/mL or 10μM), first in 20 μL of PBS for 30 minutes at room temperature and then diluted into 200 μL of complete medium for cell stimulation. In some experiments, suspensions were treated for 1 hour with DNase I (Roche Diagnostics, 600 U/mL). In other experiments, CpG-containing plasmid (pBR322) was methylated with the use of CpG Methyltransfarase (M0226L; New England Biolabs). According to the standard protocol provided by the manufacturer, poly (dC:dG) ̇ poly (dG:dC) DNA (B-type DNA) was brominated to induce the formation of Z-type DNA following the protocol described previously.24 Crude viral lysates were made following 1-hour treatment with Proteinase K.
Isolation and stimulation of human primary cells
This study was approved by the Institutional Review Board for Human Research at the MD Anderson Cancer Center and conducted in accordance with the Declaration of Helsinki. Human blood pDCs were isolated from buffy coats obtained from the blood of healthy donors at the Gulf Coast Regional Blood Center, Houston, TX. After separation of mononuclear cells by Ficoll centrifugation, pDCs were enriched by using anti-BDCA4–conjugated magnetic microbeads (Miltenyi Biotec) and further purified as lineage−CD11c−CD4+ cells with use of FACS performed with a FACSAria Cell Sorting System (BD Biosciences) to reach 99% purity. CD14+ blood monocytes obtained from PBMCs using anti-CD14 microbeads (Miltenyi Biotec), followed by pDC depletion with the use of BDCA-4 microbeads. The purity of monocytes was confirmed by FACS staining as being > 99%. CD16-positive monocytes were isolated using the CD16 monocyte isolation kit (Miltenyi Biotec). CD16-negative monocytes were subsequently isolated from the negative fraction using anti-CD14 conjugated microbeads (Miltenyi Biotec). The purity of the isolated monocyte subsets was consistently > 97%. For siRNA experiments, untouched monocytes were obtained by using Monocyte Isolation Kit II (Miltenyi Biotec). Blood myeloid DCs (mDCs) were isolated from PBMC using anti-CD1c+ microbeads (Miltenyi Biotec). T cells, B cells, and NK cells were isolated by magnetic separation using the appropriate isolation kit (Miltenyi Biotec). Adult human dermal fibroblasts (C-013–5C) and adult human epidermal keratinocytes (C-005–5C) were purchased by Invitrogen. Purified human primary cells were seeded into 96-well round-bottom plates at 5 × 104/well (pDCs) or 2 × 105/well (all other cell types) in 200 mL RPMI 1640 (Invitrogen) supplemented with 10% FCS (Atlanta Biologicals). Supernatants of stimulated human primary cells were collected after overnight culture. Cytokine levels in the supernatants were determined by using ELISA kits for human IFN-α (PBL Biomedical Laboratories), IL-6, TNF-α, ΙL-1β, IL-12, and IL-23 (R&D Systems).
To visualize the self-DNA in human primary cells, human DNA (huDNA; BioChain) was labeled with Alexa488 using the Ulysis Nucleic Acid Labeling Kit (Invitrogen), according to the standard protocol provided by the manufacturer as previously described.18 Purified human primary cells were stimulated at different time points with DNAAlexa488-LL37 containing complexes, washed, and analyzed by flow cytometry with an oil-immersion objective (Leica; 63×/1.4 numeric aperture) using identical gain settings at room temperature. Images were collected using LCS V2.61 software (Leica Microsystems) and processed with Adobe Photoshop CS2.
Analysis of self-DNA-LL37 complex uptake by confocal microscopy
For immunofluorescence imaging, human primary cells were seeded on coverslips pretreated with poly-lysine and stimulated as described above with DNAAlexa488-LL37 containing complexes. At the indicated time points, cells were gently washed, fixed in 4% paraformaldehyde, permeabilized by using 0.1% saponin (Sigma-Aldrich), and stained with an Alexa647 anti–HLA-DR antibody (Invitrogen). After the washing steps, slides were mounted in Prolong Gold antifade media (Invitrogen) and examined with use of a Leica SP2 RS SE confocal microscope.
Real-time PCR for mRNA expression.
Total RNA was extracted with the QIAGEN RNeasy mini protocol and was converted to cDNA using oligo-dT, random hexamers, and SuperScript II RT (Invitrogen). Real-time PCR was performed using a sequence detector (model ABI PRISM 9600; PerkinElmer) and target mixes (Applied Biosystems): Ifn-α2 (Hs00265051_s1), IFN-β (Hs00277188_s1), TBK1 (Hs00179410_m1), MyD88 (Hs00182082_m1), and GAPDH (Hs99999905_m1). RNAi knockdown efficiency of STING mRNA in primary human monocytes was detected by real-time RT-PCR using the following primers: STING-S, 5′-ACTGTGGGGTGCCTGATAAC-3′; STING-AS, 5′-TGGCAAACAAAGTCTGCAAG-3′; GAPDH-S, 5′-GAGTCAACGGATTTGGTCGT-3′; and GAPDH-AS, 5′-TTGATTTTGGAGGGATCTCG-3′.11 Threshold cycle (Ct) values for each gene were normalized to GAPDH using the equation 2.0(GAPDH−GENE) (105), where GAPDH was the Ct obtained for GAPDH, GENE was the Ct of the gene of interest, and 105 was an arbitrarily chosen factor to bring all values above.
Silencing of TBK1 and MyD88 expression in primary human monocytes by specific siRNA
Short interfering RNA (siRNA) targeting was used to knock down TBK-1, MyD88, and STING expression in primary monocytes. Human monocyte nucleofector kit (Amaxa) and Nucleofector device (Amaxa) were used for delivering siRNA into monocytes by following the instructions provided by the company. In brief, 5 × 106 purified monocytes were suspended in 100 μL of human monocyte nucleofector solution (Amaxa) and transfected with siRNA at a final concentration of 50nM using the Y-001 program. Transfected cells were immediately diluted with prewarmed monocyte growth media (Amaxa) and cultured in 12-well plates for 48 hours. The siRNAs used in this study were as follows: ON-TARGETplus nontargeting pool (Dharmacon #D 001810–10; control-siRNA); TBK1 ON-TARGETplus SMARTpool siRNAs (Dharmacon #L-003788–00; TBK1-siRNA), MYD88 ON-TARGETplus SMARTpool siRNAs (Dharmacon #M-004769–01; MyD88-siRNA), TMEM173 siRNA (h) (Santa Cruz Biotechnology sc-92042; STING-siRNA) and control siRNA (h): sc-36869 (Santa Cruz Biotechnology, control-siRNA for STING). To confirm inhibition of TBK-1, MyD88, and STING protein expression by the specific siRNA, RT-PCR was performed.
DNA condensation assay
Human genomic DNA (10 μg/mL) or synthetic dsDNA (10 μg/mL) sequences alone or premixed with the different concentrations of LL37 or mutated LL37 peptides (Mut+1, Mut+8) for complex formation, and stained with PicoGreen (Quant-iT PicoGreen dsDNA kit, Invitrogen) according to the standard protocol provided by the manufacturer. In some experiments, the complexes were pretreated for 30 minutes with different concentrations of NaCl. Samples were excited at 480 nm, and the emission intensity was measured fluorometrically. The sudden decrease in DNA staining occurring as a result of DNA-LL37 complex formation is indicative of DNA condensation and results from both dye exclusion as well as quenching of the fluorescence.
Statistical analysis
The data were expressed as mean ± SE. Statistical significance of differences was determined by Student t test and Bonferroni t test (P < .05 was considered statistically significant). Analysis was done in GraphPad Prism Version V software.
Results
Self-DNA-LL37 complexes induce type I IFNs in monocytes
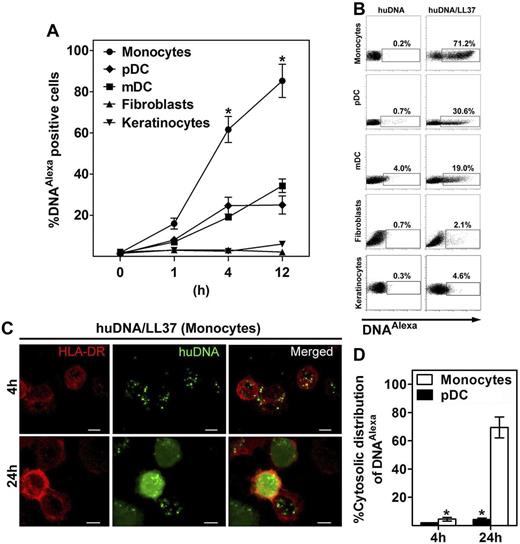
To determine whether LL37 transports self-DNA into non-pDCs for type I IFN production, we stimulated various blood-derived immune cells, including myeloid DCs, monocytes, T cells, B cells, and NK cells, as well as structural nonhematopoietic cells, including fibroblasts and keratinocytes with huDNA-LL37 complexes. We found that, in addition to pDCs, also monocytes produced significant amounts of IFN-α when stimulated with huDNA-LL37 complexes (Figure 1A). By contrast, we did not find any IFN-α production by mDCs, T cells, B cells, NK cells, fibroblasts, or keratinocytes (Figure 1A). Whereas type I IFN expression by pDCs was very abundant and its induction was rapid (peaking as early as at 6 hours after stimulation), type I IFN mRNA expression by monocytes was moderate and its induction was slower (peaking at 24 hours after stimulation; Figure 1B). Type I IFN induction in monocytes was found to be entirely DNA-dependent (Figure 1C) but to require the presence of LL37 as the huDNA alone was unable to induce it. Thus, similar to the phenomenon described for pDCs,18 LL37 converts otherwise nonstimulatory extracellular self-DNA into a trigger for type I IFN production in monocytes.
Self-DNA-LL37 complexes induce type I IFNs in human monocytes. (A) IFN-α produced by various human blood-derived immune cells (pDCs, monocytes, myeloid DCs, T cells, B cells, and NK cells), and structural nonhematopoietic cells (fibroblasts, keratinocytes) after overnight stimulation with self-DNA-LL37 complexes (105 cells/condition). Each symbol represents an independent experiment, and horizontal bars represent the mean.*P < .0001 (unpaired Student t test). (B) Fold induction of Ifna2 and Ifnb mRNA at different time points (0, 6, 12, and 24 hours) after stimulation of human monocytes and pDCs with self-DNA-LL37 complexes. Representative results are shown from 1 of 3 independent experiments. Gene expression data are normalized to human GAPDH gene. (C) IFN-α produced by monocytes and pDCs after stimulation with increasing concentrations of huDNA either alone or premixed with LL37 (105 cells/condition). Data are representative of at least 5 experiments; error bars represent the SD of triplicate wells.
Self-DNA-LL37 complexes induce type I IFNs in human monocytes. (A) IFN-α produced by various human blood-derived immune cells (pDCs, monocytes, myeloid DCs, T cells, B cells, and NK cells), and structural nonhematopoietic cells (fibroblasts, keratinocytes) after overnight stimulation with self-DNA-LL37 complexes (105 cells/condition). Each symbol represents an independent experiment, and horizontal bars represent the mean.*P < .0001 (unpaired Student t test). (B) Fold induction of Ifna2 and Ifnb mRNA at different time points (0, 6, 12, and 24 hours) after stimulation of human monocytes and pDCs with self-DNA-LL37 complexes. Representative results are shown from 1 of 3 independent experiments. Gene expression data are normalized to human GAPDH gene. (C) IFN-α produced by monocytes and pDCs after stimulation with increasing concentrations of huDNA either alone or premixed with LL37 (105 cells/condition). Data are representative of at least 5 experiments; error bars represent the SD of triplicate wells.
LL37 efficiently transports self-DNA into monocytes
We have previously found that LL37 transports extracellular self-DNA fragments into TLR9-containing endosomal compartments of pDCs leading to type I IFN production.18 To determine whether the DNA-dependent induction of type I IFNs in monocytes is also related to the ability of LL37 to internalize the DNA, we used Alexa488-labeled huDNA and tracked its internalization by flow cytometry. huDNA in complex with LL37 was very efficiently internalized by monocytes compared with other cell types, including mDCs, T cells, B cells, NK cells, fibroblasts, or keratinocytes, suggesting that the IFN induction is related to the uptake of the DNA (Figure 2A-B; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Interestingly, the efficiency of DNA-LL37 complex internalization was even higher than in pDC (98% ± 5% of monocytes vs 30% ± 10% of pDCs at 12 hours of stimulation P < .0001; n = 4 experiments; Figure 2A). Thus, huDNA-LL37 complexes are very efficiently internalized by monocytes but not into T, B, and NK cells or nonhematopoietic structural cells.
LL37 efficiently transports self-DNA into human monocytes. (A) Degree of association of Alexa488-labeled huDNA-LL37 complexes with human blood-derived immune cells (pDCs, monocytes, and myeloid DCs), and structural nonhematopoietic cells (fibroblasts, keratinocytes) at different time points (0, 1, 4, and 12 hours) after stimulation, assessed by flow cytometry. Alexa488-labeled huDNA alone showed minimal association with all cell types tested. Data are the mean ± SEM of 4 independent experiments. *P < .0001 (paired Student t test). (B) Representative FACS analysis of Alexa488 huDNA-LL37 complex association with human blood-derived immune cells (pDCs, monocytes, and myeloid DCs), and structural nonhematopoietic cells (fibroblasts, keratinocytes) 4 hours after stimulation is shown. (C) Intracellular localization of Alexa488 huDNA-LL37 complexes 4 and 24 hours after stimulation of human monocytes, assessed by confocal microscopy. Green represents Alexa488 huDNA; and red, surface staining of monocytes with Alexa647 HLA-DR. Bar represents 5 μm (D) Percentage of cytoplasmic distribution of Alexa488 huDNA-LL37 complexes 4 and 24 hours after stimulation of human pDCs, and monocytes. Data are representative of 3 independent experiments and are given as mean ± SEM of the percentage of cells with cytoplasmic DNAAlexa distribution; at least 150 cells containing Alexa488 huDNA-LL37 complexes from 10 different fields were counted for each condition. *P < .0001 (paired Student t test).
LL37 efficiently transports self-DNA into human monocytes. (A) Degree of association of Alexa488-labeled huDNA-LL37 complexes with human blood-derived immune cells (pDCs, monocytes, and myeloid DCs), and structural nonhematopoietic cells (fibroblasts, keratinocytes) at different time points (0, 1, 4, and 12 hours) after stimulation, assessed by flow cytometry. Alexa488-labeled huDNA alone showed minimal association with all cell types tested. Data are the mean ± SEM of 4 independent experiments. *P < .0001 (paired Student t test). (B) Representative FACS analysis of Alexa488 huDNA-LL37 complex association with human blood-derived immune cells (pDCs, monocytes, and myeloid DCs), and structural nonhematopoietic cells (fibroblasts, keratinocytes) 4 hours after stimulation is shown. (C) Intracellular localization of Alexa488 huDNA-LL37 complexes 4 and 24 hours after stimulation of human monocytes, assessed by confocal microscopy. Green represents Alexa488 huDNA; and red, surface staining of monocytes with Alexa647 HLA-DR. Bar represents 5 μm (D) Percentage of cytoplasmic distribution of Alexa488 huDNA-LL37 complexes 4 and 24 hours after stimulation of human pDCs, and monocytes. Data are representative of 3 independent experiments and are given as mean ± SEM of the percentage of cells with cytoplasmic DNAAlexa distribution; at least 150 cells containing Alexa488 huDNA-LL37 complexes from 10 different fields were counted for each condition. *P < .0001 (paired Student t test).
Interestingly, confocal imaging studies revealed that Alexa488huDNA-LL37 complexes were contained within organelles 4 hours after stimulation but were diffusely distributed in monocytes at 24 hours, suggesting a cytoplasmic distribution (Figure 2C). By contrast, the Alexa488huDNA-LL37 complexes were found to remain in organelles of pDCs even after 24 hours of stimulation (Figure 2D; supplemental Figure 2). These data suggest that DNA/LL37 complexes enter monocytes through the endocytic pathway. In contrast to endocytic retention of DNA in complex with LL37 in pDCs, DNA appears to be shuttled into the cytosol of monocytes.
Self-DNA-dependent type IFN production by monocytes is not induced via TLR9
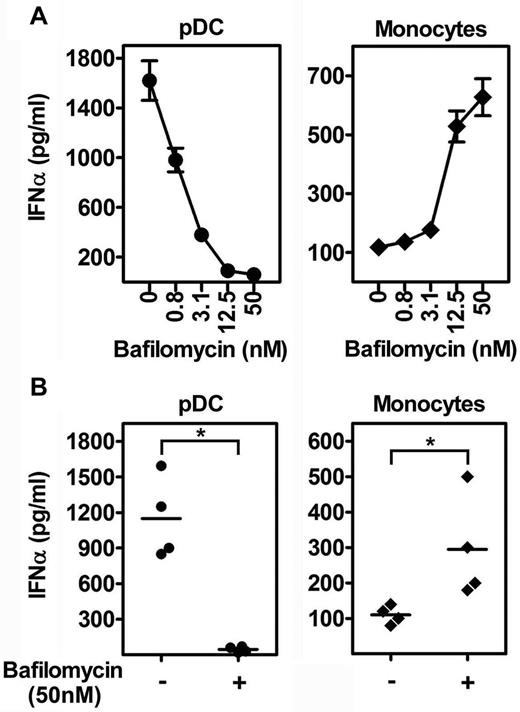
Although TLR9 appears not to be expressed by freshly isolated monocytes,25 we next sought to exclude the possibility that activation of monocytes by DNA-LL37 complexes occurs through endosomal TLRs by pretreatment of cells with bafilomycin, a specific blocker of endosomal TLR signaling.26,27 Bafilomycin efficiently blocked IFN-α induction in pDCs, confirming the requirement for endosomal TLR signaling in pDC activation (Figure 3A-B). In monocytes, bafilomycin failed to block IFN-α induction, on the contrary it significantly increased its production in a dose-dependent manner (Figure 3A-B). Thus, human monocyte activation by DNA-LL37 complexes occurs in a TLR-independent manner.
IFN-α induction in monocytes by self-DNA-LL37 complexes is TLR-independent. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) stimulated with huDNA (huDNA)-LL37 complexes after pretreatment with increasing concentrations of bafilomycin. Data are representative of at least 3 experiments; error bars represent the SD of triplicate wells. (B) IFN-α production by pDCs and monocytes stimulated with huDNA-LL37 complexes with or without pretreatment with 50nM bafilomycin. Each symbol represents an independent experiment, and horizontal bars represent the mean. *P < .0001 (paired Student t test).
IFN-α induction in monocytes by self-DNA-LL37 complexes is TLR-independent. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) stimulated with huDNA (huDNA)-LL37 complexes after pretreatment with increasing concentrations of bafilomycin. Data are representative of at least 3 experiments; error bars represent the SD of triplicate wells. (B) IFN-α production by pDCs and monocytes stimulated with huDNA-LL37 complexes with or without pretreatment with 50nM bafilomycin. Each symbol represents an independent experiment, and horizontal bars represent the mean. *P < .0001 (paired Student t test).
TLR9 specifically recognizes unmethylated or hypomethylated CpG islets in single- or double-stranded DNA sequences.28 To further confirm the IFN response is independent of TLR9 activation, we stimulated monocytes with single-stranded and double-stranded DNA sequences, either containing or lacking CpG motifs (Table 1). All the DNA sequences were generated on a natural phosphodiester backbone, thus mimicking huDNA. LL37 was found to form complexes and condense all these sequences regardless of their sequence (supplemental Figure 3A), but dependent on their charge, as this process was reversible on desalting (supplemental Figure 3B), and secondary structure of the antimicrobial peptide (supplemental Figure 3C). In addition, mutated LL37 peptides with either reduced cationicity (Mut+1) or reduced α helical content (Mut+8) in complex with human genomic DNA (huDNA) failed to induce IFN-α production in human monocytes (supplemental Figure 4).
Information on sequences and source of synthetic oligos of a phosphodiester DNA backbone and plasmids used in studies of LL37-DNA binding affinity
| DNA type . | DNA sequence . | Source . |
|---|---|---|
| Poly A | 5′ AAA AAA AAA AAA AAA 3′ | Trilink |
| Poly T | 5′ TTT TTT TTT TTT TTT 3′ | Trilink |
| CpG DNA (sense) | 5′ XTC GTC GTT TTG TCG TTT TGT CGT TG 3′; | Trilink |
| CpG DNA (antisense) | 5′ XAA CGA CAA AAC GAC AAA ACG ACG AG 3′ | Trilink |
| GpC DNA (sense) | 5′ XTG CTG CTT TTG TGC TTT TGT GCT TG 3′; | Trilink |
| GpC DNA (antisense) | 5′ XAA GCA CAA AAG CAC AAA AGC AGC AG 3′ | Trilink |
| CpG DNA biotin (5′ end; sense) | 5′ TC GTC GTT TTG TCG TTT TGT CGTT 3′ | Sigma-Aldrich |
| CpG DNA (antisense) | 5′ AA CGA CAA AAC GAC AAA ACG ACGA 3′ | Sigma-Aldrich |
| Poly dA:dT | poly (dA:dT) ̇ poly (dT:dA), synthetic phosphodiester ds DNA polymer | Sigma-Aldrich |
| Poly dC:dG | poly (dC:dG) ̇ poly (dG:dC), synthetic phosphodiester ds DNA polymer | Sigma-Aldrich |
| CpG containing plasmid | pBR322 plasmid, 4361 base pairs, contains 297 unmethylated CG base pairs | Invitrogen |
| CpG free plasmid | pCpG-mcs G2, 3066 base pairs, does not contain any CG base pairs | Invivogen |
| DNA type . | DNA sequence . | Source . |
|---|---|---|
| Poly A | 5′ AAA AAA AAA AAA AAA 3′ | Trilink |
| Poly T | 5′ TTT TTT TTT TTT TTT 3′ | Trilink |
| CpG DNA (sense) | 5′ XTC GTC GTT TTG TCG TTT TGT CGT TG 3′; | Trilink |
| CpG DNA (antisense) | 5′ XAA CGA CAA AAC GAC AAA ACG ACG AG 3′ | Trilink |
| GpC DNA (sense) | 5′ XTG CTG CTT TTG TGC TTT TGT GCT TG 3′; | Trilink |
| GpC DNA (antisense) | 5′ XAA GCA CAA AAG CAC AAA AGC AGC AG 3′ | Trilink |
| CpG DNA biotin (5′ end; sense) | 5′ TC GTC GTT TTG TCG TTT TGT CGTT 3′ | Sigma-Aldrich |
| CpG DNA (antisense) | 5′ AA CGA CAA AAC GAC AAA ACG ACGA 3′ | Sigma-Aldrich |
| Poly dA:dT | poly (dA:dT) ̇ poly (dT:dA), synthetic phosphodiester ds DNA polymer | Sigma-Aldrich |
| Poly dC:dG | poly (dC:dG) ̇ poly (dG:dC), synthetic phosphodiester ds DNA polymer | Sigma-Aldrich |
| CpG containing plasmid | pBR322 plasmid, 4361 base pairs, contains 297 unmethylated CG base pairs | Invitrogen |
| CpG free plasmid | pCpG-mcs G2, 3066 base pairs, does not contain any CG base pairs | Invivogen |
In accordance with TLR9 activation, pDCs were found to respond to both single-stranded and double-stranded DNA sequences that contain CpG motifs, but not single-stranded and double-stranded DNA sequences lacking this motif (Figure 4A-B). By contrast, monocytes did not respond to any single-stranded DNA sequences; their responsiveness required double-stranded sequences regardless of the presence of CpG motifs. Furthermore, IFN-α induction in pDCs was entirely blocked, whereas IFN in monocytes was not inhibited or even enhanced by bafilomycin. Because methylation of DNA can mask CpG motifs and abrogate TLR9 recognition, we sought to analyze the effects of DNA methylation on IFN induction in monocytes. Whereas methylation of the double-stranded CpG-DNA sequences abrogated IFN induction by DNA/LL37 complexes in pDCs, it did not affect IFN induction in monocytes (Figure 4C). Taken together, these data demonstrate that monocyte activation by DNA-LL37 complexes does not occur via TLR9.
Type I IFN induction in monocytes by DNA-LL37 complexes requires dsDNA but is independent of DNA sequence and methylation. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with 10 μg/mL of ssCpG DNA, or dsCpG DNA sequences of phosphodiester backbone alone or in complex with LL37 (10μM), with or without pretreatment with bafilomycin (50nM). In contrast to pDCs, in monocytes ssDNA sequences in complex with LL37 failed to induce IFN-α production. Data are representative of at least 4 independent experiments; error bars represent the SD of triplicate wells. (B) IFN-α produced by pDCs and monocytes after overnight stimulation with 10 μg/mL of a CpG-containing plasmid or a CpG-free plasmid, alone or with LL37 (10μM), with or without pretreatment with 50nM bafilomycin. Each symbol represents an independent experiment, and horizontal bars represent the mean. (C) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with 10 μg/mL of unmethylated or methylated dsCpG DNA, either alone or in complex with LL37 (10μM). Data are presented as mean ± SEM of 3 independent experiments. *P < .0001 (paired Student t test).
Type I IFN induction in monocytes by DNA-LL37 complexes requires dsDNA but is independent of DNA sequence and methylation. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with 10 μg/mL of ssCpG DNA, or dsCpG DNA sequences of phosphodiester backbone alone or in complex with LL37 (10μM), with or without pretreatment with bafilomycin (50nM). In contrast to pDCs, in monocytes ssDNA sequences in complex with LL37 failed to induce IFN-α production. Data are representative of at least 4 independent experiments; error bars represent the SD of triplicate wells. (B) IFN-α produced by pDCs and monocytes after overnight stimulation with 10 μg/mL of a CpG-containing plasmid or a CpG-free plasmid, alone or with LL37 (10μM), with or without pretreatment with 50nM bafilomycin. Each symbol represents an independent experiment, and horizontal bars represent the mean. (C) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with 10 μg/mL of unmethylated or methylated dsCpG DNA, either alone or in complex with LL37 (10μM). Data are presented as mean ± SEM of 3 independent experiments. *P < .0001 (paired Student t test).
Self-DNA-dependent type I IFN production by monocytes is induced via cytosolic DNA sensors
To investigate whether the IFN induction by DNA complexes in monocytes is dependent on a specific sequence in the DNA, we next tested the ability of double-stranded poly dA:dT and poly dC:dG sequences to activate monocytes when complexed with LL37. Complexes containing both poly dA:dT and poly dC:dG dsDNA were able to induce IFNs in monocytes, a process, that was enhanced by bafilomycin pretreatment (Figure 5A). On the other hand, consistent with TLR9 activation, pDCs responded only to poly dC:dG-containing complexes, a process that was completely inhibited by bafilomycin (Figure 5A). Thus, DNA-dependent monocyte requires double-stranded DNA and is independent of its sequence. Together with the finding that DNA-LL37 complexes gain access to the cytosol of monocytes, these findings are highly suggestive of an involvement of cytosolic DNA sensors. Several cytosolic DNA receptors have been identified suggesting redundancy of cytosolic DNA sensing. However a common feature of these receptors is that they signal through the adaptor protein STING and TBK-1 kinase leading to IRF3 phosphorylation.5,14,15 To assess whether the cytosolic DNA sensing pathway is involved in type I IFN induction by self-DNA-LL37 complexes in monocytes, we performed RNAi silencing of TBK1 and STING (Figure 5B). Knockdown of both TBK-1 and STING expression in monocytes resulted in a significant reduction of IFN induction compared with monocytes transfected with control RNAi (Figure 5C). By contrast, knockdown of MyD88 (Figure 5B), the key adaptor molecule in TLR9 signaling, did not affect IFN induction in monocytes (Figure 5C). No significant off-target effects were noticed for all RNAi sequences used for silencing of reference genes (supplemental Figure 5). These findings confirm that IFN induction in monocytes is mediated by cytosolic DNA sensors. Cytosolic DNA sensors have been shown to recognize B-form but not Z-form of DNA.12,13 To confirm the involvement of cytosolic DNA sensors poly dC:dG DNA (B-type DNA) was brominated to induce a Z-type DNA24 and used to activate monocytes in complex with LL37. In contrast to B-type DNA, Z-type DNA in complex with LL37 failed to induce IFNs in human monocytes (Figure 5D). Together, these findings demonstrate that LL37 in complex with dsDNA sequences, including self-DNA, triggers type I IFN production in human monocytes via activation of cytosolic DNA sensors.
Type I IFN induction by DNA-LL37 complexes in human monocytes occurs through cytosolic DNA sensors. (A) IFN-α produced by pDCs and monocytes after overnight stimulation with 10 μg/mL of poly dA:dT or poly dC:dG DNA, either alone or in complex with LL37 (10μM), with or without pretreatment with 50nM bafilomycin. Data are representative of at least 5 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test). (B) Real-time PCR of TBK1, MyD88, and STING expression after transfection with siRNA targeting TBK1, MyD88, STING, or control nontargeting RNAi sequences (control siRNA [C RNAi]) in primary human monocytes. Data are representative of at least 3 independent experi-ments; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (C) Real-time PCR of Ifna mRNA induction in primary human monocytes stimulated with huDNA-LL37 complexes 48 hours after transfection with siRNAs targeting TBK1, MyD88, or control nonsilencing siRNAs. Data are representative of at least 4 independent experiments for TBK1 and MyD88 silencing and 2 independent experiments for STING silencing; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (D) ELISA of IFN-α in human monocytes after stimulation with poly dC:dG DNA (B-type DNA) in complex with LL37 complexes or brominated poly dC:dG DNA (Z-type DNA)-LL37. Data are representative of at least 3 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test).
Type I IFN induction by DNA-LL37 complexes in human monocytes occurs through cytosolic DNA sensors. (A) IFN-α produced by pDCs and monocytes after overnight stimulation with 10 μg/mL of poly dA:dT or poly dC:dG DNA, either alone or in complex with LL37 (10μM), with or without pretreatment with 50nM bafilomycin. Data are representative of at least 5 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test). (B) Real-time PCR of TBK1, MyD88, and STING expression after transfection with siRNA targeting TBK1, MyD88, STING, or control nontargeting RNAi sequences (control siRNA [C RNAi]) in primary human monocytes. Data are representative of at least 3 independent experi-ments; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (C) Real-time PCR of Ifna mRNA induction in primary human monocytes stimulated with huDNA-LL37 complexes 48 hours after transfection with siRNAs targeting TBK1, MyD88, or control nonsilencing siRNAs. Data are representative of at least 4 independent experiments for TBK1 and MyD88 silencing and 2 independent experiments for STING silencing; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (D) ELISA of IFN-α in human monocytes after stimulation with poly dC:dG DNA (B-type DNA) in complex with LL37 complexes or brominated poly dC:dG DNA (Z-type DNA)-LL37. Data are representative of at least 3 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test).
LL37 also transports extracellular viral DNA into monocytes for type I IFN production
Cytosolic DNA sensors are activated by dsDNA exposed by the virus during replication in the cytosol. However, certain dsDNA viruses, such as HSV, have developed mechanisms to evade immune recognition by cytosolic sensors by blocking TBK1 activation.29 Because LL37 and its mouse homolog CRAMP were shown to play an important role in antiviral immunity against HSV,30 we asked whether LL37 would promote the activation of cytosolic DNA sensors by transporting HSV-DNA of lysed viruses from the extracellular compartment into the cytosol of noninfected monocytes. To test this possibility, lysates of HSV were mixed with LL37 and added to pDCs and monocytes. While live HSV virus induced robust IFN-α production in pDCs, both live viruses and viral lysates failed to activate monocytes (Figure 6A-B). However, when the lysates were mixed with LL37, there was a significant induction of IFN-α in both monocytes and pDCs. This response was DNA-dependent, as it was abolished by pretreatment of the lysates with DNAse I (Figure 6A). IFN induction in pDCs was abolished by bafilomycin indicating signaling via TLR9, whereas IFN induction in monocytes was enhanced by bafilomycin, suggesting signaling through cytosolic DNA receptors. Like HSV lysates, also VV lysates stimulated production of type I IFNs in pDCs and monocytes when mixed with LL37. Indeed, we found that VV DNA- or HSV DNA-LL37 complexes activated monocytes and pDCs to produce IFN-α in a DNA dose-dependent way (Figure 5C). Thus, during viral infection, LL37 may stimulate TLR-dependent and -independent pathways of type I IFN induction in human pDCs and monocytes.
In addition to huDNA, LL37 complexes with free microbial/viral DNA for type I IFN induction in monocytes. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with lysates of VV and HSV viruses alone or in complex with LL37 (10μM) with or without pretreatment with 50nM bafilomycin. Data are representative of at least 5 experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test). **P < .01 (paired Student t test). (B) IFN-α produced by pDCs and monocytes after overnight stimulation with increasing titers of VV and HSV viruses. HSV failed to trigger IFN-α release in monocytes; error bars represent the SD of triplicate wells. (C) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with increasing concentration of either HSV, or VV, or human genomic DNA from U937 cells (huDNA) in complex with LL37 (10μM). Data are representative of at least 3 experiments.
In addition to huDNA, LL37 complexes with free microbial/viral DNA for type I IFN induction in monocytes. (A) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with lysates of VV and HSV viruses alone or in complex with LL37 (10μM) with or without pretreatment with 50nM bafilomycin. Data are representative of at least 5 experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test). **P < .01 (paired Student t test). (B) IFN-α produced by pDCs and monocytes after overnight stimulation with increasing titers of VV and HSV viruses. HSV failed to trigger IFN-α release in monocytes; error bars represent the SD of triplicate wells. (C) IFN-α produced by pDCs (5 × 104 cells/condition) and monocytes (2 × 105 cells/condition) after overnight stimulation with increasing concentration of either HSV, or VV, or human genomic DNA from U937 cells (huDNA) in complex with LL37 (10μM). Data are representative of at least 3 experiments.
Discussion
The role type I IFNs induced by self-nucleic acid recognition is a key event in the pathogenesis of autoimmunity. Although most evidence relates to self-DNA recognized by endosomal TLR9, a role for cytosolic DNA sensors has also been proposed. However, the mechanism underlying the transport of self-DNA from the extracellular environment into cytosolic compartments has remained unclear. Here we found that the endogenous antimicrobial peptide LL37, which has been implicated in the pathogenesis of IFN-driven autoimmunity in psoriasis and lupus erythematosus, has the ability to transport self-DNA into cytosolic compartments of monocytes leading to type I IFN induction via cytosolic sensors.
The transfer of DNA across cellular membranes into the cytosol involves 3 distinct steps. The first step involves the binding of LL37 and condensation of DNA, which leads to the formation of compacted structures that are protected from extracellular enzymatic degradation.18 This process is mediated by charge interactions between the anionic nucleic acids and the cationic charges of the amphiphatic peptide and is completely independent of the DNA sequence. The second step involves the acquisition of net positive charges by the LL37-DNA complexes, which allows attachment to anionic proteoglycans on cell membranes. This process triggers receptor-independent lipid raft-mediated endocytosis.31 Surprisingly, we observed an increased efficiency in LL37-DNA complex uptake by monocytes, raising the possibility that a specific receptor may be involved in this process. One potential candidate is FPRL1, an LL37 receptor involved in the chemotaxis of monocytes.32 However, blocking of FPRL1 using a specific antagonist (WRW4 inhibitory peptide) did not result in appreciable reduction in the uptake of self-DNA-LL37 complexes by monocytes (supplemental Figure 6). We observed that only CD16-negative human monocyte produced type I IFNs in response to LL37-DNA complexes (supplemental Figure 7). As both CD16-positive and CD16-negative monocytes have the capacity to produce type I IFNs,33 LL37 may be uptaken via specific receptors exclusively expressed in CD16-negative monocytes. Future studies will have to test this possibility. The third step involves the ability of LL37-DNA complexes to efficiently escape endosomal vesicles and enter the cytosolic compartments of monocytes. Interestingly, this process appeared to be enhanced by suppression of endosomal acidification with bafilomycin, similar to the phenomenon observed for lysine-rich cationic peptides34 Specifically, El-Sayed et al showed that complexes of DNA with lysine-rich peptides (eg, LL37) do not efficiently fuse with membranes at acidic pH.34 This is because the acidic environment of endosomes may decrease the total positive charge on lysine-rich peptides because of deprotonation of some adjacent amino groups as a result of electrostatic repulsion. Because bafilomycin inhibits endosomal acidification and promotes a neutral pH, it favors endosomal fusion and escape of DNA complexed with lysin-rich peptides. Thus, bafilomycin may as well promote endosomal fusion and escape of LL37-DNA complexes, leading to entry into the cytosol.
The cytosolic localization of huDNA-LL37 complexes along with the requirement of double-stranded Z-type DNA regardless of sequence, CpG content, and methylation status indicated that IFN production by monocytes was induced via cytosolic DNA sensors. Accordingly, IFN induction was found to be dependent on STING adaptor protein and TBK-1 kinase, master regulators in the signaling of cytosolic nucleic acid sensors. Several TBK1-dependent cytosolic DNA receptors have been identified, including DNA-dependent activator of IFN-regulatory factors,7,8 RNA polymerase III,9 IFI16,10 and helicase DDX41.11 It is therefore likely that several receptors are involved in the TBK1-dependent sensing of LL37-DNA complexes. Whether LL37-DNA complexes preferentially activate novel cytosolic DNA sensors in human monocytes remains an open question.
LL37 is normally not expressed in healthy skin. However, LL37 is rapidly induced on skin injury19,35 and is constantly overexpressed in inflamed skin of psoriasis patients18 at concentrations that exceed 1000μM.36 Furthermore, LL37-self-DNA complexes are abundantly released by neutrophils in SLE.20 In these conditions, LL37-DNA complexes may trigger pathogenic type I IFNs via endosomal TLRs in pDCs and via cytosolic DNA sensors in monocytes. Although pDCs may provide the rapid initial burst of IFN-α, the sustained IFN levels in autoimmunity may originate via cytosolic sensors of non-pDCs. Evidence for a role of cytosolic sensors in IFN-driven autoimmunity comes from DNase II-deficient mice that develop SLE-like autoimmunity that cannot be reversed in the absence of TLR9 or MyD88.21 Furthermore, circulating DNA containing immune complexes can also trigger DC activation via TLR9-independent pathways in SLE22,23
LL37 also plays an essential role in the protection against dsDNA viruses, such as VV and HSV.30,37,38 Accordingly, LL37 can lyze these viruses in vitro,30 and was found to be reduced in the skin of atopic dermatitis patients, which are at increased risk for disseminated HSV or VV infections.37,38 Interestingly, HSV is able to block TBK1 activation in monocytes,29 although it is associated with robust type I IFN induction in vivo via the TLR-independent cytosolic pathway.39 Our data show that LL37 can induce IFN by binding free viral DNA released by lysed HSV and permit TBK-1-dependent activation of the cytosolic sensors in monocytes. Thus, LL37 may allow the host to circumvent a viral immune escape mechanism by directly lysing the virus and forming an immunogenic complex with viral DNA.
In conclusion, our study identifies a novel pathway that controls the ability of free viral and self-DNA to trigger IFN responses based on the presence of LL37. This finding shed new light on the role of antimicrobial peptides in the control of immune responses at epithelial surfaces.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute (PO1 grant CA128913; M.G.), the DANA Foundation (M.G.), and the University of Texas (MD Anderson Cancer Center Core Grant CA16672; D.P.K.).
National Institutes of Health
Authorship
Contribution: G.C. designed research, performed experiments, analyzed data, and wrote the manuscript; J.G. and S.M. performed experiments; R.L., R.L.M., and D.P.K. analyzed data; and M.G. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for G.C. is Department of Medicine, University of Crete, Heraklion, Crete, Greece, and Institute of Molecular Biology and Biotechnology, Foundation for Research and Technology, Heraklion, Crete, Greece.
Correspondence: Michel Gilliet, Department of Dermatology, Centre Hospitalier Universitaire Vaudois, Av de Beaumont 29, 1011 Lausanne, Switzerland; e-mail: michel.gilliet@chuv.ch.



![Figure 5. Type I IFN induction by DNA-LL37 complexes in human monocytes occurs through cytosolic DNA sensors. (A) IFN-α produced by pDCs and monocytes after overnight stimulation with 10 μg/mL of poly dA:dT or poly dC:dG DNA, either alone or in complex with LL37 (10μM), with or without pretreatment with 50nM bafilomycin. Data are representative of at least 5 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test). (B) Real-time PCR of TBK1, MyD88, and STING expression after transfection with siRNA targeting TBK1, MyD88, STING, or control nontargeting RNAi sequences (control siRNA [C RNAi]) in primary human monocytes. Data are representative of at least 3 independent experi-ments; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (C) Real-time PCR of Ifna mRNA induction in primary human monocytes stimulated with huDNA-LL37 complexes 48 hours after transfection with siRNAs targeting TBK1, MyD88, or control nonsilencing siRNAs. Data are representative of at least 4 independent experiments for TBK1 and MyD88 silencing and 2 independent experiments for STING silencing; error bars represent the SD of triplicate wells. *P < .01 (paired Student t test). (D) ELISA of IFN-α in human monocytes after stimulation with poly dC:dG DNA (B-type DNA) in complex with LL37 complexes or brominated poly dC:dG DNA (Z-type DNA)-LL37. Data are representative of at least 3 independent experiments; error bars represent the SD of triplicate wells. *P < .0001 (paired Student t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/18/10.1182_blood-2012-01-401364/4/m_zh89991297570005.jpeg?Expires=1763471642&Signature=RO5ndYiUs2IoKmr9AYa3oUXBaJLEURZ58N8~oNMn6c-HS6NB-dmTbH~6hScv7nydVeI9ZsczSTP3zt-p1gC8rVllR8zRExGrBPO4epwxUIQVIR-YSPoWpCJvFyojc0HKQS6K5LpX10vYHOmnIm8~DyVPJPjMEPZ7HdORHs2OntiHrd8RxHB-rVHNyrPLyOHSE2OJ0a5jCe3cXV4SIgr3LHCZ5wIuhVz1ZiThi7DSe6fOJTAmnp9Yyb5DMT-TDks-qY6Rcnv6gCP1rCh-ZUhP1koH-JhQW~TyKiJ~r52tIdVq~-Q5mZpWVXZnFSARgcxcQDvIn4~N71cridO9ZmVweA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal