In this issue of Blood, Nürnberg et al show that a single nucleotide polymorphism (SNP) associated with human platelet size variation involves a megakaryocyte (Mk)–specific alternate promoter of the DNM3 gene.1
The mechanisms that determine platelet size are just beginning to be understood (for recent review, see Thon and Italiano2 ). However, much remains to be elucidated. Like other blood cell types, there is natural variation in platelet cell size (mean platelet volume [MPV]) between individuals. Previously, Nürnberg's group performed a genome-wide association study (GWAS) and identified 68 SNPs that were statistically associated with variation in human MPV and platelet number at genome-wide significance.3,4
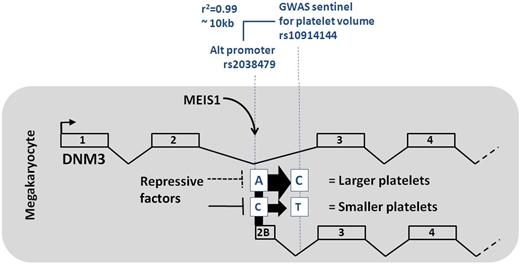
A significant challenge to following up GWAS studies is prioritizing the data to guide detailed functional studies efficiently. This is particularly true for SNPs that lie in noncoding regions. In this study, Nürnberg et al triangulate their SNP data with genome-wide Mk-specific transcription factor chromatin occupancy and Mk-specific gene expression datasets. This is based on the hypothesis that at least some SNPs act by altering key Mk cis-regulatory DNA elements. They focus on MEIS1, a homeobox transcription factor that is essential for megakaryopoiesis.5 As expected, genome-wide chromatin occupancy analysis revealed MEIS1 occupancy at genes enriched in megakaryocyte and platelet function. Importantly, one of the sentinel SNPs (and likely proxy variant SNP) identified in the prior GWAS study resides near a MEIS1 occupancy site in an intronic region of the DNM3 gene. The megakaryocyte transcription factor RUNX1 also binds to this region. Through a variety of procedures Nürnberg and colleagues show that this MEIS1/RUNX1 occupancy site marks an alternate promoter of the DNM3 gene that is selectively utilized in Mks. Transcription from this promoter results in skipping of exons 1 and 2 of the canonical DNM3 gene, and insertion of a novel exon (2b; see figure). They also show that the alternate transcript is markedly up-regulated during in vitro human Mk differentiation. Reporter gene studies as well as analysis of individuals with the different SNP genotypes provide strong evidence that the minor allele correlates with lower expression of the alternate isoform (and a lower MPV).
A prior GWAS study showed that the SNP variant rs10914144 located within the second intron of the DNM3 gene locus is associated with variation in human platelet volume. The study by Nürnberg et al shows that a second SNP variant (rs2038479), which is in high linkage disequilibrium with rs10914144, marks a megakaryocyte-specific alternate promoter. This promoter binds the Mk transcription factors MEIS1/RUNX1 and produces a shortened DNM3 transcript. The minor alleles (“C” for rs2038479 and “T” for rs10914144) are associated with decreased expression of the alternate transcript and smaller platelet size. Repressive factors are postulated to bind to the rs2038479 minor allele to a greater extent than the major allele. See Figure 6 in the article by Nürnberg et al that begins on page 4859.
A prior GWAS study showed that the SNP variant rs10914144 located within the second intron of the DNM3 gene locus is associated with variation in human platelet volume. The study by Nürnberg et al shows that a second SNP variant (rs2038479), which is in high linkage disequilibrium with rs10914144, marks a megakaryocyte-specific alternate promoter. This promoter binds the Mk transcription factors MEIS1/RUNX1 and produces a shortened DNM3 transcript. The minor alleles (“C” for rs2038479 and “T” for rs10914144) are associated with decreased expression of the alternate transcript and smaller platelet size. Repressive factors are postulated to bind to the rs2038479 minor allele to a greater extent than the major allele. See Figure 6 in the article by Nürnberg et al that begins on page 4859.
DNM3 is a member of a superfamily of genes that encode dynamin GTPases (for review, see Ferguson and De Camilli6 ). These factors play important roles in membrane remodeling via their mechanoenzymatic properties. This includes regulation of endocytic vesicle fission, linkage of cell membrane to actin cytoskeleton, and microtubule bundle associated force generation. Recent studies have provided evidence for a functional role of dynamin 3 (DNM3) in human megakaryopoiesis. Overexpression of DNM3 in cord blood CD34+ cells markedly enhanced megakaryocyte colony forming unit (Mk-Cfu) formation and increased expression of the terminal Mk transcription factor NF-E2 p45 in vitro, whereas shRNA-mediated knockdown had the opposite effect.7,8 DNM3 co-localizes to the demarcation membrane system (DMS), which serves as a large membrane reservoir during proplatelet and platelets formaton.8 DNM3 also binds to nonmuscle myosin IIA (MYH9),8 which is mutated in a series of human macrothrombocytopenia disorders. In neurons, dynamins co-localize with the Wiskott-Aldrich syndrome protein (WASP) family member N-WASP. Deficiency of WASP within the hematopoietic system leads to abnormally small platelets. In the current study, Nürnberg et al show that the pan-dynamin inhibitor DynaSore impairs Mk proplatelet production in vitro. Thus, it seems quite plausible for dynamins to be involved in platelet size determination.
The findings in this study should spur additional investigation into the role of dynamins in platelet production and membrane remodeling. Elucidation of their molecular mechanisms is likely to identify new components and regulatory pathways involved in thrombopoiesis. Another important question generated by this study is how the shorter Mk-specific DNM3 isoform influences activity of the protein in thrombopoiesis. Interestingly, the GTPase domain resides in the amino terminal portion of the molecule and would be affected by use of the alternate transcriptional start site.
It is becoming increasingly clear that much of phenotypic variation between individuals results from SNPs in or near cis-acting gene regulatory elements. The current study provides another excellent example of this. In some cases, variants in cis-regulatory elements affect transcription factor binding. Indeed, Nürnberg et al observe significant differences in protein binding between the minor and major alleles based on electromobility shift assays using synthetic oligonucleotides. Identification of the differentially associated factors should provide additional insights into DNM3 transcriptional regulation. The recent large release of data from the ENCODE project,9 which aims to annotate genome-wide cis-regulatory elements, should greatly facilitate future studies such as Nürnberg et al. Combining GWAS data with this new trove of information seems likely to provide novel mechanistic insights for some time to come.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal