Abstract
APOBEC3 proteins catalyze deamination of cytidines in single-stranded DNA (ssDNA), providing innate protection against retroviral replication by inducing deleterious dC > dU hypermutation of replication intermediates. APOBEC3G expression is induced in mitogen-activated lymphocytes; however, no physiologic role related to lymphoid cell proliferation has yet to be determined. Moreover, whether APOBEC3G cytidine deaminase activity transcends to processing cellular genomic DNA is unknown. Here we show that lymphoma cells expressing high APOBEC3G levels display efficient repair of genomic DNA double-strand breaks (DSBs) induced by ionizing radiation and enhanced survival of irradiated cells. APOBEC3G transiently accumulated in the nucleus in response to ionizing radiation and was recruited to DSB repair foci. Consistent with a direct role in DSB repair, inhibition of APOBEC3G expression or deaminase activity resulted in deficient DSB repair, whereas reconstitution of APOBEC3G expression in leukemia cells enhanced DSB repair. APOBEC3G activity involved processing of DNA flanking a DSB in an integrated reporter cassette. Atomic force microscopy indicated that APOBEC3G multimers associate with ssDNA termini, triggering multimer disassembly to multiple catalytic units. These results identify APOBEC3G as a prosurvival factor in lymphoma cells, marking APOBEC3G as a potential target for sensitizing lymphoma to radiation therapy.
Introduction
Ionizing radiation and the majority of anticancer agents inflict deleterious DNA damage on tumor cells, predominantly DNA double-strand breaks (DSBs) and covalent DNA crosslinks. DNA DSBs are highly genotoxic lesions, constituting the most disruptive form of DNA damage. Cells use an intricate set of mechanisms to repair genomic DSBs, based on nonhomologous end-joining (NHEJ) or homology-directed repair.1,2 The choice of DSB repair pathway is governed mainly by the cell-cycle phase and the nature of the DSB lesion.3-5 NHEJ operates throughout the cell cycle, resolving approximately 75% to 85% of DSBs induced by ionizing radiation (IR).6,7 Homologous recombination (HR), in contrast, functions predominantly in the S/G2 phase, after synthesis of a homologous DNA template.8,9 The complexity of the DSB lesion determines the extent of DNA end-processing required for repairing the break. Whereas simple DSBs may be repaired by direct ligation via the NHEJ machinery, complex DSBs, often introduced by IR, may require end-processing to reveal 3′ ssDNA overhangs by 5′-3′ nucleolytic end-resection.5 These ssDNA tails, which may be several kilobases long, are substrates for HR factors, such as replication protein A (RPA), RAD51, and RAD52.5,9,10
The human APOBEC3 locus encodes 7 homologous genes expanded in tandem on chromosome 22.11 APOBEC3 (A3) proteins are potent cytidine deaminases acting to restrict retroviral replication and retrotransposition.12,13 APOBEC3G (A3G) is incorporated into assembling HIV-1 virions in the cytoplasm of infected cells and leads to dC > dU hypermutation in the viral ssDNA formed during reverse transcription of the HIV-1genomic RNA in target cells.14,15 A3G is not expressed in most differentiated tissues but is highly expressed in proliferating tissues, including the testis, mitogen-activated PBMCs and various lymphoid malignancies.11,16 Whereas several nuclear A3 proteins were shown to target viral and human nuclear DNA, the predominantly cytoplasmic A3G is not implicated in processing genomic DNA.17,18 However, high expression of A3G in B cells of patients with diffuse large B-cell lymphoma treated with anthracycline-containing chemotherapy was associated with poor survival,19 suggesting that A3G may promote DNA repair. A3G was recently shown to activate ataxia-telangiectasia mutated (ATM) DNA damage checkpoint kinase in HIV-1–infected cells containing deaminated viral DNA,20 further supporting this notion. Here we show that high expression of A3G in lymphatic malignancies is associated with efficient DSB repair and enhanced cell survival after IR. A3G cytidine deaminase activity was specifically required for promoting DSB repair. These findings support a role for A3G in promoting lymphoma radioresistance by mutational-biased DNA repair.
Methods
Cell culture
T-lymphoblastic leukemia (SupT1, SupT11, CEM-SS, MOLT-4), cutaneous T-cell lymphoma (H9, Hut78), multiple myeloma (ARH-77, NCI-H929, CAG), HL-60 acute myeloid leukemia, Ly-1 diffuse large B-cell lymphoma, and Raji Burkitt lymphoma cells were maintained at 1 × 105 to 1 × 106 mL in RPMI 1640 supplemented with 2mM l-glutamine, 10% heat-inactivated FBS, 100 U/mL penicillin and 0.1 mg/mL streptomycin (Beit-haemek) complete medium. Ly-4 diffuse large B-cell lymphoma cells were maintained in complete IMDM (Beit-haemek). SupT1 and H9 cells were provided by the National Institutes of Health AIDS Research and Reference Reagent Program (AIDSP; Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health) and Ly cells by D. Ben-Yehuda (Hadassah Medical School). PBMCs were donated by anonymous healthy volunteers after given consent and isolated on a Ficoll-Hypaque gradient (Sigma-Aldrich). Cells were maintained at 2 × 106 to 4 × 106 mL in complete RPMI 1640. For induction of A3G expression, PBMCs were activated with phytohemagglutinin (5 μg/mL) for 36 hours, followed by supplement of IL-2 (20 U/mL) for 36 hours.16 Human embryonic kidney 293T adherent cell lines were grown as a subconfluent monolayer in complete DMEM (Beit-Haemek). HeLa-A3G cells stably transfected with pcDNA3.1-Apo3G expression vector encoding G418 resistance (obtained through the National Institutes of Health AIDSP) were grown in complete DMEM and with G418 (0.4 mg/mL; Invitrogen). U2OS-DR-GFP cells (obtained from S. P. Jackson, University of Cambridge) were maintained in complete DMEM not containing phenol red, and containing charcoal-treated FBS.
Immunofluorescence
Cells were irradiated by exposure to a 60Co source producing 1 Gy/second γ-radiation, or mock-irradiated. After incubation at 37°C, cells were washed with PBS, fixed with 4% paraformaldehyde/PBS, attached to glass slides by cytospin for 5 minutes at 200g, permeabilized with 0.2% Triton-X-100/PBS for 10 minutes, and blocked with 10% normal goat serum/PBS for 30 to 60 minutes. Cells were then incubated with A3G C-terminal–specific rabbit polyclonal antibody (obtained through the National Institutes of Health AIDSP from J. Lingappa) and γ-H2AX–specific monoclonal antibody (Cell Signaling, provided by E. Pikarsky, Hadassah Medical School), followed by incubation with goat anti–rabbit Cy-5–conjugated antibody, goat anti–mouse Cy-2–conjugated antibody (Jackson ImmunoResearch Laboratories) and 2 μg/mL 4′,6-diamidino-2-phenylindole (DAPI). Slides were mounted with SlowFade Gold (Invitrogen) and examined by Zeiss LSM 710 confocal microscope. Data were collected sequentially using a ×63 objective with 7-fold averaging at a resolution of 1024 × 1024 pixels. Z-sections were obtained using an optical slice of less than 1 μm. Data were analyzed with the Zen 2009 Light Edition software (Carl Zeiss).
shRNA-mediated knockdown of A3G
For short hairpin (sh) RNA expression, the following puromycin resistance-encoding vectors were used: A3G-specific pLKO.1.shA3G (TRCN0000052188, clone NM_021822.1-398s1c1; Sigma-Aldrich), and the unrelated control cyprinid herpes virus 3 (CyHV3)–specific pLKO.1.shCtrl. Lentivectors were obtained by cotransfection of 293T cells with pLKO.1.shA3G or pLKO.1.shCtrl, the packaging plasmid pCMVΔR8.91, and vesicular stomatitis virus G envelope protein expression plasmid. Culture supernatants were collected 48 and 72 hours after transfection and cleared by centrifugation for 10 minutes at 3500g to remove cell debris. Virus was pelleted by centrifugation for 1 hour at 150 000g using a swing SW-41 rotor (Beckman). Cells were transduced with the concentrated vectors by spinoculation for 1 hour at 220g and selected with puromycin (1 μg/mL) for 3 to 10 days after transduction, followed by analysis of A3G expression by Western blot.
Cell-cycle analysis
PBMCs, H9, or puromycin-resistant H9-shRNA cells were irradiated (4 Gy) or mock-irradiated. After 20-hour incubation at 37°C, cells were fixed with methanol for 1 hour at −20°C, washed with PBS, treated with 50 μg/mL RNase A (Sigma-Aldrich), and stained with 5 μg/mL propidium iodide. The total cellular DNA content was determined by flow cytometry using FACScan (BD Biosciences). Cell-cycle data were analyzed by the CellQuest Pro Version 3.3 software and include 10 000 events gated on singlet cells.
Inhibition of A3G catalytic activity and cytidine deamination assay
The inhibitory peptide Vif25-39 corresponds to amino acids 25 to 39 in HIV-1HXBII Vif.21 A fluorescein-conjugated peptide (provided by A. Friedler, Hebrew University) was used to assess the peptide uptake by H9 cells. For inhibition of endogenous A3G in H9 cells, cells were incubated with 100μM Vif25-39 or a control peptide for 2 hours at 37°C before exposure to IR. A3G deaminase activity was assessed as described.21,22 See supplemental Table 1 for oligonucleotide sequences (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
HR assay
Repair of a unique ISceI-induced DSB by HR was assessed in U2OS-DR-GFP cells stably transfected with the Cherry-ISceI-GR plasmid (HRind cells) as described.23 Cherry-ISceI-GR rapidly translocates from the cytoplasm into the nucleus on binding to triamcinolone acetonide (TA). The DR-GFP cassette enables reconstitution of GFP after ISceI-dependent HR between 2 mutated GFP sequences.24 Transfection of A3G or A3G W285A expression plasmids was performed in 24-well plates with GenJet (Signagen). Forty hours after transfection, ISceI-directed DSB was induced with 10−7M TA (Sigma-Aldrich). GFP+ cells were sorted 52 hours later by flow cytometry using FACScan as described.23
AFM
Purification of C-terminally His6-tagged A3G and A3G W285A from human embryonic kidney 293T cells (> 90% purity) and production of ssDNA by in vitro rolling circle amplification were performed as described.22 Atomic force microscopy (AFM) imaging was performed at room temperature using a multimode scanning probe microscope with a Nanoscope 3A controller (Digital Instruments/Veeco Probes). AFM images were recorded on freshly cleaved mica surfaces (Structure Probe). The mica surfaces were activated with 5mM MgCl for 1 minute and washed with distilled water. For binding assays, purified A3G and ssDNA were incubated on ice in buffer containing 25mM Tris, pH 7.0, in 10-μL reaction volume. Samples were deposited on the mica surface for 1.5 minutes and washed with distilled water. Images were taken with NSC 15 AFM tips (Mikromasch) using the tapping mode at their resonant frequency. The 4.0 images were analyzed with WSxM 4.0 Beta 2.0 SPIP software (Nanotec). Stoichiometry of bound A3G proteins was calculated by the flooding option, assuming a monomeric volume of approximately 60 nm3.
Analysis of terminal ssDNA tethering by DNA polymerase terminal extension
DNA1 and DNA2 (supplemental Table 1) were annealed to an equimolar amount of a short oligonucleotide (30 nt) complementary to their 5′-terminus by heating to 95°C followed by slow cooling to room temperature. DNA-polymerase extension reactions were performed in a total volume of 7 μL and included 100 fmol annealed oligonucleotides, 50μM dNTPs (dCTP, dGTP, dTTP, and dATP-biotin, Invitrogen; NEBuffer 1; final dilution × 0.35, NEB), 1 mg/mL BSA, 100 fmol A3G or A3G W285A, and 5 U Klenow fragment (3′ → 5′ exo(−); NEB). Reactions were incubated for 30 minutes at 37°C and terminated by heating to 95°C for 5 minutes and addition of 5 μL formamide. Samples were then heated to 80°C before loading on preheated 10% acrylamide gel containing 6M urea. Gels were washed in Tris-Boric acid-EDTA buffer, pH 8.3, for 15 minutes, transferred to a Hybond N nylon membrane (GE Healthcare) using a semidry transfer apparatus (Bio-Rad) and UV-crosslinked at 302 nm for 15 minutes. After blocking with Casein blocking buffer (Sigma-Aldrich), the membrane was treated with horseradish peroxidase conjugated streptavidin (Jackson ImmunoResearch Laboratories) for 20 minutes at room temperature, washed 6 times with TBS, pH 7.4, and visualized by enhanced chemiluminescence.
NHEJ assay
Preparation of HeLa and HeLa-A3G whole-cell extracts (WCEs) was performed as described,25 with inclusion of RNase A treatment (50 μg/mL; Sigma-Aldrich) for 20 minutes at room temperature, after homogenization. DNA substrates were prepared by linearizing the pBlueScript SK+ plasmid (Stratagene) with EcoRI, ApaI, or EcoRV (Fermentas) to produce DNA fragments (3 kb) with compatible ends or blunt ends. Restriction reaction products were purified with QIAquick PCR purification kit (QIAGEN). End joining assays were performed in 50 μL and contained 60 ng linearized plasmid, 0.3 μg WCE, 25mM Tris, pH 7.5, 100mM KCl, 5mM MgCl2, 5mM DTT, and 0.5mM ATP. Control ligations with T4 DNA ligase (NEB) were performed in the designated buffer. Reactions were incubated for 18 hours at room temperature (∼ 21°C) and terminated by adding proteinase K (0.1 mg/mL). After 1.5-hour incubation at 50°C, DNA was extracted with phenol/chloroform (1:1) and ethanol-precipitated. DNA was resuspended in H2O, separated by agarose (1.2%) gel electrophoresis and stained with SYBR Gold (Invitrogen). Gels were visualized by UV light (302 nm), captured by an Olympus C-5050 CCD, and analyzed by optical density (OD) scan using the TINA2.0 densitometry software (Raytest).
Results
A3G expression in lymphoid cell lines is associated with efficient DSB repair
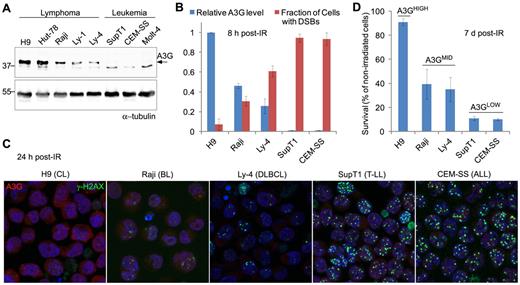
To explore a possible role for A3G in the response of lymphoma cells to genotoxic treatment,19 we used γ-radiation to generate DSBs in a panel of lymphoid cell lines expressing differential A3G protein levels (Figure 1A). DSBs were detected by probing the phosphorylated form of histone H2AX indicative of chromatin flanking DSBs. Initial DSB formation measured 30 minutes after exposure to 4 Gy γ-radiation was similar in all cells. Leukemic cell lines not expressing A3G had high cellular DSB incidence 8 hours after IR, encompassing more than 90% of cultured cells (Figure 1B). In contrast, lymphoma cell lines expressing A3G had lower cellular DSB incidence after IR, encompassing approximately 5% to 60% of cultured cells, inversely dependent on A3G expression level. This correlation persisted 24 hours after IR and was observed also in other lymphoid cell lines (Figure 1C; supplemental Figure 1A). A3G expression level in these cells also correlated with overall cell survival 7 days after irradiation (Figure 1D; supplemental Figure 1B). Thus, DSB repair efficiency and survival of various irradiated lymphoid cell lines assessed here correlate with A3G expression level.
A3G expression correlates with DSB repair efficiency and survival after IR. (A) Western blot showing A3G protein level in lymphoma and leukemia cell lines. α-tubulin was used as a loading control. Molecular weights (kDa) are indicated on the left. (B) Cells were exposed to γ-radiation (4 Gy) and stained after 8 hours with anti-A3G and anti–γ-H2AX antibodies. The fraction of cells containing DSBs according to γ-H2AX staining is plotted against relative A3G expression measured in panel A. Values represent mean ± SD from 3 independent experiments and at least 10 different fields for each cell line analyzed. (C) Cells were exposed to γ-radiation and stained after 24 hours with anti-A3G and anti–γ-H2AX antibodies. Nuclei were counterstained with DAPI (original magnification ×630). CL indicates cutaneous lymphoma; BL, Burkitt lymphoma; DLBCL, diffuse large B-cell lymphoma; T-LL, T lymphoblastic leukemia; and ALL, acute lymphocytic leukemia. (D) Cells were exposed to γ-radiation (4 Gy) or mock and counted after 7 days. Cell survival was calculated as percentage of trypan blue-negative irradiated cells out of nonirradiated cells. Values represent mean ± SD from 2 independent experiments.
A3G expression correlates with DSB repair efficiency and survival after IR. (A) Western blot showing A3G protein level in lymphoma and leukemia cell lines. α-tubulin was used as a loading control. Molecular weights (kDa) are indicated on the left. (B) Cells were exposed to γ-radiation (4 Gy) and stained after 8 hours with anti-A3G and anti–γ-H2AX antibodies. The fraction of cells containing DSBs according to γ-H2AX staining is plotted against relative A3G expression measured in panel A. Values represent mean ± SD from 3 independent experiments and at least 10 different fields for each cell line analyzed. (C) Cells were exposed to γ-radiation and stained after 24 hours with anti-A3G and anti–γ-H2AX antibodies. Nuclei were counterstained with DAPI (original magnification ×630). CL indicates cutaneous lymphoma; BL, Burkitt lymphoma; DLBCL, diffuse large B-cell lymphoma; T-LL, T lymphoblastic leukemia; and ALL, acute lymphocytic leukemia. (D) Cells were exposed to γ-radiation (4 Gy) or mock and counted after 7 days. Cell survival was calculated as percentage of trypan blue-negative irradiated cells out of nonirradiated cells. Values represent mean ± SD from 2 independent experiments.
A3G is recruited to DSB repair foci
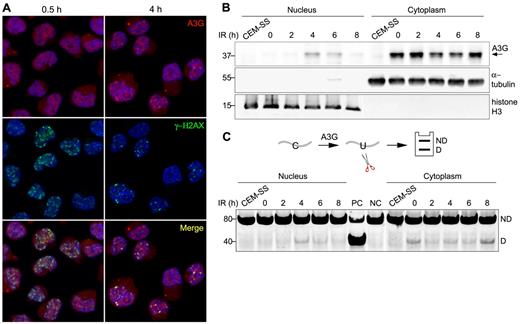
A3G localizes predominantly to the cytoplasm of PBMCs and H9 T cells16 (supplemental Figure 2A). To determine whether A3G is recruited to genomic DSBs, A3G and γ-H2AX subcellular localization was probed in H9 cells exposed to 4 Gy γ-radiation. At 30 minutes after IR, A3G was diffused throughout the cell, and multiple IR-induced DSBs were evident by the formation of γ-H2AX nuclear foci (Figure 2A). After 1 to 4 hours, A3G formed distinct nuclear foci, which colocalized with γ-H2AX (Figure 2A; supplemental Figure 2B). A3G direct association with genomic DSBs subsided 8 hours after IR concurrently with γ-H2AX foci (supplemental Figure 2B). Consistently, A3G was also detected in H9 nuclear fractions 4 to 6 hours after IR, coinciding with a parallel reduction of A3G levels in the cytoplasm (Figure 2B). These nuclear fractions were associated with cytidine deaminase activity measured on an oligunocleotide substrate (Figure 2C). The reduction in cytoplasmic deaminase activity in the absence of comparable increase in nuclear activity 2 hours after IR may reflect sequestration of A3G activity by cytoplasmic RNA induced in the initial stage of the cellular response to IR.26-28 Furthermore, we cannot exclude low level of nuclear deaminase activity 2 hours after IR, which is below our assay detection threshold. After peak nuclear deaminase activity 4 to 6 hours after IR, A3G activity was reduced in the nuclear fraction and increased in the cytoplasmic fraction 8 hours after IR to comparable levels as in nonirradiated cells, in line with A3G transient nuclear localization after IR. These results indicate that catalytically active A3G is transiently recruited to the nucleus and that A3G localizes to DSB repair foci in response to IR.
A3G is recruited to DSB repair foci. (A) H9 cells were irradiated (4 Gy) and probed with anti-A3G and anti-γ-H2AX antibodies at the indicated times. Nuclei were counterstained with DAPI. (B) Irradiated cells were harvested at the indicated times after IR and A3G content in nuclear and cytoplasmic fractions was assessed by Western blotting. α-tubulin and histone H3 were used as cytoplasmic and nuclear markers, respectively. CEM-SS cells not expressing A3G were used as a negative control. Molecular sizes (kDa) are indicated. (C, top): Scheme of the cytidine deamination assay in which C > U deamination in an oligonucleotide forms a restriction enzyme cleavage site after PCR amplification. ND indicates not deaminated; and D, deaminated. (Bottom): Cytidine deaminase activity in extracts of nuclear and cytoplasmic fractions was assessed by incubation with an oligonucleotide substrate containing a single A3G CCC target site (SEco, 80 nt) for 30 minutes at 37°C. DNA sizes (bp) are indicated on the left. PC indicates positive control oligonucleotide; and NC, negative control oligonucleotide.
A3G is recruited to DSB repair foci. (A) H9 cells were irradiated (4 Gy) and probed with anti-A3G and anti-γ-H2AX antibodies at the indicated times. Nuclei were counterstained with DAPI. (B) Irradiated cells were harvested at the indicated times after IR and A3G content in nuclear and cytoplasmic fractions was assessed by Western blotting. α-tubulin and histone H3 were used as cytoplasmic and nuclear markers, respectively. CEM-SS cells not expressing A3G were used as a negative control. Molecular sizes (kDa) are indicated. (C, top): Scheme of the cytidine deamination assay in which C > U deamination in an oligonucleotide forms a restriction enzyme cleavage site after PCR amplification. ND indicates not deaminated; and D, deaminated. (Bottom): Cytidine deaminase activity in extracts of nuclear and cytoplasmic fractions was assessed by incubation with an oligonucleotide substrate containing a single A3G CCC target site (SEco, 80 nt) for 30 minutes at 37°C. DNA sizes (bp) are indicated on the left. PC indicates positive control oligonucleotide; and NC, negative control oligonucleotide.
A3G directly promotes DSB repair
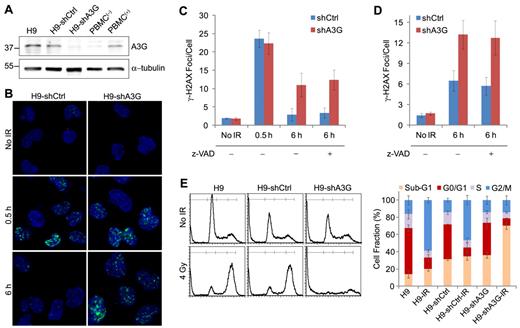
A3G accumulation at the breakage sites coincided with reduction in the number and magnitude of γ-H2AX foci, suggesting that A3G is involved in DSB repair. In that case, knocking down A3G expression in cells should result in impaired DSB repair and possibly cell death. A3G knockdown or control H9 cells were generated by expression of specific A3G-directed shRNA (H9-shA3G) or control shRNA (H9-shCtrl). A3G expression in H9-shA3G cells was reduced by approximately 70% to 80% compared with H9-shCtrl cells, and was comparable with nonstimulated PBMCs (Figure 3A). A3G levels in H9-shCtrl cells resembled those in activated PBMCs stimulated with phytohemagglutinin.16 To determine whether A3G is involved in DSB repair, we compared the dynamics of γ-H2AX phosphorylation in IR-exposed H9-shCtrl versus H9-shA3G cells. Gamma-H2AX foci formation in the first 30 minutes after IR was similar in both H9-shCtrl and H9-shA3G cells and resembled the parental H9 cells (Figure 3B-C). At 6 hours after IR, most H9-shCtrl cells were negative for γ-H2AX staining and the average number of γ-H2AX foci per cell was only approximately 50% higher than in nonirradiated cells, reflecting efficient DSB repair. However, H9-shA3G cells did not exhibit a similar decrease in γ-H2AX foci but had on average 3.8 ± 0.7-fold more γ-H2AX foci per cell compared with H9-shCtrl cells, signifying impaired DNA repair (Figure 3B-C). To test whether A3G indirectly reduces the cellular DSB load by inhibiting apoptosis, cells were preincubated with the caspase inhibitor z-VAD-fmk and probed for γ-H2AX 6 hours after IR. DSB incidence in the presence of z-VAD-fmk was comparable with cells treated with mock, suggesting that A3G has a direct role in DSB repair (Figure 3C). Depletion of A3G in ARH-77 multiple myeloma cells also resulted in higher DSB incidence 6 hours after IR, indicating that A3G activity is not restricted to H9 cells (Figure 3D). Similar to H9 cells, the elevated DSB load in ARH-77-shA3G cells was caspase-independent (Figure 3D). To verify the defect in DSB repair in A3G knockdown cells, the DNA content of H9, H9-shCtrl, and H9-shA3G cells was measured 20 hours after 4 Gy IR by flow cytometry. Both H9 and H9-shCtrl cells were arrested at G2/M, accounting for a comparable decrease in G0/G1 and S-phase cells (Figure 3E). In contrast, H9-shA3G cells did not undergo G2/M arrest but instead had an increased sub-G1 fraction, indicating increased nuclear fragmentation or cell death in the absence of A3G. Thus, A3G directly promotes DSB repair independently of inhibition of apoptosis; however, deficient DSB repair in the absence of A3G may result in apoptosis.
A3G is required for DSB repair in H9 lymphoma cells. (A) Western blot showing A3G cellular protein level. PBMC(−) and PBMC(+), naive, and phytohemagglutinin-activated peripheral blood mononuclear cells, respectively. α-tubulin was used as a loading control. Molecular weights (kDa) are indicated on the left. (B) Cells were irradiated (4 Gy) or mock-irradiated and stained with anti–γ-H2AX antibody. A3G knockdown or control H9 (C) or ARH-77 (D) cells were preincubated with 20μM z-VAD-fmk or mock for 1 hour and treated as in panel B. Values represent mean ± SD from 3 independent experiments and at least 10 different fields for each time point analyzed. (E) Cell-cycle analysis after IR. (Left): Irradiated (4 Gy) or mock-irradiated (No IR) cells were stained after 20 hours with propidium iodide, and DNA content was determined by FACS (10 000 acquired events). (Right): Values represent mean ± SD from 3 independent experiments.
A3G is required for DSB repair in H9 lymphoma cells. (A) Western blot showing A3G cellular protein level. PBMC(−) and PBMC(+), naive, and phytohemagglutinin-activated peripheral blood mononuclear cells, respectively. α-tubulin was used as a loading control. Molecular weights (kDa) are indicated on the left. (B) Cells were irradiated (4 Gy) or mock-irradiated and stained with anti–γ-H2AX antibody. A3G knockdown or control H9 (C) or ARH-77 (D) cells were preincubated with 20μM z-VAD-fmk or mock for 1 hour and treated as in panel B. Values represent mean ± SD from 3 independent experiments and at least 10 different fields for each time point analyzed. (E) Cell-cycle analysis after IR. (Left): Irradiated (4 Gy) or mock-irradiated (No IR) cells were stained after 20 hours with propidium iodide, and DNA content was determined by FACS (10 000 acquired events). (Right): Values represent mean ± SD from 3 independent experiments.
A3G-mediated DSB repair is cytidine deaminase dependent
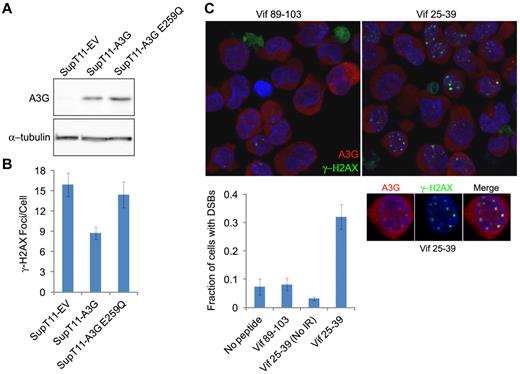
To assess whether A3G cytidine deaminase activity is required for DSB repair, we asked whether IR-induced γ-H2AX focus formation occurred differentially in SupT11 cells stably expressing A3G, an E259Q catalytic mutant or empty vector (EV).29 The SupT11 cell line is a single-cell subclone of SupT1 that is nearly devoid of all endogenous APOBEC3s, including A3G29,30 (Figure 4A). The average DSB load as assessed by γ-H2AX focus formation was approximately 2-fold lower in wild-type A3G-expressing cells compared with the EV control cells 24 hours after IR, demonstrating that reconstitution of A3G expression in these cells enhances DSB repair (Figure 4B). In contrast, enhanced repair was not observed in A3G E259Q-expressing cells. To corroborate these data, we attempted to inhibit A3G catalytic activity in H9 cells. We have recently shown that the HIV-1 Vif protein and Vif-derived peptides inhibit A3G catalytic activity.21 The Vif25-39 peptide specifically inhibited the deaminase activity of purified A3G with an IC50 of approximately 0.8μM, unlike Vif89-103, which did not inhibit A3G at a concentration of 100μM (supplemental Figure 3). H9 cells were preincubated with these Vif-derived peptides before IR exposure, and DSB repair was determined 8 hours after IR. Cells preincubated with Vif89-103 exhibited efficient DSB repair (< 8% of cells containing DSBs), similar to cells incubated with mock or nonirradiated cells preincubated with Vif25-39 (Figure 4C). In contrast, 32% plus or minus 4.3% of irradiated cells preincubated with Vif25-39 contained DSBs, suggesting that cytidine deamination is involved in A3G-mediated DSB repair. Importantly, Vif25-39 did not prevent recruitment of A3G to DSBs, as A3G colocalized with γ-H2AX nuclear foci at unrepaired DSBs in pretreated cells (Figure 4C inset). This is consistent with the uncompetitive inhibition mechanism of Vif25-39, which binds the enzyme-substrate complex.21 Overall, these observations indicate that the catalytic activity of A3G is required to facilitate the repair of IR-induced DSBs.
A3G-mediated DSB repair is cytidine deaminase dependent. (A) Immunoblot of A3G in SupT11 cells stably expressing an EV control, wild-type A3G, or A3G E259Q catalytic mutant. α-tubulin was used as a loading control. (B) Quantification of γ-H2AX foci in SupT11 cells 24 hours after IR (4 Gy). Values represent mean ± SD from 2 independent experiments and at least 10 different fields. (C) H9 cells were incubated for 2 hours with the indicated Vif-derived peptides, irradiated (4 Gy) or mock-irradiated (No IR), and stained after 8 hours with anti-A3G and anti–γ-H2AX antibodies. Nuclei were counterstained with DAPI. (Bottom) Values represent mean ± SD from 2 independent experiments and at least 10 different fields. (Inset) Magnification of an irradiated cell preincubated with Vif25-39.
A3G-mediated DSB repair is cytidine deaminase dependent. (A) Immunoblot of A3G in SupT11 cells stably expressing an EV control, wild-type A3G, or A3G E259Q catalytic mutant. α-tubulin was used as a loading control. (B) Quantification of γ-H2AX foci in SupT11 cells 24 hours after IR (4 Gy). Values represent mean ± SD from 2 independent experiments and at least 10 different fields. (C) H9 cells were incubated for 2 hours with the indicated Vif-derived peptides, irradiated (4 Gy) or mock-irradiated (No IR), and stained after 8 hours with anti-A3G and anti–γ-H2AX antibodies. Nuclei were counterstained with DAPI. (Bottom) Values represent mean ± SD from 2 independent experiments and at least 10 different fields. (Inset) Magnification of an irradiated cell preincubated with Vif25-39.
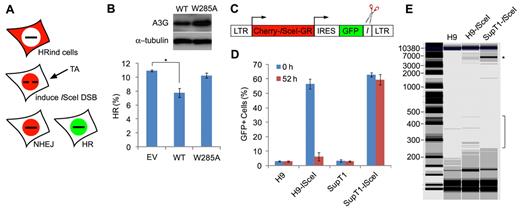
A3G mediates deletional repair of a persistent ISceI-induced DSB
Deamination of cytidines in resected DSB ssDNA overhangs may result in cleavage of ssDNA in abasic sites, mediated by the base excision repair mechanism or other repair factors, such as the MRN complex.31,32 To test whether A3G deaminates resected ssDNA, we expressed A3G or an A3G W285A catalytic mutant, which retains wild-type DNA binding properties (supplemental Figure 4) in U2OS cells stably carrying a DR-GFP HR reporter cassette24 and expressing an inducible ISceI-Cherry endonuclease (HRind cells).23 Western blot analysis confirmed that these cells do not express endogenous A3G (results not shown). Induction of ISceI generates a unique DSB within the DR-GFP cassette that produces functional GFP only if repaired by HR (Figure 5A). We assumed that cytidine deamination in resected ssDNA flanking the ISceI restriction site may lead to truncation of the DR-GFP DNA, resulting in reduced GFP reconstitution. Whereas 10.9% plus or minus 1.0% of the cells underwent repair by HR 52 hours after ISceI induction, expression of wild-type A3G reduced HR by 30.2% plus or minus 5.6%. In contrast, expression of A3G W285A resulted in only 6.2% plus or minus 3.3% reduction in HR (Figure 5B). This may indicate that A3G targets resected ssDNA flanking ISceI-induced DSB. To further test the activity of endogenous A3G, we cloned a Cherry-ISceI-GR gene23 into a lentiviral expression vector and inserted an ISceI target site directly downstream to an IRES-GFP gene (Figure 5C). After transduction of cells with the lentiviral vector, induction of the ISceI enzyme leads to a unique DSB directly downstream to the GFP gene. In case A3G targeted ssDNA overhangs flanking the DSB after resection, we expected that C > U mutations and/or deletion of the GFP coding sequence would result in inactivation of the GFP gene. H9 (A3G-high) and SupT1 (A3G-low) cells were infected with lentiviruses containing the ISceI vector (H9-ISceI and SupT1-ISceI cells) or mock, and analyzed for GFP+ cells by FACS before and after induction of Cherry-ISceI-GR with TA. Assessment of GFP+ cells 48 hours after lentiviral infection indicated that transduction efficiency was 54% to 64% (Figure 5D). Whereas the percentage of GFP+ SupT1-ISceI cells was only marginally reduced 52 hours after ISceI induction, the percentage of GFP+ H9-ISceI cells was reduced 9-fold. Notably, microscopic examination of the cells 7 days after ISceI induction revealed poor survival of SupT1-ISceI cells but high viability of H9-ISceI cells (supplemental Figure 5). To test whether the reduction in GFP+ H9-ISceI cells is attributed to deletion of the GFP coding sequence, total genomic DNA was extracted from H9-ISceI and SupT1-ISceI cells 52 hours after ISceI induction, and lentivector-specific sequence flanking the ISceI target site was amplified by PCR (Figure 5E). The 5900-bp DNA band corresponding to the parental lentiviral cassette was markedly reduced in H9-ISceI compared with SupT1-ISceI cells. In addition, several PCR products in the range of 250 to 450 bp were detected after amplification of H9-ISceI DNA but not SupT1-ISceI or H9-mock DNA, suggesting that these PCR products represent truncated DNA junctions. Thus, repair of a persistent ISceI-induced DSB in H9 cells involves mutagenic deaminase-dependent processing of genomic sequences flanking the break.
A3G mediates deletional repair of a persistent ISceI-induced DSB. (A) Scheme of the DR-GFP HR reporter assay.23 Repair of ISceI-induced DSB by HR reconstitutes the expression of functional GFP. (B) HRind cells were transfected with an EV, A3G (WT), or A3G W285A expression plasmids and induced with TA for 52 hours. GFP expression was measured by FACS. Transfection yield was 55% to 60%, as determined by cotransfection with DsRed expression plasmid. Expression of A3G and A3G W285A was evaluated by Western blot (top). Values represent mean ± SD from 3 independent experiments. *P = .007 (unpaired t test). (C) ISceI-expressing lentiviral vector containing long terminal repeats (LTRs), internal ribosomal entry site (IRES), and ISceI target sequence (I) adjacent to GFP. (D) H9 or SupT1 cells were infected with lentiviruses containing the ISceI vector or mock and assessed 48 hours later for GFP+ cells by FACS (0 hours). Cells were sorted again 52 hours after induction of ISceI with TA (52 hours). Values represent mean ± SD from 3 independent experiments. (E) Genomic DNA was extracted from mock- or ISceI lentivirus-infected H9 or SupT1 52 hours after ISceI induction with TA. *Analysis of PCR amplification of a 5900-bp fragment using vector specific primers was performed by a Bioanalyzer. DNA marker (M) sizes are indicated (kb).
A3G mediates deletional repair of a persistent ISceI-induced DSB. (A) Scheme of the DR-GFP HR reporter assay.23 Repair of ISceI-induced DSB by HR reconstitutes the expression of functional GFP. (B) HRind cells were transfected with an EV, A3G (WT), or A3G W285A expression plasmids and induced with TA for 52 hours. GFP expression was measured by FACS. Transfection yield was 55% to 60%, as determined by cotransfection with DsRed expression plasmid. Expression of A3G and A3G W285A was evaluated by Western blot (top). Values represent mean ± SD from 3 independent experiments. *P = .007 (unpaired t test). (C) ISceI-expressing lentiviral vector containing long terminal repeats (LTRs), internal ribosomal entry site (IRES), and ISceI target sequence (I) adjacent to GFP. (D) H9 or SupT1 cells were infected with lentiviruses containing the ISceI vector or mock and assessed 48 hours later for GFP+ cells by FACS (0 hours). Cells were sorted again 52 hours after induction of ISceI with TA (52 hours). Values represent mean ± SD from 3 independent experiments. (E) Genomic DNA was extracted from mock- or ISceI lentivirus-infected H9 or SupT1 52 hours after ISceI induction with TA. *Analysis of PCR amplification of a 5900-bp fragment using vector specific primers was performed by a Bioanalyzer. DNA marker (M) sizes are indicated (kb).
A3G multimers interact with ssDNA termini leading to multimer disassembly
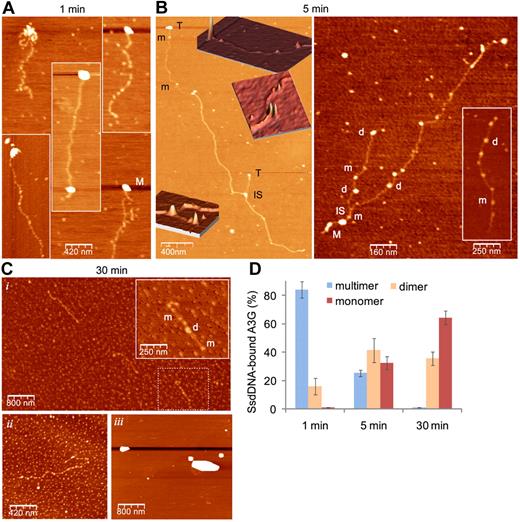
DSB resection is estimated to generate an average of 2- to 4-kb-long ssDNA in each side of the break.10 To simulate A3G association with long ssDNA overhangs, we generated approximately 1.5- to 7-kb-long (average, 3-kb-long) ssDNA containing a GGCCC site at 100-nt intervals (LC DNA) by in vitro rolling circle amplification. Single-molecule interactions of highly purified A3G-His6 and rolling circle amplification ssDNA were assessed by atomic force microscopy (AFM). We have previously shown that human 293T cell–derived A3G resides mostly in high-order multimers,22 consistent with multimerization of both endogenous and recombinant A3G expressed in various systems.27,33 Figure 6A shows that A3G multimers rapidly bind ssDNA preferentially at its terminus or within tens to few hundreds of bases from the ssDNA terminus. After 5-minute incubation on ice, an overall reduction in the size of bound A3G multimers was accompanied by A3G dimers and monomers occupying internal DNA domains (Figure 6B and supplemental Figure 6). End-bound A3G multimers were engaged in interstrand synapsis between the terminus of one strand and an internal domain in a second strand, in line with our previous results demonstrating that A3G is able to tether 2 ssDNA segments.22 After 30-minute incubation, virtually all A3G multimers were reduced to monomers and dimers (Figure 6Ci,ii). This process was DNA-dependent, as in the absence of DNA, A3G remained in the form of high-order multimers (Figure 6Ciii). The fraction of ssDNA-associated multimeric, dimeric, and monomeric A3G at each time point is summarized in Figure 6D. Thus, A3G multimers bind preferentially to ssDNA termini, facilitating interstrand synapsis and A3G multimer disassembly to dimers and monomers that are able to move freely on ssDNA.
A3G multimers interact with ssDNA termini and mediate interstrand synapsis. (A-C) AFM images showing binding of purified 293T cell–derived A3G-His6 to LC ssDNA (A3G multimer:ssDNA molar ratio ∼ 1:1) after 1 minute (A), 5 minutes (B), and 30 minutes (Ci-ii) incubation on ice. (B) Insets: Three-dimensional representations of parts of the 2D image (orientation has been tilted for clarity). (Ci) Inset: magnification of dashed box area. (Ciii) A3G was incubated in the absence of ssDNA. M indicates A3G multimer; d, dimer; m, monomer; T, A3G multimer associated with ssDNA terminus; and IS, ssDNA interstrand synapse. (D) Quantitation of A3G-ssDNA complexes. Data are represented as mean ± SD from at least 10 different fields for each time point.
A3G multimers interact with ssDNA termini and mediate interstrand synapsis. (A-C) AFM images showing binding of purified 293T cell–derived A3G-His6 to LC ssDNA (A3G multimer:ssDNA molar ratio ∼ 1:1) after 1 minute (A), 5 minutes (B), and 30 minutes (Ci-ii) incubation on ice. (B) Insets: Three-dimensional representations of parts of the 2D image (orientation has been tilted for clarity). (Ci) Inset: magnification of dashed box area. (Ciii) A3G was incubated in the absence of ssDNA. M indicates A3G multimer; d, dimer; m, monomer; T, A3G multimer associated with ssDNA terminus; and IS, ssDNA interstrand synapse. (D) Quantitation of A3G-ssDNA complexes. Data are represented as mean ± SD from at least 10 different fields for each time point.
A3G promotes end joining of ssDNA overhangs
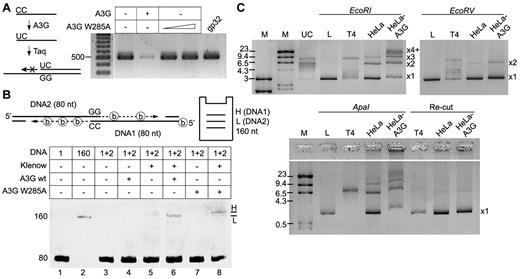
Association of A3G with ssDNA ends together with its ability to mediate interstrand synapsis suggests that it may promote ssDNA end joining. To validate that A3G is able to associate with the terminal base of ssDNA, a primer with 3′-terminal CC was incubated with A3G and then used for extension and PCR amplification of a target sequence. Should A3G deaminate the 3′-terminal C, Taq DNA polymerase will not be able to extend a 3′-incompatible CU terminus. Incubation of the primer with A3G resulted in marked inhibition of the PCR reaction (∼ 90%), in contrast to normal PCR amplification in the presence of the A3G W285A catalytic mutant or recombinant T4 gp32 ssDNA binding protein (Figure 7A). Hence, A3G interacts with the extreme base of the ssDNA 3′-terminus and is able to perform terminal cytidine deamination.
A3G promotes end joining of ssDNA overhangs. (A) Terminal cytidine deamination by purified A3G was determined by a DNA polymerase primer extension assay. A primer with 3′-terminal CC was incubated with A3G or A3G W285A mutant and used for PCR amplification of a target sequence. T4 gp32 ssDNA binding protein was used as control. DNA size is indicated on the left (bp). (B) Terminal ssDNA joining was assessed in a polymerase extension assay. (Top) Schematic depiction of the assay, including expected product migration by PAGE. Thirty biotinylated ATP molecules (shown as circled “b”) are incorporated in the DNA1 extension product (H, high), and only 21 in the DNA2 extension product (L, low). (Bottom) Denaturing urea-PAGE image showing ssDNA extension products using oligonucleotides with GG/CC terminal microhomology. ssDNA sizes (nt) and lane numbers are indicated to the left and below the image. (C) Plasmid-based NHEJ assay. HeLa or HeLa-A3G whole-cell extracts were incubated with pBluescript plasmid linearized with EcoRI, EcoRV, or ApaI restriction enzymes. (Bottom) End-joining products were restricted with ApaI (Re-Cut). Product sizes of joint linear plasmid (L) are indicated to the right of each PAGE image, as described in the text. UC indicates uncut plasmid; and T4, T4 DNA ligase-positive control. DNA marker (M) sizes are indicated (kb).
A3G promotes end joining of ssDNA overhangs. (A) Terminal cytidine deamination by purified A3G was determined by a DNA polymerase primer extension assay. A primer with 3′-terminal CC was incubated with A3G or A3G W285A mutant and used for PCR amplification of a target sequence. T4 gp32 ssDNA binding protein was used as control. DNA size is indicated on the left (bp). (B) Terminal ssDNA joining was assessed in a polymerase extension assay. (Top) Schematic depiction of the assay, including expected product migration by PAGE. Thirty biotinylated ATP molecules (shown as circled “b”) are incorporated in the DNA1 extension product (H, high), and only 21 in the DNA2 extension product (L, low). (Bottom) Denaturing urea-PAGE image showing ssDNA extension products using oligonucleotides with GG/CC terminal microhomology. ssDNA sizes (nt) and lane numbers are indicated to the left and below the image. (C) Plasmid-based NHEJ assay. HeLa or HeLa-A3G whole-cell extracts were incubated with pBluescript plasmid linearized with EcoRI, EcoRV, or ApaI restriction enzymes. (Bottom) End-joining products were restricted with ApaI (Re-Cut). Product sizes of joint linear plasmid (L) are indicated to the right of each PAGE image, as described in the text. UC indicates uncut plasmid; and T4, T4 DNA ligase-positive control. DNA marker (M) sizes are indicated (kb).
We next developed a DNA polymerase extension assay designed to assess functional juxtaposing of two 3′ ssDNA termini with minimal terminal microhomology (Figure 7B). To simulate a resected DSB, the 5′-termini of 2 ss-oligonucleotides (DNA1 and 2, 80 nt) were annealed to complementary oligonucleotides (30 nt), leaving 3′ ssDNA overhangs (50 nt). Only the two 3′-terminal bases in each overhang are complementary (CC in DNA1 and GG in DNA2) so that juxtaposing the 2 overhangs may enable each terminus to serve as a primer for DNA polymerase extension using the opposite strand as a template. To distinguish between the 2 possible extension products, biotinylated dATP was used in the polymerase reaction and the sequence of DNA1 was designed to contain less dT than DNA2. Thus, the DNA1 extension product, which uses DNA2 as a template will contain more dATP-biotin compared with the DNA2 extension product and will therefore migrate slower in gel electrophoresis. Incubation of DNA1 + 2 with Klenow fragment yielded only background level products (Figure 7B lane 5). However, a discernible extension product, which migrated slightly above the 160-nt marker, was apparent in the presence of A3G (Figure 7B lane 6). Extension occurred approximately 9-fold more efficiently in the presence of A3G than with Klenow fragment alone. Interestingly, incubation with A3G W285A catalytic mutant yielded a higher extension product with similar efficiency (Figure 7B lane 8). These results indicate that A3G mediated the extension of DNA2 from the terminal GG, using DNA1 as a template, whereas the W285A mutant mediated the extension of DNA1 from the terminal CC, using DNA2 as a template. The fact that A3G did not extend the CC terminus of DNA1 while the W285A mutant did suggests that A3G deaminated the terminal cytidine, creating a U:G base pair that could not be used as a primer under these conditions. To confirm that the extension products resulted from the specific annealing of ssDNA termini, the terminal GG in DNA2 were substituted with TT, which cannot base pair with the terminal CC of DNA1. As expected, no extension products were detected in the presence or absence of wild-type or mutant A3G.
A3G activity in ssDNA end synapsis may promote DSB end joining. The A3G ability to promote NHEJ was assessed in a plasmid-based assay25 using WCEs of HeLa and HeLa-A3G cells stably expressing A3G. Specifically, we measured joining efficiency of linearized plasmids with compatible ends (EcoRI- or ApaI-linearized) and blunt ends (EcoRV-linearized). HeLa WCE supported end joining of EcoRI compatible DNA ends, promoting joining of 2 (× 2) and 3 (× 3) linear DNA molecules and using approximately 17% of the substrate (Figure 7C). However, the DNA end joining activity of HeLa-A3G WCE was 4- to 5-fold more efficient, producing × 2, × 3 and higher forms of joint DNA (× 4+), and using more than 80% of the substrate. Joining of blunt-ended substrate was less efficient in general and was enhanced only marginally by HeLa-A3G WCEs. To determine whether terminal cytidines affect joining efficiency, we used ApaI, which produces 5′-GGCC overhangs. Whereas end joining of ApaI DNA ends by HeLa WCE was comparable with EcoRI DNA ends, end joining activity of HeLa-A3G WCE was at least 2-fold more efficient and produced high molecular weight DNA as well as × 2- × 4+ joint DNA forms. Interestingly, cleavage of end joining products of both cell extracts with ApaI reformed most of the linear plasmid, indicating that end joining was accurate and that terminal cytidines were not deaminated. The apparent discrepancy with terminal deamination observed on longer ssDNA overhangs (Figure 7A-B) is probably the result of the minimal length of ssDNA required for A3G activity.33 We conclude that A3G promotes ssDNA end synapsis and joining of ssDNA overhangs in vitro.
Discussion
Combination chemotherapy and radiotherapy are effective treatments for non-Hodgkin lymphoma, however, more so in the case of localized disease (stage I or II) rather than in advanced disease (stage III or IV).34-36 The response of various cancers to genotoxic agents generally reflects cells ability to repair or tolerate DNA damage. Unrepaired persistent DSBs in human cells pose a prominent threat to genomic integrity and cause cell death or senescence.37,38 Notably, the rate of DSB repair is a critical determinant of radiosensitivity in human hematopoietic cell lines.39 Unlike most other mammalian cells that are radioresistant in a resting state and radiosensitive while activated, lymphocytes are radiosensitive when resting and radioresistant while activated.40,41 Considering that lymphocytes are uniquely destined for activation after differentiation, it is therefore probable that lymphocytes entail mechanisms to avert accumulation of persistent DSBs to ensure proliferation after activation.
An underlying mechanism may involve recruitment of A3G, whose expression is induced in activated lymphocytes, leading to cytidine deamination of resected DSBs ssDNA overhangs. Analogous to generation of DSBs during immunoglobulin class switch recombination by activation induced deaminase,42 we suggest that A3G-induced cytidine deamination may introduce a single-strand break in resected ssDNA. Cytidine deamination is likely to recruit the base-excision repair complex containing UNG2, which preferentially deglycosylates dU residing on ssDNA, forming an abasic site.43,44 Further processing by the base-excision repair factor apurinic/apyrimidinic endonuclease-1 or the MRN complex will create a nick in the phosphodiester backbone at abasic sites,31,32 leading to cleavage of the ssDNA overhang. ATM is activated by interaction with the MRN complex associated with a ssDNA terminus.45 Thus, A3G-mediated cytidine deamination of resected ssDNA may facilitate MRN-ssDNA terminus-dependent activation of ATM, promoting DSB repair. This may explain A3G-mediated activation of ATM in HIV-1–infected cells containing integrated hypermutated viral sequences, and the reduced genomic stability of these cells.20 Alternatively, truncation of resected ssDNA may promote a transition from HR to direct end-joining by A3G or NHEJ factors. Based on its role in ssDNA synapsis, it is tempting to speculate that A3G may facilitate microhomology-mediated end joining, a deletional repair mechanism involving short homologous sequences in both sides of the break after DSB resection.
Survival of cancer cells in the face of genotoxic treatment may accelerate tumor progression by forcing clonogenic selection of radioresistant and chemoresistant cells in advanced tumors.34-36 Our results suggest that A3G has a dual role in promoting survival of lymphoma cells in vivo19 : (1) by enhancing DSB repair after genotoxic treatment, thus preventing cell death; and (2) by promoting a mutator phenotype, driving tumor progression.46 Hence, strategies aimed at inhibiting A3G expression or catalytic activity may improve the outcome of genotoxic lymphoma therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Dina Ben-Yehuda and Nuha Higazi for Ly lymphoma cells, Judd Hultquist for SupT11 cell lines, Joy Lengyel and Ming Li for A3G plasmid DNA constructs and sharing unpublished EMSA data, and Drs Eli Pikarsky, Marc Wold, and Amikam Cohen and Ms Tamar Shiloach for critical reading of the manuscript.
The following reagents were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health: anti-APOBEC3G C-terminal from Dr Jaisri Lingappa, anti-APOBEC3G from Dr Warner C. Greene, HeLa-A3G cells from Dr Klaus Strebel and Consensus HIV-1 B VIF (15-mer) peptides (complete set). This work was carried out in the Peter A. Krueger laboratory with the generous support of Nancy and Lawrence Glick and Pat and Marvin Weiss. This work was supported by the National Institutes of Health (grant P01 GM091743; R.S.H.; subaward to M.K.), Israel Ministry of Health, and the United States–Israel Binational Science Foundation. R.N. is a fellow of the Clore Scholars Program.
National Institutes of Health
Authorship
Contribution: R.N. and M.K. designed the research and analyzed the data; R.N. wrote the paper; R.N., O.I.W., O.C., O.D.S., E.K., and L.B. performed the experiments; and E.B.-R., A.N., R.S.H., M.G., and I.W. contributed reagents and provided assistance.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Moshe Kotler, Department of Pathology and Immunology, Hebrew University–Hadassah Medical School, Jerusalem, 91120, Israel; e-mail: moshek@ekmd.huji.ac.il.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal