Abstract
Ribosomal protein (RP) mutations in diseases such as 5q− syndrome both disrupt hematopoiesis and increase the risk of developing hematologic malignancy. However, the mechanism by which RP mutations increase cancer risk has remained an important unanswered question. We show here that monoallelic, germline inactivation of the ribosomal protein L22 (Rpl22) predisposes T-lineage progenitors to transformation. Indeed, RPL22 was found to be inactivated in ∼ 10% of human T-acute lymphoblastic leukemias. Moreover, monoallelic loss of Rpl22 accelerates development of thymic lymphoma in both a mouse model of T-cell malignancy and in acute transformation assays in vitro. We show that Rpl22 inactivation enhances transformation potential through induction of the stemness factor, Lin28B. Our finding that Rpl22 inactivation promotes transformation by inducing expression of Lin28B provides the first insight into the mechanistic basis by which mutations in Rpl22, and perhaps some other RP genes, increases cancer risk.
Introduction
In addition to their role as structural components of ribosomes, ribosomal proteins (RPs) are increasingly understood to play critical roles in development and disease, in some cases from outside of the ribosome. These include roles in regulation of cell-cycle progression, apoptosis,1 and translation, through direct interactions with mRNA.2 Mutations in RPs cause diseases collectively termed ribosomopathies, which include myelodysplastic syndromes (MDS) and diamond blackfan anemia (DBA). DBA is caused by mutations in a variety of RPs, with approximately one-half of all cases resulting from mutations in RPS19, RPS26, RPL5, and RPL11, whereas a type of myelodysplastic syndrome known as 5q− syndrome has been attributed to the monoallelic loss of RPS14.3,4 RPS14 haploinsufficiency in 5q− syndrome, as well as the ribosome dysfunction observed in other bone marrow failure syndromes, is associated with increased risk in patients for the development of hematologic malignancies.5 Observations in animal models have similarly linked RP gene mutations with alterations in cancer risk because loss of one copy of numerous, essential RP genes increased susceptibility to tumor formation in zebrafish,6 suggesting that some RPs may serve as haploinsufficient tumor suppressors. Nevertheless, neither the basis by which RP function as tumor suppressors nor the way RP mutations predispose to malignancy has been explained.
The ribosomal protein L22 (Rpl22) is an RNA-binding component of the 60S ribosomal subunit that is not thought to be required for global cap-dependent translation, but its normal physiologic role is poorly understood. We have determined that despite the ubiquitous expression of Rpl22, its germline ablation in mouse is not lethal, unlike ablation of most RP genes.7,8 Instead, mice in which the Rpl22 gene is biallelically inactivated in the germline are viable, fertile, and grossly normal, with the only striking defect being an exquisitely specific block in the development of αβ lineage T cells.9 Because genes that are required for the normal development of a particular cell or tissue often regulate its transformation10 and because Rpl22 is essential for the development of T lymphocytes, we address here whether Rpl22 regulates T-cell transformation. We present evidence that Rpl22 functions as a haploinsufficient tumor suppressor and provide the first mechanistic insights into how mutations in an RP gene predispose cells to transformation.
Methods
Patient samples
Patient samples were collected with informed consent in accordance with the Declaration of Helsinki and Institutional Review Board approval from children with T-acute lymphoblastic leukemia (T-ALL) treated in clinical trials at the Children's Oncology Group or Dana-Farber Cancer Institute. Microarray-based comparative genomic hybridization (aCGH) was performed with the use of genomic DNA on Agilent Human Genome CGH 244A Microarrays (Agilent Technologies), and circular binary segmentation was performed with the DNAcopy package of BioConductor (http://www.bioconductor.org/packages/2.2/bioc/html/DNAcopy.html), as described.11 Color plots of the segmented Log2 copy number data were generated with dChip software (http://biosun1.harvard.edu/complab/dchip). aCGH data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession no. GSE14959 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14959) and no. GSE7615 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE7615). Sequencing of the RPL22 coding exons in primary T-ALL, T-ALL cell lines, and T-ALL isolates from relapsed patients was performed by Agencourt Inc.
Animal studies
Mice were maintained in the Association for Assessment and Accreditation of Laboratory Animal Care–accredited Laboratory Animal Facility at Fox Chase Cancer Center and were handled in compliance with guidelines established by the Institutional Animal Care and Use Committees. Transgenic myristoylated Akt2 (MyrAkt2 Tg) Rpl22+/+ and Rpl22+/− littermate mice bearing a mixed 129-C57BL/6 background were used in all experiments. To evaluate the effect of Rpl22 inactivation on development of thymic lymphoma, Myr-Akt2;Rpl22+/+ and +/− littermates were monitored daily and killed when they began to manifest signs of disease, after which the thymic lymphomas were excised for further analysis. All analysis of premalignant phenotypes was performed on mice when they were 4-6 weeks of age.
Flow cytometry
Explanted thymic lymphomas were purified for subsequent analysis by cell sorting using the FACSVantageSE (Becton Dickinson). For phenotypic analysis of thymocytes, single-cell suspensions were stained with the indicated antibodies as described.9 BrdU staining was performed according to manufacturer's specifications after a 4-hour pulse with BrdU (intraperitoneal injection of 100 μg/mouse). Where applicable, dead cells were excluded from analysis via the use of propidium iodide.
Metabolic labeling
Thymocyte single-cell suspensions were labeled with [35S] methionine at 1 mCi/mL for 30 minutes at 37°C, after which counts incorporated were determined by TCA precipitation on detergent extracts as described.9 The spectrum of proteins synthesized was assessed by SDS-PAGE and visualized after fluorography.
Cell culture and analysis of thymic lymphomas
Explanted thymic lymphomas were adapted to growth in vitro by serial passage in IMDM (Mediatech) with standard supplements and 20% FBS (Hyclone). To determine whether p53 responsiveness was disabled in the thymic lymphomas, genomic DNA from fresh explanted lymphoma cells was analyzed by Southern blotting, as described.12 In addition,tissue culture adapted lymphomas were treated with 0.5 μg/mL doxorubicin for 4 hours, after which expression of the p53 target, PUMA, was measured by real-time PCR with the use of primers and probe from Applied Biosystems, as described.13 All analyses were performed in triplicate and normalized to β-actin. Primary MEFs isolated from embryonic day 13.5 Rpl22+/+, Rpl22+/−, and Rpl22−/− embryos were maintained in IMDM supplemented with 10% FBS and cultured no more than 8 passages. Immortalized MEF lines were generated by use of the 3T3 method. Human T-ALL lines were maintained in RPMI containing standard supplements and 10% FBS. NF-κB inhibitor IMD-0350 and nuclear export inhibitor LMB were obtained from Sigma-Aldrich.
Viral transduction and cellular transformation assays
The following retroviral constructs were used: (1) pBabe-12S-E1A65 (Addgene); (2) pBabe-Puro-H-RASG12V, pBabe-Neo-H-RASG12V, and pBabe-Neo were described previously14 ; (3) pLKO.1-puro lentitroviral shRNA constructs targeting murine Rpl24, murine Rpl11, murine and human Lin28B15 ; and green fluorescent protein were purchased from Sigma-Aldrich; (4) Rpl22 and control shRNA were expressed in murine stem cell virus–based retroviral vectors LMS or LMP, obtained from Dr Scott Lowe (Cold Spring Harbor), as described.9 Retrovirus was produced by transient calcium phosphate transfection of phoenix-ecotropic packaging cells, as described.16 Lentivirus was produced by transfection of HEK293T with both packaging (delta8.2 and VSV-G) and pLKO.1 shRNA vectors using FuGENE 6 (Roche). Virus infected cells were drug selected for at least 5 days before the experiments. For soft agar transformation assays, immortalized MEFs were infected with Ras virus alone, and primary MEFs were coinfected with both E1A and Ras, after which drug selected cells were mixed with an equal volume of 0.7% agar (Difco; BD Biosciences), plated on the top of 0.5% agar layer, and cultured for the indicated time.
Measurement of protein and RNA
NP-40 detergent extracts were resolved by SDS-PAGE and immunoblotted with antibodies reactive with the following proteins: (1) Rpl229 ; (2) GAPDH (Abcam); (3) Lin28 (Abcam); (4) Lin28B (Cell Signaling Technology); (5) Ras (BD Biosciences); (6) c-myc (Cell Signaling Technology); (7) NF-κB p65 (Santa Cruz Biotechnology Inc); (8) Rpl24 (Sigma-Aldrich); and (9) Rpl11 (Sigma-Aldrich). For measurements of RNA, total RNA was extracted from cells with the RNeasy Mini Kit (QIAGEN) and reverse transcribed to cDNA before real-time PCR quantification on the ABI 7500 system with the use of TaqMan FAM-probes from ABI. Probe numbers will be supplied on request.
EMSA
We performed an EMSA analysis of NF-κB activity as described previously.17 To summarize, after hypotonic lysis, high-salt nuclear extracts were prepared and assessed for NF-κB activity by mixing with end-labeled oligonucleotides comprising an NF-κB consensus sequence (5′-AGTTGA GGGGACTTTCCC AGGC-3′; Santa Cruz Biotechnology Inc) or a mutant oligonucleotide that abrogates p65 binding (5′-AGTTGA GGCGACTTTCCC AGGC-3′). Complexes were then subjected to 5% nondenaturing PAGE, vacuum dried, and subjected to autoradiography.
Results
RPL22 is inactivated in a subset of patients with T-ALL
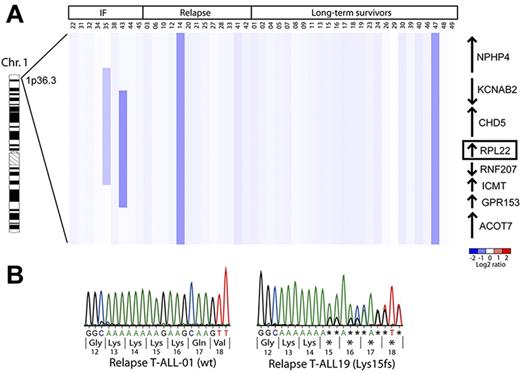
Perturbations in ribosome biogenesis and mutations in RP genes have been reported in animal models and in humans predisposed to malignant transformation.5,6 Because Rpl22 is essential for the development of the T-lineage progenitors from which T-ALL derives, we sought to determine whether RPL22 inactivation affected the development of T-ALL.9 To explore this possibility, aCGH analysis was performed on primary human T-ALL samples to determine whether the RPL22 gene (1p36.3-p36.2) exhibited copy number alternations. As shown in Figure 1A, 4 of the 47 (∼ 9%) samples exhibited deletion of 1 RPL22 allele. Among the samples in which the RPL22 locus was deleted, 3 occurred in patients who succumbed to disease, either through induction failure or relapse. Two of 9 (22%) induction failure samples exhibited focal deletions encompassing the RPL22 locus whereas 2 of 38 patients in whom induction chemotherapy was successful harbored large deletions on the p arm of chromosome 1 (n = 2 of 38, or 5%). Because the deletions that monoallelically inactivated RPL22 also eliminated other genes, we performed sequence analysis of RPL22 in the T-ALL samples to determine whether specific mutations were present. Although no specific point mutations in RPL22 were found in primary T-ALL samples collected at the time of diagnosis, 6 of 19 (∼ 30%) T-ALL cell lines (CEM, Dnd41, Koptk1, Molt13, Molt16, and Supt7), and 1 of 20 primary patient samples collected at relapse exhibited single adenine nucleotide deletions in the 5′ end of the coding region of 1 RPL22 allele, causing a frame-shift predicted to truncate Rpl22 protein at amino acid 18 (Figure 1B). These data indicate that monoallelic, focal RPL22 genetic alterations are observed in some patients with aggressive disease and frequently in cell lines derived from relapsed patients. Moreover, several microarray studies have revealed that Rpl22 mRNA levels were reduced in adult T-cell lymphoma/leukemia,18 invasive breast carcinoma,19 and lung adenocarcinoma.20 Taken together, the mutation and down-regulation of Rpl22 expression in various cancers is consistent with a potential tumor suppressor role for Rpl22.
Deletions encompassing the RPL22 locus are observed in approximately 10% of primary T-ALL samples. (A) aCGH copy number analysis of primary T-ALL samples. Genomic DNA from primary T-ALL samples was subjected to copy number analysis as described.11 Vertical dark blue bars denote the position of the deletion. Adjacent genes and their orientation relative to the RPL22 locus are indicated on the right. (B) Sequence analysis of the RPL22 alleles of T-ALL relapse patient samples and cell lines. Representative DNA sequencing chromatograms from T-ALL cell lines with wild-type (left) or mutant (right) RPL22 alleles are depicted. The loss of a single A nucleotide in the stretch of 8 consecutive A on the mutant allele causes a shift in the translational reading frame that truncates Rpl22 after 18 amino acids.
Deletions encompassing the RPL22 locus are observed in approximately 10% of primary T-ALL samples. (A) aCGH copy number analysis of primary T-ALL samples. Genomic DNA from primary T-ALL samples was subjected to copy number analysis as described.11 Vertical dark blue bars denote the position of the deletion. Adjacent genes and their orientation relative to the RPL22 locus are indicated on the right. (B) Sequence analysis of the RPL22 alleles of T-ALL relapse patient samples and cell lines. Representative DNA sequencing chromatograms from T-ALL cell lines with wild-type (left) or mutant (right) RPL22 alleles are depicted. The loss of a single A nucleotide in the stretch of 8 consecutive A on the mutant allele causes a shift in the translational reading frame that truncates Rpl22 after 18 amino acids.
Rpl22 haploinsufficiency accelerates the development of T lymphoma
To test whether the targeted inactivation of Rpl22 alters development of T-cell malignancy, we used a mouse model of T-cell lymphoma driven by enforced expression of constitutively active MyrAkt2 in T-cell progenitors.21 The MyrAkt2 Tg model was selected because the most frequently observed mutations in T-ALL (ie, Notch activation or PTEN loss) activate AKT.11,22 Non-Tg mice did not develop disease during the time of the study; however, MyrAkt2;Rpl22+/+ mice developed thymic lymphoma with a median latency of 19 weeks (Figure 2A). Importantly, MyrAkt2;Rpl22+/− mice developed thymic lymphoma much more rapidly, with a median latency of 11 weeks (Figure 2A). This development was not accompanied by alterations in either thymic cellularity or in thymic subsets defined by CD4/8 expression (Figure 2B), consistent with our previous report that Rpl22+/− mice had no overt phenotype.9 The accelerated development of disease in MyrAkt2;Rpl22+/− mice was accompanied by greater proliferation among CD4+CD8+ double positive (DP) thymocytes, as evidenced by their increased incorporation of BrdU (Figure 2C) and increased Ki-67 staining of MyrAkt2;Rpl22+/− lymphomas (Figure 2D). Neither the increased proliferation nor the enhanced transformation potential of MyrAkt2;Rpl22+/− thymocytes was accompanied by changes in global protein synthesis (Figure 2E). These data demonstrating that Rpl22 haploinsufficiency accelerates the development of T-cell lymphoma in a mouse model suggest that the loss of RPL22 in human T-ALL may also contribute to the aggressiveness of disease in those cases. Furthermore, the remaining allele of Rpl22 was found to be intact in the MyrAkt2;Rpl22+/− thymic lymphomas, as evidenced by both sequence analysis of 14 tumors (data not shown) and immunoblotting (supplemental Figure 1A, available at the Blood Web site; see the Supplemental Materials link at the top of the online article). Taken together, these data demonstrate that loss of a single Rpl22 allele is sufficient to accelerate the development of thymic lymphoma and suggest that Rpl22 is functioning as a haploinsufficient tumor suppressor.
Rpl22 haploinsufficiency accelerates the development of cancer in a mouse model of T-cell malignancy. (A) Kaplan-Meier curves of mice of the indicated genotypes, which were killed on manifestation of outward signs of disease. Myr-Akt2;Rpl22+/+, n = 10; Myr-Akt2;Rpl22+/− n = 14; (B) Distribution of CD4/8 subpopulations in thymi of mice with the indicated genotypes. Single-cell suspensions of thymocytes from young adult mice (4-6 weeks) were stained with antibodies reactive CD4 and CD8. Absolute numbers of thymocytes were determined and the mean ± SD are depicted graphically to the right. Analysis was performed on a minimum of 3 mice per group and is representative of 3 experiments performed. (C) Proliferation of explanted thymocytes from MyrAkt2 Tg mice measured by BrdU incorporation. Proliferation of the indicated populations was assessed flow cytometrically by determining the extent of BrdU incorporation after a 4-hour pulse. The mean ± SD of the fraction of BrdU+ cells for a representative experiment is depicted graphically. Each bar represents an individual experiment involving at least 3 mice. Three experiments were performed. *P < .05. (D) Assessment of the extent of proliferation of Rpl22+/+ and +/− thymic lymphoma cells by Ki-67 staining in situ. Thymic sections from the indicated mice were either stained with hematoxylin and eosin (H&E) or with anti-Ki67 antibodies to detect the number of proliferating cells. The micrograph was generated using the ×20 objective (×200 total magnification) of a Nikon Eclipse 50i microscope and a Digital Sight DS-Fi1 camera. Mean ± SEM of the thymic organ weight relative to body weight from Rpl22+/+ (n = 6) and Rpl22+/− (n = 9) mice at the time of sacrifice is depicted graphically below. *P < .05. Representative thymi are shown on the left. (E) Evaluation of the rate of protein synthesis in thymocytes measured by metabolic labeling. Thymocyte suspensions from mice of the indicated genotypes were metabolic labeling for 30 minutes with [35S]methionine after which the counts incorporated were quantified by TCA precipitation of aliquots of the detergent lysates. Data were derived from triplicate values from 2 independent experiments. In addition, extracts were resolved directly by SDS-PAGE and visualized by fluorography (right).
Rpl22 haploinsufficiency accelerates the development of cancer in a mouse model of T-cell malignancy. (A) Kaplan-Meier curves of mice of the indicated genotypes, which were killed on manifestation of outward signs of disease. Myr-Akt2;Rpl22+/+, n = 10; Myr-Akt2;Rpl22+/− n = 14; (B) Distribution of CD4/8 subpopulations in thymi of mice with the indicated genotypes. Single-cell suspensions of thymocytes from young adult mice (4-6 weeks) were stained with antibodies reactive CD4 and CD8. Absolute numbers of thymocytes were determined and the mean ± SD are depicted graphically to the right. Analysis was performed on a minimum of 3 mice per group and is representative of 3 experiments performed. (C) Proliferation of explanted thymocytes from MyrAkt2 Tg mice measured by BrdU incorporation. Proliferation of the indicated populations was assessed flow cytometrically by determining the extent of BrdU incorporation after a 4-hour pulse. The mean ± SD of the fraction of BrdU+ cells for a representative experiment is depicted graphically. Each bar represents an individual experiment involving at least 3 mice. Three experiments were performed. *P < .05. (D) Assessment of the extent of proliferation of Rpl22+/+ and +/− thymic lymphoma cells by Ki-67 staining in situ. Thymic sections from the indicated mice were either stained with hematoxylin and eosin (H&E) or with anti-Ki67 antibodies to detect the number of proliferating cells. The micrograph was generated using the ×20 objective (×200 total magnification) of a Nikon Eclipse 50i microscope and a Digital Sight DS-Fi1 camera. Mean ± SEM of the thymic organ weight relative to body weight from Rpl22+/+ (n = 6) and Rpl22+/− (n = 9) mice at the time of sacrifice is depicted graphically below. *P < .05. Representative thymi are shown on the left. (E) Evaluation of the rate of protein synthesis in thymocytes measured by metabolic labeling. Thymocyte suspensions from mice of the indicated genotypes were metabolic labeling for 30 minutes with [35S]methionine after which the counts incorporated were quantified by TCA precipitation of aliquots of the detergent lysates. Data were derived from triplicate values from 2 independent experiments. In addition, extracts were resolved directly by SDS-PAGE and visualized by fluorography (right).
Acceleration of lymphoma development in Rpl22+/− mice does not depend on inactivation of p53
It is well documented that impaired ribosomal biosynthesis activates p53 by inducing a nucleolar stress response.23-26 Whereas we also have observed that Rpl22 deficiency results in translational derepression of p53 in developing αβ lineage T cells, this was not observed in Rpl22 haploinsufficiency. Nevertheless, we wished to determine whether p53 responsiveness might be disabled during development of T-cell lymphoma in Rpl22-haploinsufficient mice. Southern blotting revealed that neither the Cdkn2a locus encoding p19Arf, which can induce p53 in response to oncogene activation,27 nor the Trp53 locus itself exhibited genomic alterations in thymic lymphomas from MyrAkt2;Rpl22+/+ or Rpl22+/− mice (supplemental Figure 1B). Moreover, induction of DNA damage by doxorubicin treatment of thymic lymphomas from MyrAkt2;Rpl22+/+ and +/− mice increased expression of the direct p53 target, PUMA, indicating that the p53 pathway was intact (supplemental Figure 1C).13,28 These data demonstrate that inactivation of the p53 pathway does not play a significant role in the acceleration of thymic lymphoma in MyrAkt2;Rpl22+/− mice.
Rpl22 serves as a haploinsufficient tumor suppressor in acute transformation assays
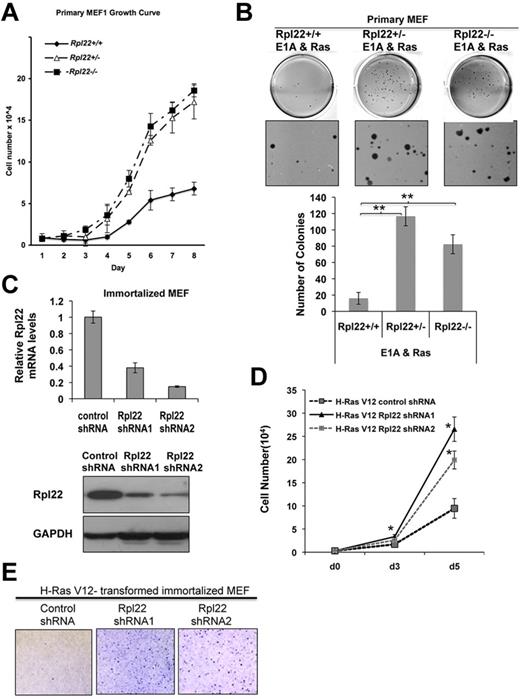
To determine whether Rpl22 haploinsufficiency might also accelerate transformation in other cell types or disease models, we isolated primary mouse embryonic fibroblasts (MEFs) from Rpl22+/+, +/−, and −/− mice. Interestingly, both Rpl22+/− and −/− primary MEFs exhibited a faster growth rate than MEFs from Rpl22+/+ littermates (Figure 3A). Rpl22+/− and −/− MEFs also displayed increased transformation potential in a soft-agar colony formation assay after transduction with oncogenic Ras (Ha-RasV12) and E1A (Figure 3B). Similar results were obtained with immortalized Rpl22+/− and −/− MEFs transformed with oncogenic Ras alone (supplemental Figure 2). Importantly, these findings were recapitulated in MEFs in which Rpl22 expression was diminished by shRNA knockdown because knockdown of Rpl22 both increased cell growth (Figures 3C-D) and soft agar colony formation (Figure 3E). These data indicate that in addition to accelerating the development of thymic lymphoma in a mouse model, Rpl22 inactivation also enhances the transformation potential of both primary and immortalized MEFs in response to a distinct oncogenic insult, altogether providing strong support for Rpl22's tumor suppressor function.
Rpl22 haploinsufficiency and deficiency promote growth and transformation in cell models in vitro. (A) Effect of Rpl22 inactivation on growth of primary MEFs. Primary Rpl22+/+, +/−, and −/− MEFs were seeded in triplicate, cultured in 3% O2 at 37°C, and counted at the indicated intervals for 8 days. Mean cell number ± SD at each time point is represented graphically. Results are representative of 3 independent experiments. (B) Effect of Rpl22 inactivation on transformation of primary MEFs. Primary Rpl22+/+, +/−, and −/− MEFs were transduced with oncogenes E1A and H-RasV12, followed by drug selection for 1 week, and plating in 0.7% agar. After 3 weeks, colonies were stained with crystal violet and enumerated. Images of representative wells were captured using an EPSON Perfection V700 Photo scanner and are depicted in the top panels. Mean colony number per well ± SD for each genotype is represented graphically in the bottom panel. **P < .005. Data are representative of 3 independent experiments performed in triplicate. (C) Knockdown of Rpl22 expression in immortalized MEFs. Immortalized Rpl22+/+ MEFs were transduced with control or Rpl22 shRNA constructs, after which the effect on Rpl22 mRNA and protein expression was evaluated by real-time PCR (top) and immunoblotting (bottom). (D-E) Evaluation of immortalized MEF growth and transformation after Rpl22 knockdown. MEFs stably expressing control or 2 Rpl22 shRNA constructs were transformed by oncogenic H-RasV12, after which their growth rate was assessed by counting (panel D; *P > .05 vs control shRNA) and their transformation by colony formation in soft agar (E) as in panel B.
Rpl22 haploinsufficiency and deficiency promote growth and transformation in cell models in vitro. (A) Effect of Rpl22 inactivation on growth of primary MEFs. Primary Rpl22+/+, +/−, and −/− MEFs were seeded in triplicate, cultured in 3% O2 at 37°C, and counted at the indicated intervals for 8 days. Mean cell number ± SD at each time point is represented graphically. Results are representative of 3 independent experiments. (B) Effect of Rpl22 inactivation on transformation of primary MEFs. Primary Rpl22+/+, +/−, and −/− MEFs were transduced with oncogenes E1A and H-RasV12, followed by drug selection for 1 week, and plating in 0.7% agar. After 3 weeks, colonies were stained with crystal violet and enumerated. Images of representative wells were captured using an EPSON Perfection V700 Photo scanner and are depicted in the top panels. Mean colony number per well ± SD for each genotype is represented graphically in the bottom panel. **P < .005. Data are representative of 3 independent experiments performed in triplicate. (C) Knockdown of Rpl22 expression in immortalized MEFs. Immortalized Rpl22+/+ MEFs were transduced with control or Rpl22 shRNA constructs, after which the effect on Rpl22 mRNA and protein expression was evaluated by real-time PCR (top) and immunoblotting (bottom). (D-E) Evaluation of immortalized MEF growth and transformation after Rpl22 knockdown. MEFs stably expressing control or 2 Rpl22 shRNA constructs were transformed by oncogenic H-RasV12, after which their growth rate was assessed by counting (panel D; *P > .05 vs control shRNA) and their transformation by colony formation in soft agar (E) as in panel B.
Rpl22 inactivation enhances transformation potential through dysregulation of Lin28B
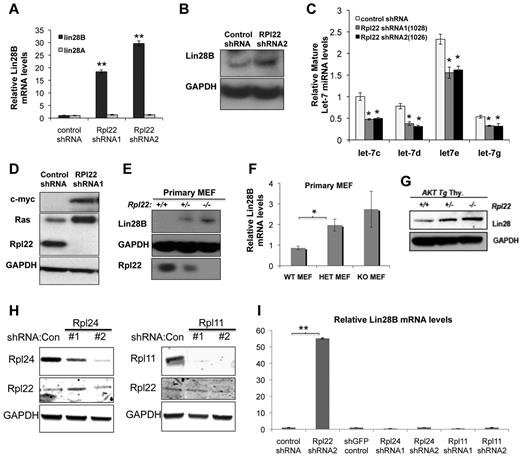
To explore the molecular mechanism by which Rpl22 inactivation enhances growth and transformation, we performed microarray analysis on immortalized MEF lines lacking Rpl22 as a result of either gene targeting (Rpl22−/−) or shRNA knockdown (supplemental Table 1). Among the genes most differentially expressed in Rpl22-deficient cells was Lin28B, whose expression was increased nearly 20-fold. There are 2 genes in the Lin28 family, Lin28A and Lin28B,29 both of which have been implicated in human malignancy.15,30 Real-time PCR analysis verified that expression of Lin28B, but not Lin28A, was increased in cells with reduced Rpl22 expression (Figure 4A). Rpl22 knockdown increased Lin28B mRNA levels from 20- to 30-fold relative to cells transduced with control shRNA, whereas Lin28A expression was unchanged (Figure 4A). Likewise, Rpl22 knockdown in MEF increased Lin28B protein levels (Figure 4B).
Increased transformation potential associated with of Rpl22 loss or inactivation is accompanied by induction of Lin28B. Effect of Rpl22 knockdown on Lin28 expression in immortalized MEF. MEF lines stably expressing control or 2 different Rpl22 shRNA constructs were harvested for RNA and protein, after which Lin28A and Lin28B mRNA levels were evaluated by real-time PCR (A). **P < .005 for Lin28B expression in Rpl22 shRNA relative to controls. Protein levels were measured by blotting (B). Data are representative of 2 independent experiments. (C) Let-7 miRNA levels in immortalized MEFs where Rpl22 expression was suppressed by shRNA. Expression of Let-7 family miRNA was evaluated by real-time PCR in immortalized MEFs stably expressing control or Rpl22 shRNA constructs. Expression levels were normalized to sno202 RNA and to the expression level in cells transduced with control shRNA. Mean expression levels of triplicate measurements ± SD are represented graphically. Data are representative of 3 independent experiments. P < .05 for Let-7 miRNA levels in Rpl22 shRNA compared with control shRNA. (D) Effect of Rpl22 knockdown on expression of Let-7 targets. Expression of Ras and Myc was assessed by immunoblotting in control or Rpl22 knockdown MEFs. GAPDH served as a loading control. Expression of Lin28B in primary MEFs. Lin28B protein and mRNA levels were measured in primary MEFs of the indicated genotypes by immunoblotting (E) and real-time PCR (F), respectively. Mean ± SD of Lin28B mRNA expression levels are depicted graphically. Results are representative of at least 3 experiments performed. *P < .05. (G) Lin28B expression in primary thymocytes. The expression of Lin28B in thymocytes from mice with the indicated genotypes was evaluated by immunoblotting. GAPDH served as a loading control. Lin28B expression in MEFs after knockdown of Rpl11 and Rpl24. Immortalized MEFs were transduced with 2 different shRNA constructs targeting Rpl24 or Rpl11, after which protein levels were evaluated by immunoblotting (H). GAPDH served as loading control. (I) The level of Lin28B mRNA expressed by these cells was measured by real time PCR as in panel F. **P < .005.
Increased transformation potential associated with of Rpl22 loss or inactivation is accompanied by induction of Lin28B. Effect of Rpl22 knockdown on Lin28 expression in immortalized MEF. MEF lines stably expressing control or 2 different Rpl22 shRNA constructs were harvested for RNA and protein, after which Lin28A and Lin28B mRNA levels were evaluated by real-time PCR (A). **P < .005 for Lin28B expression in Rpl22 shRNA relative to controls. Protein levels were measured by blotting (B). Data are representative of 2 independent experiments. (C) Let-7 miRNA levels in immortalized MEFs where Rpl22 expression was suppressed by shRNA. Expression of Let-7 family miRNA was evaluated by real-time PCR in immortalized MEFs stably expressing control or Rpl22 shRNA constructs. Expression levels were normalized to sno202 RNA and to the expression level in cells transduced with control shRNA. Mean expression levels of triplicate measurements ± SD are represented graphically. Data are representative of 3 independent experiments. P < .05 for Let-7 miRNA levels in Rpl22 shRNA compared with control shRNA. (D) Effect of Rpl22 knockdown on expression of Let-7 targets. Expression of Ras and Myc was assessed by immunoblotting in control or Rpl22 knockdown MEFs. GAPDH served as a loading control. Expression of Lin28B in primary MEFs. Lin28B protein and mRNA levels were measured in primary MEFs of the indicated genotypes by immunoblotting (E) and real-time PCR (F), respectively. Mean ± SD of Lin28B mRNA expression levels are depicted graphically. Results are representative of at least 3 experiments performed. *P < .05. (G) Lin28B expression in primary thymocytes. The expression of Lin28B in thymocytes from mice with the indicated genotypes was evaluated by immunoblotting. GAPDH served as a loading control. Lin28B expression in MEFs after knockdown of Rpl11 and Rpl24. Immortalized MEFs were transduced with 2 different shRNA constructs targeting Rpl24 or Rpl11, after which protein levels were evaluated by immunoblotting (H). GAPDH served as loading control. (I) The level of Lin28B mRNA expressed by these cells was measured by real time PCR as in panel F. **P < .005.
Because Lin28 negatively regulates the processing of Let-7 miRNAs,31 we next determined whether expression of Let-7 family miRNA was altered. In accordance with the increased expression of Lin28B, MEF lines in which Rpl22 was knocked down also exhibited reduced expression of several Let-7 miRNA family members (Figure 4C). The reduction in Let-7 miRNA expression, in turn, was accompanied by increased expression of oncogene targets Myc and Ras, which they negatively regulate (Figure 4D). The increased expression of Lin28B was not restricted to immortalized MEFs in which Rpl22 expression had been knocked down because Lin28B levels also were elevated in Rpl22+/− and −/− primary MEFs, as well as in thymocytes from MyrAkt2;Rpl22+/− and −/− mice (Figure 4E-G). To determine whether the link between Rpl22 inactivation and Lin28B induction was unique to Rpl22 or was perhaps observed on loss of other RP, we assessed the level of Lin28B expression in MEF in which Rpl24 and Rpl11 had been knocked down using shRNA. Nevertheless, despite effective knockdown of Rpl24 and Rpl11, Lin28B mRNA levels were unchanged (Figure 4H-I). These data indicate that the Lin28B induction that accompanies inactivation of Rpl22 is not a general cellular response to loss of RP and implicates Lin28B induction as a potential mechanism by which Rpl22 haploinsufficiency and deficiency increases transformation potential.
Enhanced growth and transformation potential accompanying Rpl22 inactivation is dependent on Lin28B
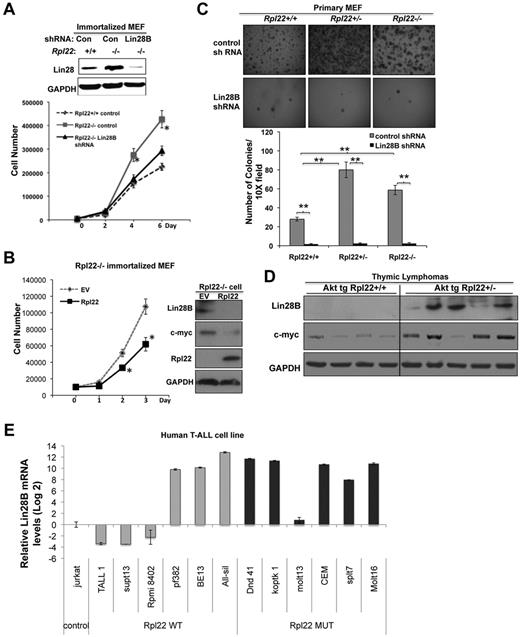
Lin28B has been shown to increase cell proliferation and promote tumor growth.15,30 Because Lin28B induction is correlated with the enhanced proliferation and transformation potential of Rpl22 mutant cells, we used shRNA knockdown to determine whether Lin28B was responsible for these behaviors. ShRNA knockdown of Lin28B in immortalized Rpl22−/− MEFs decreased the growth rate of immortalized Rpl22−/− MEFs to that observed in Rpl22+/+ MEFs (Figure 5A) and abrogated the spontaneous formation of soft agar colonies by immortalized Rpl22−/− MEFs (supplemental Figure 3). Importantly, the increased Lin28B expression and enhanced growth exhibited by Rpl22-deficient immortalized MEF was reversed on ectopic expression of Rpl22, as was the increased expression of the Let-7 target, c-myc (Figure 5B).
Lin28B is necessary for the enhancement of growth and transformation by Rpl22 inactivation. (A) Effect of knockdown of Lin28B on the growth of immortalized Rpl22−/− MEFs. Immortalized MEFs of the indicated genotypes were transduced with control or Lin28B shRNA. The effect on expression of Lin28B was assessed by immunoblotting. Growth of triplicate wells of MEFs stably expressing control shRNA or Lin28B shRNA was determined by counting and then plotted as the mean cell number ± SD *P < .05, for Rpl22−/− compared with Rpl22−/− in which Lin28B was knocked down. (B) Effect on growth of reintroducing Rpl22 into Rpl22−/− MEF. Immortalized Rpl22−/− MEFs were transfected with Rpl22 or empty vector (EV) control, after which we assessed cell growth by counting triplicate wells and depicting the mean ± SD graphically. The expression of Lin28B, c-myc, Rpl22, and GAPDH (loading control) were evaluated by immunoblotting. (C) Dependence of soft agar colony formation on expression of Lin28B. Primary MEFs of the indicated genotypes were transduced with Lin28B shRNA followed by E1A and Ras, and then plated in triplicate in soft agar as Figure 3B. Representative images were captured with a Digital Slight DS-Fi1 camera and NIS Element AR3.0 imaging software at 1× with 3× zoom (×30 total magnification) using a Nikon SMZ1500 stereomicroscope and are shown in the top panels. The mean colony number ± SD is represented graphically beneath. **P < .005 for colonies in Rpl22+/− and −/− relative to Rpl22+/+. (D) Lin28B expression in Rpl22-haploinsufficient thymic lymphomas. Explanted thymic lymphomas from MyrAkt2;Rpl22+/+ and MyrAkt2;Rpl22−/− mice were evaluated for Lin28B and c-myc expression by immunoblotting. GAPDH served as a loading control. (E) Lin28B mRNA levels in RPL22+/+ and Rpl22+/− human T-ALL lines. Lin28B mRNA levels in the indicated T-ALL cell lines were quantified by real-time PCR. Data are presented as Log2 value relative to Jurkat cells (control).
Lin28B is necessary for the enhancement of growth and transformation by Rpl22 inactivation. (A) Effect of knockdown of Lin28B on the growth of immortalized Rpl22−/− MEFs. Immortalized MEFs of the indicated genotypes were transduced with control or Lin28B shRNA. The effect on expression of Lin28B was assessed by immunoblotting. Growth of triplicate wells of MEFs stably expressing control shRNA or Lin28B shRNA was determined by counting and then plotted as the mean cell number ± SD *P < .05, for Rpl22−/− compared with Rpl22−/− in which Lin28B was knocked down. (B) Effect on growth of reintroducing Rpl22 into Rpl22−/− MEF. Immortalized Rpl22−/− MEFs were transfected with Rpl22 or empty vector (EV) control, after which we assessed cell growth by counting triplicate wells and depicting the mean ± SD graphically. The expression of Lin28B, c-myc, Rpl22, and GAPDH (loading control) were evaluated by immunoblotting. (C) Dependence of soft agar colony formation on expression of Lin28B. Primary MEFs of the indicated genotypes were transduced with Lin28B shRNA followed by E1A and Ras, and then plated in triplicate in soft agar as Figure 3B. Representative images were captured with a Digital Slight DS-Fi1 camera and NIS Element AR3.0 imaging software at 1× with 3× zoom (×30 total magnification) using a Nikon SMZ1500 stereomicroscope and are shown in the top panels. The mean colony number ± SD is represented graphically beneath. **P < .005 for colonies in Rpl22+/− and −/− relative to Rpl22+/+. (D) Lin28B expression in Rpl22-haploinsufficient thymic lymphomas. Explanted thymic lymphomas from MyrAkt2;Rpl22+/+ and MyrAkt2;Rpl22−/− mice were evaluated for Lin28B and c-myc expression by immunoblotting. GAPDH served as a loading control. (E) Lin28B mRNA levels in RPL22+/+ and Rpl22+/− human T-ALL lines. Lin28B mRNA levels in the indicated T-ALL cell lines were quantified by real-time PCR. Data are presented as Log2 value relative to Jurkat cells (control).
As was true for immortalized MEF, knockdown of Lin28B in primary MEFs reversed the enhanced colony formation observed on transduction of Rp22+/− and −/− primary MEFs with E1A/Ras (Figure 5C). Rpl22 haploinsufficiency also was linked to Lin28B and c-myc induction in the thymic lymphomas that developed more rapidly in MyrAkt2Tg:Rpl22+/− mice (Figure 5D). Finally, we determined that all of the 6 T-ALL cell lines bearing RPL22 mutations exhibited greater Lin28B mRNA levels than Jurkat (RPL22+/+), whereas this was true for only half of RPL22+/+ T-ALL cell lines (Figure 5E). The induction of Lin28B in some of the RPL22+/+ lines may have resulted from elevated NF-κB activity because Lin28B is a direct transcriptional target of NF-κB,32 and NF-κB can be activated by Notch signaling,33 which is frequently dysregulated in T-ALL.34 Together, these data provide evidence that Rpl22 haploinsufficiency leads to Lin28B induction in both MEF and T-cell malignancies. Moreover, the ability of knockdown of Lin28B to reverse both the enhanced growth and transformation potential of Rpl22-deficient cells strongly suggests that these behaviors are dependent on increased Lin28B expression in Rpl22+/− and −/− cells.
Induction of Lin28B expression in Rpl22 mutant cells is dependent on NF-κB signaling
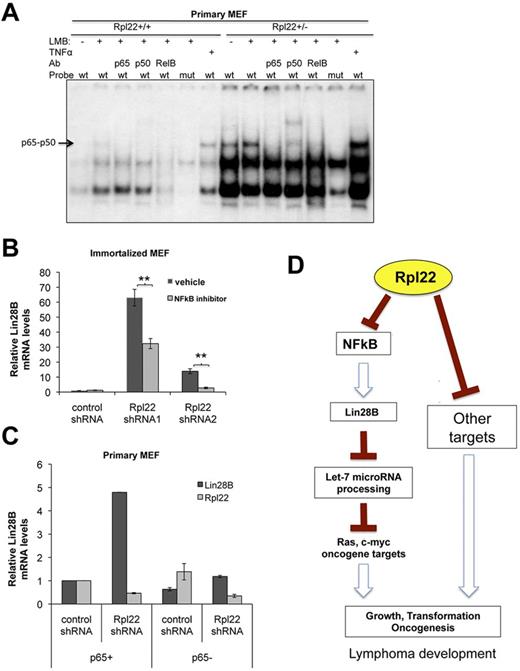
Because the enhanced predisposition of Rpl22-haploinsufficient and deficient cells to transformation is dependent on induction of Lin28B, we wished to determine how Rpl22 inactivation increases Lin28B expression. Lin28B is a direct target of NF-κB signaling.32 Accordingly, we sought to determine whether Rpl22 inactivation induces Lin28B by up-regulation of NF-κB signaling. To address this possibility, we performed EMSA analysis by using equal quantities of nuclear extract from primary Rpl22+/+ and +/− MEF. We found that NF-κB complexes comprising p65/p50 heterodimers (defined by antibody supershift) were increased in Rpl22+/− primary MEFs (Figure 6A). NF-κB activity was increased further upon treatment with leptomycin B, which traps NF-κB in the nucleus by blocking nuclear exit, and still further upon treatment with TNFα (Figure 6A). Moreover, binding was abrogated by mutation of the consensus p65 binding motif (Figure 6A). Increased NFκB activity and Lin28B expression also were observed upon knocking down Rpl22 in a thymic lymphoma line (supplemental Figure 4).
The increased Lin28B expression that results from Rpl22 loss or inactivation is dependent on NF-κB activity. (A) Measurement of NF-κB activity in Rpl22-haploinsufficient primary MEF. EMSA analysis was performed using equal quantities of nuclear extract protein from primary MEFs of the indicated genotypes using both an intact (wt) and p65 binding mutant (mut) NF-κB probe. NF-κB activity was measured in untreated cells, cells pretreated with leptomycin B to trap NF-κB in the nucleus, and after TNFα stimulation (positive control). The composition of the NF-κB complexes was evaluated using supershift analysis using the indicated Abs. Effect of NF-κB inhibition on Lin28B expression. (B) Lin28B mRNA levels were quantified by real-time PCR on RNA extracted from immortalized MEFs stably expressing 2 different Rpl22 shRNAs, in which NF-κB activity had been pharmacologially inhibited by treatment with 1μM NF-κB inhibitor, IMD-350. **P < .01 for IMD-350 treated compared with control treated. (C) Rpl22 was knocked down by shRNA in primary MEF from p65 wild-type (p65+) or Rela−/−, p65 knockout mice (p65−). Lin28B induction was blocked in p65 knockout cells in which Rpl22 was knocked down. Lin28B and Rpl22 mRNA levels were quantified by real-time PCR, and data are plotted as the average of 2 experiments. (D) Model of Rpl22 function in transformation. The model proposes that Rpl22 normally acts to restrain NF-κB activity by an unknown mechanism. However, when Rpl22 expression is diminished either by shRNA knockdown or mutation, NF-κB activity is increased, resulting in increased expression of Lin28B. Lin28B, in turn, promotes transformation at least in part by repressing Let-7 MiR processing, which results in derepression of oncogenic targets such as c-myc. Rpl22 is also likely to regulate additional targets that contribute to transformation.
The increased Lin28B expression that results from Rpl22 loss or inactivation is dependent on NF-κB activity. (A) Measurement of NF-κB activity in Rpl22-haploinsufficient primary MEF. EMSA analysis was performed using equal quantities of nuclear extract protein from primary MEFs of the indicated genotypes using both an intact (wt) and p65 binding mutant (mut) NF-κB probe. NF-κB activity was measured in untreated cells, cells pretreated with leptomycin B to trap NF-κB in the nucleus, and after TNFα stimulation (positive control). The composition of the NF-κB complexes was evaluated using supershift analysis using the indicated Abs. Effect of NF-κB inhibition on Lin28B expression. (B) Lin28B mRNA levels were quantified by real-time PCR on RNA extracted from immortalized MEFs stably expressing 2 different Rpl22 shRNAs, in which NF-κB activity had been pharmacologially inhibited by treatment with 1μM NF-κB inhibitor, IMD-350. **P < .01 for IMD-350 treated compared with control treated. (C) Rpl22 was knocked down by shRNA in primary MEF from p65 wild-type (p65+) or Rela−/−, p65 knockout mice (p65−). Lin28B induction was blocked in p65 knockout cells in which Rpl22 was knocked down. Lin28B and Rpl22 mRNA levels were quantified by real-time PCR, and data are plotted as the average of 2 experiments. (D) Model of Rpl22 function in transformation. The model proposes that Rpl22 normally acts to restrain NF-κB activity by an unknown mechanism. However, when Rpl22 expression is diminished either by shRNA knockdown or mutation, NF-κB activity is increased, resulting in increased expression of Lin28B. Lin28B, in turn, promotes transformation at least in part by repressing Let-7 MiR processing, which results in derepression of oncogenic targets such as c-myc. Rpl22 is also likely to regulate additional targets that contribute to transformation.
To determine whether the increased expression of Lin28B was in fact dependent on NF-κB activity, we used a pharmacologic inhibitor of IκB kinase β, IMD-0354, which inhibits NF-κB activation by blocking the phosphorylation of IκBα that is normally responsible for translocation of NF-κB to the nucleus. IκBα phosphorylation is increased on knockdown of Rpl22 in immortalized MEF (supplemental Figure 5). Moreover, treatment of Rpl22 knockdown MEFs with IMD-0354 partially reduced Lin28B expression levels (Figure 6B). Activation of IκB kinase β as part of canonical NF-κB signaling leads to the translocation of p65/p50 complexes to the nucleus and transactivation of target genes.35
To determine whether canonical NF-κB complexes comprising p65 were required for the induction of Lin28B on Rpl22 knockdown, we knocked Rpl22 down in matched p65-sufficient and p65-deficient (Rela−/−) littermate MEF.17 p65-deficiency abrogated the ability of Rpl22 knockdown to induce Lin28B expression (Figure 6C). p65 deficiency blocked Lin28B induction more effectively than the pharmacologic inhibitor, presumably because IMD-0354 failed to completely block NF-κB activation. Together, these data indicate that the induction of Lin28B expression that occurs on loss or inactivation of Rpl22 is dependent on NF-κB signaling.
Discussion
Although numerous reports in both model organisms and humans indicate that monoallelic inactivation of RP genes can contribute to the development of cancer, the mechanistic basis by which RP mutations do so has remained unexplained.4,6 We provide here the first insights into how haploinsufficiency of an RP, Rpl22, contributes to transformation. The RPL22 locus is monoallelically deleted in approximately 10% of primary T-ALL in humans. Moreover, the monoallelic inactivation of Rpl22 enhanced development of disease in a mouse model of T-cell malignancy as well as in acute in vitro models of transformation using MEFs. The predisposition to transformation or tumor progression afforded by Rpl22 haploinsufficiency is associated with and dependent on induction of the stemness factor Lin28B, which recently has been found to be increased in late-stage, aggressive cancers in humans.15 Finally, the induction of Lin28B that occurs accompanies repression or inactivation of Rpl22 is dependent on elevated canonical NF-κB signaling (Figure 6D). Together, these observations provide insight into the mechanistic basis for how Rpl22, which plays a critical role in normal T lymphocyte development, also contributes to T-cell oncogenesis when haploinsufficiency occurs.9
Although the basis by which RP haploinsufficiency promotes transformation is poorly understood, MacInnes et al recently demonstrated that monoallelic inactivation of numerous RP genes in zebrafish increased the development of malignant peripheral nerve sheath tumors and that this was associated with failure to activate p53 in response to DNA damage.36 Effects of RP gene mutations on p53 activation have been known for some time because inactivation of RPs has been shown to activate p53 by inhibiting p53 degradation mediated by the E3 ubiquitin ligase, MDM2.23,26,37 Interestingly, MacInnes et al found that the failure of the resulting malignant peripheral nerve sheath tumors to induce p53 did not result from mutations in p53 or effects on stability; rather, it resulted from an inability to translate p53 mRNA.36 Nevertheless, it remained unclear whether the inability to translate intact p53 mRNA was responsible for the increased susceptibility to transformation or was instead an indirect effect of selection by the tumor cells after formation. We have previously found that Rpl22 deficiency, but not haploinsufficiency, blocks development of αβ T lineage progenitors because of selective translational derepression of p53 in those progenitors9 ; however, mutations disabling p53 do not appear to play a role in the susceptibility to transformation associated with Rpl22 haploinsufficiency (supplemental Figure 1).
The inactivation of Rpl22, as well as and some other RP (eg, RPS14), predisposes cell transformation; however, it is important to note that inactivation of still other RP genes actually antagonizes cancer development and progression. For example, haploinsufficiency of Rpl24 was found to delay development of B and T lymphoma in mouse models.38,39 The delay in development of B and T lymphoma was associated with impairment of the core ribosome function of CAP-dependent protein synthesis,38,39 presumably in a similar manner to the way inactivation of other RP that are required for ribosome biogenesis or core function impairs erythrocyte development in DBA.3,5
Interestingly, Rpl38 haploinsufficiency also delayed the development of B lymphoma in a mouse model but apparently did so without grossly altering ribosome biogenesis or global protein synthesis.38 Rpl38 was recently shown to exert selective control over the translation of a cluster of Hox genes, suggesting that Rpl38 can act either in the context of a specialized subset of ribosomes with functions influenced by their complement of RP, or alternatively, in an extraribosomal fashion.40 It would appear that Rpl22 may also represent a RP that plays a more specialized, regulatory role because it is not essential for core ribosome functions (Figure 2E) yet clearly influences the predisposition of cells to transformation by regulating the expression of Lin28B (Figures 2,Figure 3,Figure 4–5). Accordingly, although Rpl22 haploinsufficiency predisposes to transformation through the induction of Lin28B expression because Rpl22 is dispensable for core ribosome function, it is possible, if not likely, that it influences transformation by a mechanism that is distinct from other RP whose inactivation affects global, CAP-dependent translation (Figure 4H), and activates p53 through nucleolar stress.3,5
The Lin28 RNA binding proteins are highly enriched in stem cells and are capable, along with Oct4, Sox2, and Nanog, of reprogramming somatic cells into inducible pluripotent stem cells.41 In recent reports investigators also have revealed that both Lin28A and Lin28B are overexpressed in approximately 15% of human primary tumors, with elevations in Lin28B expression being associated with advanced disease across multiple tumor types.15,30 We found that the enhanced transformation potential of Rpl22-haploinsufficient cells was not only associated with but was also dependent on increased expression of Lin28B (Figures 4 and 5). Importantly, the elevation of Lin28B expression was already evident in premalignant primary thymocytes as well as in primary MEFs (Figure 4), indicating that Lin28B induction predisposed these cells to transformation rather than being a consequence of transformation.
Although the mechanism by which Lin28B promotes transformation is incompletely understood, a consensus is emerging that its function in antagonizing the processing of Let-7 miRNAs is likely to play an important role.31,32 Consistent with this view, we found that the induction of Lin28B that occurs on repression of Rpl22 also results in reduced expression of multiple Let-7 miRNA family members. Let-7 family miRNAs, in turn, negatively regulate expression of the critical oncogenes, Myc and Ras, as we observed (Figures 4D, 5B,D).42 In addition, a recent report has indicated that enforced expression of Lin28B in adult HSCs reprograms the adult HSC to acquire fetal-like characteristics, which may also contribute to the susceptibility to transformation.43
Accordingly, we hypothesize that induction of Lin28B in Rpl22+/− T-lineage progenitors predisposes them to transformation at least in part through effects on Let-7 miRNA expression. In support, enforced expression of Lin28b in HSC is also sufficient to promote the development of T lymphoma.44 Although it is clear that Lin28B promotes transformation and this is likely to depend on inhibition of Let-7 processing, it remains possible that Lin28B might also have important Let-7–independent functions because Let-7–independent roles of Lin28 in both normal development and transformation have recently been described.30,45 Further studies are required to distinguish these possibilities.
The induction of Lin28B that we observed on knockdown of Rpl22 expression was not observed on knockdown of other RP (Rpl11 and Rpl24; Figure 4H-I). That the induction of Lin28B is selectively associated with Rpl22 loss raises 2 interesting questions. First, how does repression or inactivation of Rpl22 increase Lin28B expression? Because the induction of Lin28B expression was associated with changes in mRNA levels, we reasoned that Rpl22 inactivation might increase expression by activating NF-κB, because NF-κB has been shown to directly bind to and transactivate the Lin28b locus.32 We found that NF-κB activity was increased by Rpl22 inactivation and that the increase in Lin28B expression was dependent on the activity of canonical p65-containing NFκB complexes (Figure 6A-C). The mechanistic basis by which Rpl22 loss increases canonical NF-κB signaling is unclear but could be direct or indirect. An indirect mechanism might include increasing the expression of a cytokine, such as TNF or IL-6, which could trigger NF-κB activation in either an autocrine or paracrine fashion.32 Alternatively, Rpl22 might play a more direct, proximal role in regulating NF-κB activity. Indeed, a recent report demonstrating that Rps3 associates with p65 and is an integral part of the NF-κB complex has provided precedent for such a mechanism.46
Rps3 was found to directly bind p65 through its KH domain, and this interaction appears to be required for transactivation of target genes.46 Rpl22 lacks a KH domain and its loss does not inhibit NF-κB activity, suggesting that Rpl22 is likely to be functioning by a distinct mechanism. Efforts are currently in progress to determine at which level Rpl22 regulates NF-κB activity. The second question of interest is whether Rpl22 is exerting its effect on NF-κB activity from within or outside of the ribosome. There are several examples of extraribosomal functions for RP, but one the best understood is the interferon-induced release of Rpl13a from ribosomes. On release, Rpl13a associates with mRNAs containing GAIT elements and regulates their translation.2 Insight into whether Rpl22 functions from within or outside of the ribosome was recently provided by the crystal structure of the eukaryotic ribosome,47 which revealed that Rpl22 was represented in monomeric form and was positioned well away from the 40S/60S interface as well as the mRNA entry and peptide exit tunnels. This finding suggests that Rpl22 is unlikely to be exerting its effect on NF-κB activity from within the ribosome and that it, like Rpl13a, may be acting in an extraribosomal manner.
Our observation that germline ablation of the Rpl22 locus was not lethal, as most RP knockouts are,7,8 but instead caused a selective block in development of αβ T-lineage progenitors, prompted us to investigate whether Rpl22 mutations might predispose T cells to transformation. To do so, we focused on the human cancer that arises from those cells, T-ALL. We found that Rpl22-haploinsufficiency in a mouse model of T-cell malignancy accelerated the development of thymic lymphoma. Rpl22 haploinsufficiency was also observed in approximately10% of primary T-ALL isolates and one-third of T-ALL cell lines. All of the T-ALL cell lines bearing RPL22 mutations exhibited Lin28B mRNA levels in excess of those observed in Jurkat (RPL22+/+), compared with approximately one-half of the lines with intact RPL22 alleles (Figure 5E). Finally, meta-analysis of gene expression data from primary T-ALL revealed that of the 10 isolates with reduced Rpl22 mRNA levels, 70% exhibited elevated Lin28B expression levels.48 If, as our in vitro analysis suggests, increased NF-κB activity underlies the increased Lin28B expression observed in T-ALL either bearing RPL22 mutations or in which Rpl22 expression is reduced, then the NF-κB pathway may represent an opportunity for therapeutic intervention in aggressive, Rpl22-haploinsufficient T-ALL (Figure 1), as the targeting NF-κB has shown some efficacy in T-ALL.49,50
Our identification of the link between Rpl22 haploinsufficiency and Lin28B induction represents the first insight into how inactivation of an RP can predispose to transformation and contribute to tumorigenesis. Although in this initial analysis we focused on the role of RPL22 inactivation in pediatric T-ALL, it is possible that Rpl22 may play a role in transformation of other tissues as well. Indeed, in a human breast cancer study, mRNA encoding Rpl22 was found to be substantially down-regulated in invasive breast carcinoma compared with normal breast.19 Down-regulation of Rpl22 mRNA also has been observed in other cancer types, including lung adenocarcinoma, small squamous cell lung carcinoma,20 and adult T-cell leukemia.18 Finally, it will be particularly important to determine whether RPL22 inactivation is observed in other diseases and whether the link between RPL22 inactivation and Lin28B induction is unique to RPL22 or if it might also observed on inactivation of other RP that appear to be dispensable for global, CAP-dependent translation and instead appear to function more selectively in controlling target gene expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Dietmar Kappes and Maureen Murphy for helpful discussion and critical review of the manuscript. They gratefully acknowledge the assistance of the following core facilities of the Fox Chase Cancer Center: Cell Culture, DNA Sequencing, Flow Cytometry, Genomics, Histopathology, and Laboratory Animal.
This work was supported by National Institutes of Health grants R01-AI073920, R01-CA77429, and R21-CA141194; National Institutes of Health core grant P01CA06927; Center grant P30-DK-50306; an appropriation from the Commonwealth of Pennsylvania; and support from the Blood Cell Development and Cancer Keystone. This work was supported in part by grants to the COG, including U10 CA98543 (COG Chair's grant), U10 CA98413 (COG Statistical Center), and U24 CA114766 (COG Specimen Banking), as well as by grant 5P01CA068484 (to the Dana-Farber Cancer Institute and A.T.L.). S.Y.L. was supported by both the Greenwald and Plain & Fancy Fellowships. J.P. is a Merck fellow of the Life Sciences Research Fellowship and was a fellow of the National Institutes of HealthT32 CA009035 training grant. A.G. is a scholar of the American Society of Hematology–Amos Faculty Development Program and is supported by National Institutes of Health grant 5K08CA133103.
National Institutes of Health
Authorship
Contribution: S.R. performed all in vitro transformation analysis and investigation of the role of Lin28 in transformation; S.-Y.L. performed all analysis of the murine T-cell malignancy model; A.G. and A.T.L. assembled and analyzed primary T-ALL samples; J.P. produced primary MEF lines and performed growth analysis; R.J.T. and S.B. performed EMSA studies on NF-κB; Z.T. and R.Z. generated oncogenic Ras and E1A retrovirus; J.R.J. and G.P.Z. performed Southern blot analysis on the Tp53 and Cdkn2a loci; M.R. evaluated the p53 pathway in thymic lymphoma lines; S.A. and T.O. generated the Rpl22 mutant mice; S.P.H. provided relapse patient samples; R.A.T. and J.R.T. produced and maintained MyrAkt2 Tg mice; and S.-Y.L., S.R., and D.L.W. conceived of the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: S.A. and T.O. are or have been employees of, and received stock options from, Lexicon Pharmaceuticals Inc. The remaining authors declare no competing financial interests.
Correspondence: David L. Wiest, Fox Chase Cancer Center, 333 Cottman Ave, Philadelphia, PA 19111; e-mail: david.wiest@fccc.edu.
References
Author notes
S.R. and S.Y.-L. contributed equally to this work.

![Figure 2. Rpl22 haploinsufficiency accelerates the development of cancer in a mouse model of T-cell malignancy. (A) Kaplan-Meier curves of mice of the indicated genotypes, which were killed on manifestation of outward signs of disease. Myr-Akt2;Rpl22+/+, n = 10; Myr-Akt2;Rpl22+/− n = 14; (B) Distribution of CD4/8 subpopulations in thymi of mice with the indicated genotypes. Single-cell suspensions of thymocytes from young adult mice (4-6 weeks) were stained with antibodies reactive CD4 and CD8. Absolute numbers of thymocytes were determined and the mean ± SD are depicted graphically to the right. Analysis was performed on a minimum of 3 mice per group and is representative of 3 experiments performed. (C) Proliferation of explanted thymocytes from MyrAkt2 Tg mice measured by BrdU incorporation. Proliferation of the indicated populations was assessed flow cytometrically by determining the extent of BrdU incorporation after a 4-hour pulse. The mean ± SD of the fraction of BrdU+ cells for a representative experiment is depicted graphically. Each bar represents an individual experiment involving at least 3 mice. Three experiments were performed. *P < .05. (D) Assessment of the extent of proliferation of Rpl22+/+ and +/− thymic lymphoma cells by Ki-67 staining in situ. Thymic sections from the indicated mice were either stained with hematoxylin and eosin (H&E) or with anti-Ki67 antibodies to detect the number of proliferating cells. The micrograph was generated using the ×20 objective (×200 total magnification) of a Nikon Eclipse 50i microscope and a Digital Sight DS-Fi1 camera. Mean ± SEM of the thymic organ weight relative to body weight from Rpl22+/+ (n = 6) and Rpl22+/− (n = 9) mice at the time of sacrifice is depicted graphically below. *P < .05. Representative thymi are shown on the left. (E) Evaluation of the rate of protein synthesis in thymocytes measured by metabolic labeling. Thymocyte suspensions from mice of the indicated genotypes were metabolic labeling for 30 minutes with [35S]methionine after which the counts incorporated were quantified by TCA precipitation of aliquots of the detergent lysates. Data were derived from triplicate values from 2 independent experiments. In addition, extracts were resolved directly by SDS-PAGE and visualized by fluorography (right).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/18/10.1182_blood-2012-03-415349/4/m_zh89991298690002.jpeg?Expires=1765073134&Signature=nPb66AEHyEag6f~XAGilgsIPZAbb4yPUlw9NtzwYNwBY~DHy3NN2BFVxsvONLo9NCw05rFT2MKONwjatmC5PgwcN5xaS5qBTfmKM4nQ9ftUNQSN3aqxB5xx6rcefkfh7Y58CA9fzzrnpVvaJckoB-Tj~p--Am8j6O3GVCDx3D6AnrcdeVErmmtOg2PJyktQJkewPDiV6zT3FS0bMItsbvQY5p6gF~97MBxw4xXkgn-IxtOfAQvUM0jLNJabSh7vvNOCa1F6JfRf40~JxKnQKRiowhacsyKciVjXTFX-bkKKIqNJByDwlUHDtHPZ2Ko6fvsY3UUCffW58lreimCgnTQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal