Abstract
We have analyzed the role of the REL family members in Hodgkin lymphoma (HL). shRNA targeting of each REL member showed that HL was uniquely dependent on relB, in contrast to several other B-cell lymphomas. In addition, relA and c-rel shRNA expression also decreased HL cell viability. In exploring relB activation further, we found stable NF-κB inducing kinase (NIK) protein in several HL cell lines and that NIK shRNA also affected HL cell line viability. More importantly, 49 of 50 HL patient biopsies showed stable NIK protein, indicating that NIK and the noncanonical pathway are very prevalent in HL. Lastly, we have used a NIK inhibitor that reduced HL but not other B-cell lymphoma cell viability. These data show that HL is uniquely dependent on relB and that the noncanonical pathway can be a therapeutic target for HL. Furthermore, these results show that multiple REL family members participate in the maintenance of a HL phenotype.
Introduction
Hodgkin lymphoma (HL) is a rare lymphoma characterized by the hallmark Reed-Sternberg cell (RS) containing bilobed nuclei and prominent nucleoli.1,2 It appears likely that HL is a germinal center-derived lymphoma arising mostly from a B cell.1,2 Indeed, RS cells were found in most cases to contain Ig rearrangements and mutations indicative of a germinal center B-cell (GCB) origin.1,3,4
Many B-cell lymphomas, including HL, are dependent on activating mutations in the NF-κB pathways.5 NF-κB signaling consists of the canonical and noncanonical pathway (NCP).6-10 On activation of the canonical pathway, the IKK complex is activated (specifically IKKβ), which in turn phosphorylates the IκB complex, specifically IκBα. The IκB complex is bound up with most notably relA/p50 and c-rel/p50 heterodimers. On phosphorylation and degradation of IκBα, these heterodimers translocate to the nucleus to activate NF-κB target genes. The activation of the NCP is mediated through the stabilization of NF-κB inducing kinase (NIK) protein, which is constitutively degraded by a TRAF2/TRAF3/cIAP1/2 ubiquitin ligase complex in the absence of ligands. Ligand binding to receptors inactivates the TRAF2/TRAF3/cIAP complex, thus negating the degradation of NIK. Stable NIK in turn phosphorylates IKKα, and both NIK and IKKα phosphorylate the N-terminus of p100. Subsequently, p100 is processed to p52, and the p52/relB heterodimers translocate to the nucleus.
HL cell lines consistently show constitutive activity of NF-κB, and several genetic lesions that affect the classic NF-κB pathway have been identified in HL cell lines. c-rel amplifications are present in almost 50% of classic HL cases (cHL), and mutations in IκBα and IκBϵ have also been reported.11-16 More recently, mutations in the tumor suppressor A20 protein, a negative regulator of the canonical NF-κB pathway, were found in 30%-40% of cases.17 As far as the NCP is concerned, nuclear relB has been reported in 3 HL cell lines, HDLM-2, L540, and L428.18,19 The significance of these results is unclear as the first 2 lines are of T-cell origin and T-cell HLs represent only a few percent of all HLs, if they exist at all.1 Second, the conclusion that relB is present in HL cell lines is disputed.20 Nevertheless, these cell lines do express NIK and have phosphorylated p100, suggesting that the NCP is indeed activated.18,19 More recently, several groups have described mutations in TRAF3 (a negative regulator of the NCP) and have identified increased copies of NIK, whose stable expression activates the NCP.21,22
We set out to address the functional contributions of relA, c-rel, and relB to both HL and aggressive B-cell lymphomas for several reasons. First, and this applies to all B-cell lymphomas, the relative contributions of each REL to the lymphoma phenotype have not been explored. It is not clear which REL family members participate in these lymphoma phenotypes. Second, there is no evidence for a functional requirement of c-rel in HL despite the documented amplifications of c-rel in ∼ 50% of HL patients. Third, there is little functional evidence for the role of the NCP in many of these lymphomas, except for multiple myeloma (MM).23,24
Here we have used REL shRNAs to target relA, relB, and c-rel in HL and relB in several other B-cell lymphoma cell lines modeling the most frequent subtype of non-HLs, diffuse large B-cell lymphoma (DLBCL) and mantle cell lymphoma cell lines. DLBCL consist of 3 subtypes described by gene expression profiling: GCB-DLBCL, activated B-cell (ABC)–DLBCL, and primary mediastinal B-cell lymphomas (PMBL).25 Our results show that relB is unique among the REL family in B-cell lymphomas, playing a very specific and unique role in HL and not in any NHLs. In addition, we show that HL requires all 3 REL family members where each is supplying essential functions in cell viability. We explore the requirements for the NCP in HL and show that HL cell lines are dependent on NIK expression and that all HL patient biopsies we have assayed contain stable NIK protein. We also provide evidence that relB positively autoregulates its own expression by repressing a negative regulator of the NCP, TRAF2. Lastly, we show that a small-molecule inhibitor of NIK specifically targets NIK and kills only HL cell lines. These data show unequivocally that relB and the noncanonical NF-κB pathway are essential features of the HL phenotype.
Methods
Cell lines
Cell lines were maintained in the corresponding growth medium supplemented with 20% FBS (Hyclone, Defined) and 1% antibiotics as follows: mantle cell lymphoma (MCL) cell lines HBL-2, Jeko, and UPN1, PMBL cell lines MedB1 and K1106, germinal center like-diffuse large B cell lymphoma (GCB-DLBCL) cell lines BJAB, SUDH4, and HT, HL cell lines L1236, U-H01, KM-H2, L428, and L540, and the MM cell lines UTMC2 and L363 were maintained in RPMI. Activated B-cell like DLBCL (ABC-DLBCL) U2932, HBL-1, OCI-Ly3, OCI-Ly10 cell lines were grown in IMDM. Each lymphoma and MM cell line used in the toxicity screens was first transduced with a feline endogenous virus expressing the ecotropic retroviral receptor and then secondarily infected with a retrovirus expressing the bacterial tetracycline repressor (TetR). The engineering process of the different cell lines used in the toxicity assays was described in details in.26
Retroviral production
The packaging 293-T cells were transfected with the pRSMX PG-eGFP-Puro vector delivering the shRNA for relA, relB, and c-rel and the empty retrovirus backbone-PG-eGFP-Puro as a control; along with the following helper vectors: the mutant ecotropic envelope-expressing plasmid EA6 × 3* and the pHIT60 Gag/Pol expressing plasmid using Lipofectamine 2000. Virus supernatant was collected at 48 and 72 hours after transfection. Doxycycline inducible lymphoma cells were spin-infected twice on consecutive days at 2500 rpm for 90 minutes. Virus was produced and lymphoma cells were infected as described.26
Toxicity assays
ABC-DLBCL, GCB-DLBC, MCL, PMBL, HL, and MM engineered cell lines were transduced with the retrovirus delivering the shRNA sequences to knock down the expression of relA, relB, and c-rel as well as the empty retrovirus backbone as a control as described in “Retroviral production.” Doxycycline (20 ng/mL) was added to the culture medium 2 days after the second round of spin infection. Cells expressing the shRNA sequences or the vector control were monitored by flow cytometry for a coexpressed GFP marker. The GFP+ cell population was measured by FACS every 2 days during 12 days. For each time point and shRNA sequence, the data were normalized to the percentage of GFP+ cells at day 2 after induction of the hairpin expression. Then the GFP+ fraction of shRNA-induced cells was normalized to the GFP+ fraction in parallel cultures transduced with the corresponding mock-infected control. Toxicity assays were performed as described by Staudt et al.26-29 Toxicity assays were initially done with 6 different shRNAs that targeted each REL, then twice with 2 different shRNAs targeting each REL.
RelB rescue experiments
RelB (Invitrogen; IOH11686) was cloned into the doxycycline inducible retrovirus PGK-CMV-puro. Silent mutations were introduced in the relB cDNA used for these rescue experiments. Point mutations within the coding region of relB that was targeted by the shRNA hairpin used in this experiment were introduced by PCR using the QuickChange Site-Directed Mutagenesis Kit (#200519) from Stratagene following the manufacturer's instructions. The purpose was to prevent the exogenous relB from being targeted by the shRNA for relB. L1236, U-H01, and KM-H2 HL cell lines were first transduced with the expression vector delivering the relB cDNA or the corresponding control. Transduced cells (mutated relB and control infected) were selected with puromycin for a week. These HL cell lines were then infected with the relB shRNA-containing virus, followed by FACS analysis to identify infected cells by GFP expression. Data were analyzed as described in “Toxicity assays.”
Immunoblot analysis
Infected and control cells (10 × 106) were lysed at indicated time points for each experiment in buffer consisting of 50mM Tris-HCL pH 7.5, 200mM NaCl, 50mM β-glycerophosphate, 1% Tween-20, 0.2% Nonidet P-40, and a cocktail of protease inhibitors (Roche Diagnostics). Proteins were resolved by 10% SDS-PAGE gels after immunoblot. Membranes were incubated with the primary antibodies (supplemental Tables, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) 1 hour at room temperature, except for anti-NIK and anti-TRAF2, which were incubated overnight at 4°C. Peroxidase-conjugated goat anti–rabbit (111-035-003; Jackson ImmunoResearch Laboratories) was used as second antibody.
Immunohistochemistry
NIK stains were performed on formalin-fixed, paraffin-embedded tissue sections of HL, composing cHL, and nodular lymphocyte predominant HL (NLPHL), cell pellets of cHL cell lines (L1236 and U-H01), a NHL cell line (SUDH4), and reactive tonsils as controls. Sections were deparaffinized and rehydrated in graded alcohol and then placed in a low pH antigen retrieval solution (Dako North America) and steamed for 30 minutes. Sections were blocked with Tris-goat (5%) solution, incubated for 60 minutes at room temperature with NIK polyclonal antibody (Abcam). Dako Envision + System-HRP labeled anti–rabbit and 3,3′-diaminobenzidine + chromogen were used for detection. The NIK stains were scored 0 to 3+ (1 indicates rare positive; 2+, a subset; and 3+, the majority) as either RS or lymphocyte predominant or background cells. Images were taken with Olympus Bx41 microscope: objective UPlanFI 40×/0.75 ∞/0.17, adaptor U-TV0.5xC, digital camera Q-imaging Micropublisher Version 5.0 RTV. The images were captured with “Q-Capture Version 3.1” and imported to Adobe Photoshop Version 7.0. A routine immunohistochemical panel was used to characterize 50 biopsy samples diagnosed as either cHL (31 cases) or NLPHL (19 cases) as outlined in the WHO classification.30 Cases of cHL were classified as mixed cellularity,9 nodular sclerosis,14 lymphocyte-rich,4 and not otherwise specified.4 In situ hybridization for EBV with the EBER probe was performed as previously described.31
Treatment with a NIK inhibitor
Cells were plated at 1 × 106 in 6-well plates in duplicates and treated with 28nM compound 5 [1,3 (2H, 4H)-isoquinolinedione]32 or DMSO 100% as a control each 48 hours. Total cell viability using trypan blue was assayed every other day.
Nuclear and cytoplasmic extracts
To determine the noncanonical NF-κB pathway components that are affected by the NIK Inhibitor, we prepared nuclear and cytoplasmic extracts from L1236 and U-H01 HL cell lines and performed Western blots for Ser176-IKKα, NF-κB2/p100, p52, relB, and NIK. HL cell lines were treated with the NIK inhibitor (28nM) and DMSO as a control every other day. Extracts were prepared at 48, 72, and 96 hours after treatment following a published method.33 Cells were pelleted, washed in PBS, resuspended in buffer A (10mM HEPES-KOH, pH 7.9, at 4°C, 1.5mM MgCl2, 10mM KCl, 0.5mM DTT, 0.2mM PMSF, 50mM β-glycerophosphate, protease inhibitor cocktail; Roche Applied Science) and allowed to swell on ice for 10 minutes. The pellet was resuspended in buffer C (20mM MgCl2, 0.2mM EDTA, 0.5mM DTT, 0.2mM PMSF, 50mM β-glycerophosphate, protease inhibitor cocktail; Roche Diagnostics) and incubated on ice for 20 minutes.
Results
HL, but not NHL, cell lines require relB for viability
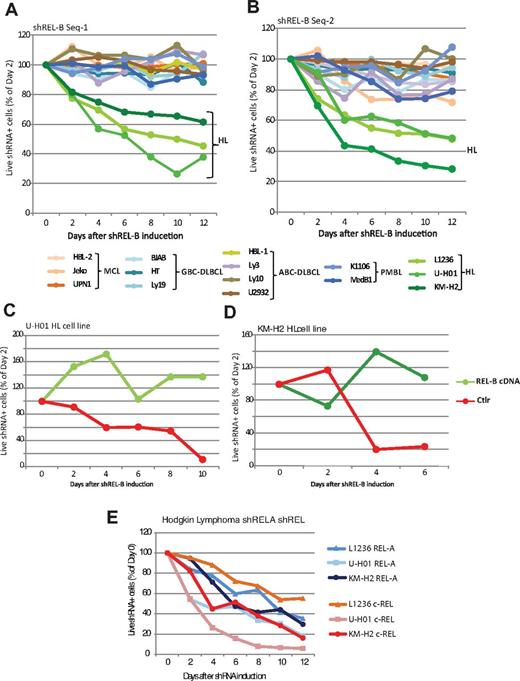
To determine the requirements for members of the NCP in NHL and HL, we constructed retrovirus expression systems containing inducible REL shRNAs fused to GFP. The virus also contained the puromycin selectable marker to initially select for infected cells. Toxicity assays were performed as described by Staudt et al.26-29 Toxicity assays were initially done with 6 different shRNAs that targeted each REL; then the assay of each cell line was performed twice with 2 different shRNAs targeting each REL. On treatment with doxycycline to induce shRNA expression, the GFP-positive cells (containing the shRNA) were segregated by FACS and the shRNA induced cells were examined for viability starting 2 days after transduction for a 2-week period (see “Methods” for description of the data analysis).
We first infected NHL cell lines representing MCL, GCB-DLBCL, and ABC-DLBCL, PMBL, and HL (Figure 1A). To our surprise, we found that, of all of those cell lines, only the viability of the HL lines was affected by the relB shRNA. A reduction in relB mRNA was confirmed for all cell lines tested (data not shown). We observed these effects with a second relB shRNA as well (Figure 1B).
HL cell lines require relA, relB, and c-rel for maintenance of cell viability. (A-B) shRNA knockdown of RelB in the HL and NHL cell lines as indicated underneath the graphs. Each panel represents a different relB shRNA. (C-D) Ectopic relB expression (with PCR-introduced silent mutations that evades being targeted by the relB small hairpin) rescued the relB shRNA toxicity in HL cells. The line labeled “Ctrl” represents expression of the relB shRNA. The relB rescue is shown for 2 HL cell lines KM-H2 (C) and U-H01 (D). (E) shRNA knockdown of RelA and c-Rel in HL cell lines.
HL cell lines require relA, relB, and c-rel for maintenance of cell viability. (A-B) shRNA knockdown of RelB in the HL and NHL cell lines as indicated underneath the graphs. Each panel represents a different relB shRNA. (C-D) Ectopic relB expression (with PCR-introduced silent mutations that evades being targeted by the relB small hairpin) rescued the relB shRNA toxicity in HL cells. The line labeled “Ctrl” represents expression of the relB shRNA. The relB rescue is shown for 2 HL cell lines KM-H2 (C) and U-H01 (D). (E) shRNA knockdown of RelA and c-Rel in HL cell lines.
To control for the specificity of the relB shRNA and to eliminate potential nonspecific toxic effects, we expressed a relB cDNA during relB shRNA induction. We first infected KM-H2 or U-H01 HL cells with a virus expressing relB cDNA. Silent mutations were introduced into the relB cDNA to avoid being targeted by the relB shRNA. After selecting for stable cell lines, we infected the cells with the relB shRNA-containing virus and treated the cells with doxycycline to induce shRNA expression. We found that the relB expression vector rescued HL cell lines from the relB shRNA lethality, confirming that the decrease in viability was a specific response to relB depletion (Figure 1C-D). We further assayed for any toxicity effects of the shRNAs by infecting the human cervical carcinoma HeLa cell line with relA, relB, or c-rel shRNAs (data not shown). We did not observe any loss of viability.
As expected from literature indicating mutations in the canonical pathway, we found that both relA and c-rel shRNAs led to significant loss of cell viability for the 3 HL lines tested (Figure 1E). Thus, HL displays a unique requirement for both the canonical and NCPs.
Stabilization and accumulation of NIK maintain relB signaling in HL cell lines
We next wished to address the causes of sustained relB signaling in HL cell lines. This pathway is dependent on the stabilization and accumulation of NIK.9 To determine whether the NCP was indeed active in the HL cell lines, we assayed for both positive and negative regulators of this pathway by Western blot. Under normal conditions, NIK undergoes constitutive degradation, preventing it from phosphorylating and activating IKKα.9,34 As such, NIK is usually undetectable by Western blot. However, we found that HL cell lines L1236, U-H01, L428, and L540 expressed significant amounts of NIK, whereas typical GCB-DLBCL, ABC-DLBCL, and PMBL cell lines did not (Figure 2B). We also found truncated NIK species in the KM-H2 HL cell line (data not shown). The only other cell line expressing NIK was the NIK-dependent MM line L363.23 Stabilized NIK suggests that there is a defect in the TRAF2/TRAF3/c-IAP-1/c-IAP-2 complex, which is responsible for NIK degradation.9 Indeed, we observed a lack of TRAF2 expression at the protein level in the L1236 cell line and little TRAF3 protein in the U-H01, L428, and L540 HL cell lines (Figure 2A). U-HO1 cells also contain inactivating mutations in TRAF3 genes.35 Lastly, the HL cell lines SUPHD1 and HMD2 were negative for both TRAF2 and TRAF3 (data not shown).
Stable NIK expression occurs in all assayed HL cell lines. (A) Western blot analysis of NIK, TRAF2, and TRAF3 components of the NCP in HL cell lines KM-H2, L1236, U-H01, L428, and L540. (B) Western blot analysis of NIK protein levels in GCB-DLBCL lines HT and SUDH4, ABC-DLBCL lines Ly3, Ly10, and HBL-1, MM cell line L33, and PMBL cell lines MedB1 and K1106. (C) Western blot analysis of relB, NIK TRAF2, and TRAF3 levels on induction of relB shRNA in U-HO1 HL cell line, from 1 to 4 days after shRNA induction.
Stable NIK expression occurs in all assayed HL cell lines. (A) Western blot analysis of NIK, TRAF2, and TRAF3 components of the NCP in HL cell lines KM-H2, L1236, U-H01, L428, and L540. (B) Western blot analysis of NIK protein levels in GCB-DLBCL lines HT and SUDH4, ABC-DLBCL lines Ly3, Ly10, and HBL-1, MM cell line L33, and PMBL cell lines MedB1 and K1106. (C) Western blot analysis of relB, NIK TRAF2, and TRAF3 levels on induction of relB shRNA in U-HO1 HL cell line, from 1 to 4 days after shRNA induction.
RelB autoregulation by repression of TRAF2
After finding that the L1236 HL cell line was deficient in TRAF2 protein, we reexamined the relB shRNA-induced L1236 cell line to look at the behavior of NIK and TRAF2. We found that the induction of relB shRNA induced TRAF2 protein expression and down-regulated NIK protein levels (Figure 2C). This is consistent with the known negative regulatory properties of TRAF2 on NIK expression.36-38 Furthermore, the TRAF2 derepression in HL cells (Figure 2C) is also observed in the induction of a NIK shRNA in HL cell lines (see Figure 4B). These data show that nuclear relB positively autoregulates itself by repressing TRAF2 expression, thus preventing the degradation of NIK.
Stabilization and accumulation of NIK also occur in primary HL tumors
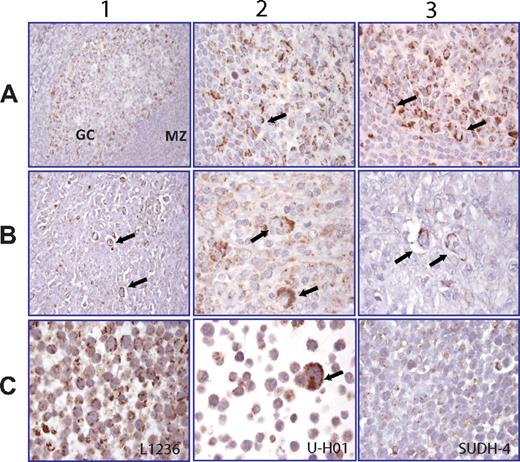
Having observed significant levels of NIK protein in HL cell lines, we next asked whether stable NIK protein existed in primary HL tumors. We checked this by NIK immunohistochemistry of HL patient biopsies, using normal tonsil as a control. In normal tonsil, the GCB cells, especially centroblasts, showed NIK expression, in contrast to the surrounding mantle zone cells (Figure 3A).
Stable NIK expression can be detected in both HL cell lines and primary HL biopsies. (A) Row 1: Normal human tonsil. NIK expression is restricted to the germinal center (GC), particularly centroblasts appearing positive, whereas the mantle zone cells (MZ) are negative. Rows 2-3: High magnification of A1 showing a cytosolic granular staining in centroblasts. (B) Rows 1-3: Lymph node biopsies of 3 HL patients. Arrows indicate the granular cytosolic NIK staining in RS cells. (C) Rows 1-3: HL cell lines L1236 (C1) and U-H01 (C2) with NIK expression in the cytosol of tumor cells. (C3) SUDH-4, a DLBCL lymphoma cell line, is negative.
Stable NIK expression can be detected in both HL cell lines and primary HL biopsies. (A) Row 1: Normal human tonsil. NIK expression is restricted to the germinal center (GC), particularly centroblasts appearing positive, whereas the mantle zone cells (MZ) are negative. Rows 2-3: High magnification of A1 showing a cytosolic granular staining in centroblasts. (B) Rows 1-3: Lymph node biopsies of 3 HL patients. Arrows indicate the granular cytosolic NIK staining in RS cells. (C) Rows 1-3: HL cell lines L1236 (C1) and U-H01 (C2) with NIK expression in the cytosol of tumor cells. (C3) SUDH-4, a DLBCL lymphoma cell line, is negative.
We next assayed for NIK expression in HL patient biopsies. A total of 50 biopsies were stained for NIK, including 31 cHL and 19 NLPHL samples (see Table 1 for details). RS cells showed a specific granular pattern for NIK expression localized to one pole of the cell in 30 of 31 (96.8%) cHL biopsies, irrespective of histologic subtype or prior treatment status (Table 1). In 16 of 19 (84.2%) NLPHLs, specifically the lymphocyte predominant cells (large tumor cells that usually resemble RS cells) showed NIK expression with the same pattern as described in Table 1; the other 3 NLPHL cases showed NIK expression in cells from the environment. Three representative patient samples are shown with NIK positive RS cells (Figure 3B rows 1-3). There was no correlation between EBV positivity of the neoplastic cells and the expression of NIK. Finally, the HL cell lines (L1236 and U-H01) and the NHL cell line SUDHL-4 were analyzed by immunohistochemistry (Figure 3C rows 1-3). The Hodgkin cell lines were positive for NIK, whereas SUDHL-4, a GCB-DLBCL cell line, is negative for NIK staining (Figure 3C row 3). These data show that both the HL cell lines and primary HL tumors have a constitutively active noncanonical NF-κB pathway.
NIK expression in HL patient biopsies in the neoplastic cells (RS cells and LP cells)
| Diagnosis . | No. of cases . | Score 3+ . | Score 2+ . | Score 1+ . | 0 . |
|---|---|---|---|---|---|
| cHL* | 31 | 9 | 8 | 13 | 1 |
| Subtype MC | 9 | 3 | 2 | 4 | 0 |
| Subtype NS | 14 | 2 | 3 | 8 | 1 |
| Subtype LR | 4 | 1 | 3 | 0 | 0 |
| NOS | 4 | 3 | 0 | 1 | 0 |
| NLPHL† | 19 | 9 | 7 | 3 | 0 |
| Total | 50 | 18 | 15 | 16 | 1 |
| Diagnosis . | No. of cases . | Score 3+ . | Score 2+ . | Score 1+ . | 0 . |
|---|---|---|---|---|---|
| cHL* | 31 | 9 | 8 | 13 | 1 |
| Subtype MC | 9 | 3 | 2 | 4 | 0 |
| Subtype NS | 14 | 2 | 3 | 8 | 1 |
| Subtype LR | 4 | 1 | 3 | 0 | 0 |
| NOS | 4 | 3 | 0 | 1 | 0 |
| NLPHL† | 19 | 9 | 7 | 3 | 0 |
| Total | 50 | 18 | 15 | 16 | 1 |
A total of 55% of the cHL RS cells showed NIK positivity, and the positivity shows no association with cHL subtypes, whereas 84% of NLPHL LP cells were NIK positive.
LP indicates lymphocyte predominant; MC, mixed cellularity; NS, nodular sclerosis; LR, lymphocyte rich; and NOS, not otherwise specified.
≥ 2+ indicates 17 of 31 (55%).
≥ 2+ indicates 16 of 19 (84%); and ≥ 1+, NIK expression in both cHL and NLPHL: 49 of 50 (98%).
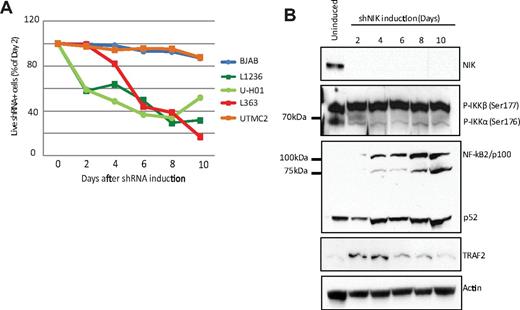
In accordance with the HL dependence on relB and presence of stabilized NIK protein, the L1236 and U-H01 HL cell lines are sensitive to NIK protein down-regulation induced by a NIK-specific shRNA (Figure 4A). NIK knockdown also reduced cell viability of the NIK-dependent L363 MM cell line.23 Conversely, UTMC2, a NIK-independent MM cell line and the BJAB GCB-DLBCL cell line are not affected by NIK shRNA (Figure 4A).
NIK shRNA adversely affects HL cell line viability. (A) The depletion of NIK with a NIK-specific shRNA kills HL cell lines L1236 and U-HO1 and the NIK-expressing L363 MM cell line.23 BJAB and the UTMC2 cells (a NIK-independent MM cell line23 ) are used as negative controls. (B) Western blot analysis of NIK, phospho-IKKα, p100/p52, and TRAF2 levels after induction of the NIK shRNA. Actin levels as a loading control.
NIK shRNA adversely affects HL cell line viability. (A) The depletion of NIK with a NIK-specific shRNA kills HL cell lines L1236 and U-HO1 and the NIK-expressing L363 MM cell line.23 BJAB and the UTMC2 cells (a NIK-independent MM cell line23 ) are used as negative controls. (B) Western blot analysis of NIK, phospho-IKKα, p100/p52, and TRAF2 levels after induction of the NIK shRNA. Actin levels as a loading control.
We further tested for the effects of the NIK shRNA on molecular events downstream of NIK. If NIK activity is being reduced, we would expect to see reductions in phospho-IKKα and the accumulation of p100. Western blot analysis of NIK shRNA-induced HL cell line L1236 clearly showed a reduction in phospho-IKKα Ser-176 levels and the accumulation of p100 levels. In addition, we also noted a dramatic rise in expression of TRAF2, which we showed in Figure 2C to be under negative regulation by relB. Hence, both reductions in relB and NIK result in the expression of TRAF2 and the loss of NIK protein, shutting down the NCP.
A NIK inhibitor specifically kills HL but not NHL cell lines
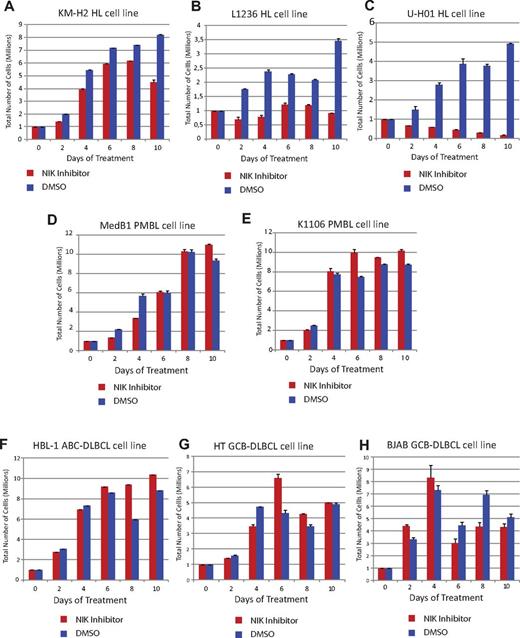
We next examined the efficacy of a small-molecule inhibitor of NIK.32 We treated cells with 4H-isoquinoline-1,3-dione (at a concentration of 26nM for 48 hours) or the vehicle (DMSO) as a control. 4H-isoquinoline-1,3-dione specifically killed the KM-H2, L1236, and U-H01 HL cell lines (Figure 5A-C). The compound did not affect the survival of PMBL lymphomas (MedB and K1106; Figure 5D-E), DLBCL subtypes ABC-DLBCL (HBL-1), or GCB-DLBCL (HT and BJAB) cell lines (Figure 5F-H). All of these negative control cell lines are negative for stable NIK expression (Figure 2B; and data not shown).
Small-molecule drug targeting NIK protein specifically reduces HL cell line viability. (A-C) Treatment of L1236, KM-H2, and U-H01 HL cell lines with the NIK inhibitor 4H-isoquinoline-1,3-dione. (D-E) Med-B1 and K1106 cell line survival is not affected by 4H-isoquinoline-1,3-dione. (F-H) Neither HBL-1 (F) nor HT (G) nor BJAB (H) cell lines were sensitive to the 4H-isoquinoline-1,3-dione. Error bars represent the SD of quadruplicates from a representative experiment.
Small-molecule drug targeting NIK protein specifically reduces HL cell line viability. (A-C) Treatment of L1236, KM-H2, and U-H01 HL cell lines with the NIK inhibitor 4H-isoquinoline-1,3-dione. (D-E) Med-B1 and K1106 cell line survival is not affected by 4H-isoquinoline-1,3-dione. (F-H) Neither HBL-1 (F) nor HT (G) nor BJAB (H) cell lines were sensitive to the 4H-isoquinoline-1,3-dione. Error bars represent the SD of quadruplicates from a representative experiment.
We next wished to determine whether the loss of NIK affected events downstream of NIK in the NCP. As with the NIK shRNA, we found by Western blot analysis that, in addition to the loss of NIK protein, we also could not detect any phospho-IKKα in cytoplasmic extracts from 4H-isoquinoline-1,3-dione–treated L1236 and U-H01 HL cell lines (Figure 6A-B). Furthermore, we also saw significant reductions in nuclear extracts of both relB and p52 (Figure 6C-D). All of these data indicate that the NCP is severely hampered by the effects of 4H-isoquinoline-1,3-dione.
Evidence that the NIK inhibitor 4H-isoquinoline-1,3-dione specifically targets the NCP. (A) Western blot analysis of 4H-isoquinoline-1,3-dione–treated L1236 HL cell line S100 cytoplasmic extracts for NIK and phospho-IKKα protein levels. (B) Western blot analysis of 4H-isoquinoline-1,3-dione–treated U-H01 HL cell line for NIK and phospho-IKKα protein levels. (C) Western blot analysis of 4H-isoquinoline-1,3-dione–treated L1236 HL cell line nuclear extracts for relB and p52 levels compared with TBP levels. (D) Western blot analysis of 4H-isoquinoline-1,3-dione–treated U-H01 HL cell line nuclear extracts for relB and p52 levels compared with TBP levels.
Evidence that the NIK inhibitor 4H-isoquinoline-1,3-dione specifically targets the NCP. (A) Western blot analysis of 4H-isoquinoline-1,3-dione–treated L1236 HL cell line S100 cytoplasmic extracts for NIK and phospho-IKKα protein levels. (B) Western blot analysis of 4H-isoquinoline-1,3-dione–treated U-H01 HL cell line for NIK and phospho-IKKα protein levels. (C) Western blot analysis of 4H-isoquinoline-1,3-dione–treated L1236 HL cell line nuclear extracts for relB and p52 levels compared with TBP levels. (D) Western blot analysis of 4H-isoquinoline-1,3-dione–treated U-H01 HL cell line nuclear extracts for relB and p52 levels compared with TBP levels.
Discussion
Our studies have yielded several significant insights into the functions of the REL family members in HL. It is important to note that, although other studies have provided evidence of NF-κB involvement in HL, none has shown functional requirements for relA, relB, or c-rel. IκB experiments have shown that the canonical pathway is involved in HL, but no distinctions between relA and c-rel could be made. Second, there has been no functional evidence directly supporting a role of relB and the NCP in HL. Our shRNA analysis fills in several important gaps in our knowledge of NF-κB functions in HL.
A role for the NCP and relB in HL
Prior evidence for a role of the NCP in HL can be inferred from several observations. The first is the correlation between EBV and HL. It is thought that the expression of EBV proteins LMP1 and LMP2 mimics the CD40 signaling that can activate both the canonical and NCPs.39 A second indication of the involvement of the NCP in HL is the expression of NIK in the T cell–derived HL cell line L54019 and nuclear relB in 2 T cell–derived HL lines and one B cell–derived HL line.18,19 In addition, recent genetic evidence showing TRAF3 deficiencies and NIK amplifications supports a functional role of the NCP in HL.21,35
Most interestingly, we show that relB and the NCP are unequivocally active and required in HL. RelB shRNA experiments show a clear loss of HL cell line viability (Figure 1). Activation of the NCP, as exemplified by stable NIK protein expression,37,40-43 is clearly occurring in HL cell lines and in 49 of 50 HL tumor biopsies (Figures 2 and 3; Table 1). Supporting this is NIK shRNA targeting, which, like relB shRNA, results in significant decreases in HL cell line viability (Figure 4). Furthermore, 4-isoquinolinedione, an NIK inhibitor, specifically affects HL cell line viability while not affecting any other B-cell lymphoma cell lines, which do not express stable NIK protein (Figures 5 and 6).
We also noted that several HL cell lines were deficient for either TRAF2 or TRAF3 (Figure 2). Both TRAF2 and TRAF3 are negative regulators of the NCP. These proteins form a complex with either cIAP1 or cIAP2 and function as ubiquitin ligases necessary for the constitutive degradation of NIK.44 Loss of either protein in mouse knockout experiments stabilizes NIK protein and activate the NCP.36-38 That we see such losses in several HL cell lines suggests that this is a mechanism used in HL etiology. This is supported by genetic studies of HL tumors and cell lines showing TRAF deficiencies and NIK amplifications.21,35
Our experiments using the relB shRNA support this conclusion. They also show a novel mechanism of activation of the NCP by other than DNA mutations. We found that targeted ablation of relB or NIK resulted in the expression of TRAF2 in the L1236 HL cell line (Figures 2 and 4). TRAF2 was not expressed in the wild-type L1236 cell line. It is clear then that relB has established a positive autoregulatory loop by repressing TRAF2 expression.
The role of relA and c-rel in HL
The activity of the canonical pathway, but not relA and c-rel themselves, has been shown extensively by others.2 As we have already mentioned, it was not possible from those experiments to make any conclusions as the respective roles of c-rel and relA in HL, as both are activated in the canonical pathway. DNA analysis of HL cell lines and tumor samples, however, did suggest that c-rel was specifically involved in HL because of c-rel amplifications in ∼ 40% of tumors.14,16,21,45 However, it is difficult to ascertain whether the c-rel amplifications played a role in the etiology of the HL and/or in the maintenance of the HL phenotype. The targeted ablation of relA and c-rel in HL cell lines shows that HL is dependent on both relA and c-rel (Figure 1). Our data show that c-rel is probably playing an important functional role in HL and show that the c-rel amplifications in HL are functionally important.
From a global perspective, it has never been clear what the relative contributions of each REL member are to a lymphoma. Because of the lack of any apparent differences in DNA binding sites for the various NF-κB dimers (with some exceptions46 ), it is possible to think that all 3 RELs are somehow functionally redundant, binding interchangeably to any NF-κB DNA site. Our REL shRNA experiments suggest that this is not the case: in HL, the RELs are not redundant and each contributes uniquely to maintaining HL cell viability. If there had been redundancy, then we would have expected that any one REL shRNA ablation would have had little effect. We expect to further understand the extent of this redundancy with ChIP-seq analysis of REL distributions in the genome.
A candidate NIK inhibitor specifically affects the NCP
Lastly, we found that a known NIK small-molecule inhibitor 4H-isoquinoline-1,3-dione32 selectively killed HL but not other B-cell lymphoma cell lines (Figures 4 and 5). This specificity is consistent with our data showing that these cell lines do not need relB for viability and do not express stabilized NIK protein. The drug itself or derivatives of it offer the possibility of targeted therapeutic intervention in cases of refractory or recurring HL and other NIK-dependent tumors, such as MM.23
Model of NF-κB function in HL
We therefore suggest a model of HL in which HL requires the constitutive activation of the NCP. Furthermore, HL requires the activity of all 3 REL family members. We hypothesize that this is because of unique roles that each REL factor plays in establishing genetic regulatory networks. The REL family members clearly are not redundant, at least in the case of a viability phenotype. The requirement for all 3 RELs is an essential event in the etiology and maintenance of the HL phenotype, whether this state is attained by an EBV infection or other events. We hypothesize further that infections (eg, EBV) and inflammatory responses are potentially oncogenic in HL by activating the NCP.47,48 In any case, it is clear that each REL is contributing to and playing an important role in the phenotype of HL cells.
The NF-κB pathway is dysregulated in a variety of human disorders, including many chronic inflammatory diseases and cancers. As such, an understanding of the molecular details of NF-κB–dependent gene networks has implications for improved disease diagnoses and pharmacology interventions. The data herein shed light on unexplored aspects of the HL and NF-κB biology. This translates into new candidate targets in the NF-κB signaling pathway, focusing on targeting NIK function in HL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Tom Waldmann, Lou Staudt, Alfonso Fernandez, and Michael McDevitt for critical reading of the manuscript, Lou Staudt for advice on NIK expression, and Tom Waldmann and Lou Staudt for support and interest in the work.
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health.
National Institutes of Health
Authorship
Contribution: S.M.R. designed and performed research, analyzed data, and wrote the manuscript; S.P., M.O.E., and E.S.J. performed NIK staining of patient biopsies, interpreted IHC data, and discussed data and manuscript preparation; and B.A.L. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian A. Lewis, Metabolism Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bldg 10, Rm 6B05, 9000 Rockville Pike, Bethesda, MD 20852; e-mail: lewisbri@mail.nih.gov.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal