Abstract
Myelodysplastic syndromes (MDS) are clonal disorders of hematopoietic stem cells characterized by ineffective hematopoiesis. The DNA-hypomethylating agents 5-azacytidine and 5-aza-2′-deoxycytidine are effective treatments for patients with MDS, increasing the time to progression to acute myelogenous leukemia and improving overall response rates. Although genome-wide increases in DNA methylation have been documented in BM cells from MDS patients, the methylation signatures of specific gene promoters have not been correlated with the clinical response to these therapies. Recently, attention has been drawn to the potential etiologic role of decreased expression of specific ribosomal proteins in MDS and in other BM failure states. Therefore, we investigated whether rRNA expression is dysregulated in MDS. We found significantly decreased rRNA expression and increased rDNA promoter methylation in CD34+ hematopoietic progenitor cells from the majority of MDS patients compared with normal controls. Treatment of myeloid cell lines with 5-aza-2′-deoxycytidine resulted in a significant decrease in the methylation of the rDNA promoter and an increase in rRNA levels. These observations suggest that an increase in rDNA promoter methylation can result in decreased rRNA synthesis that may contribute to defective hematopoiesis and BM failure in some patients with MDS.
Introduction
The DNA-hypomethylating drugs 5-azacytidine (5AC) and 5-aza-2′-deoxycytidine (DAC) are in clinical use for the treatment of myelodysplastic syndrome (MDS)1,2 and have been shown to improve overall response rates and increase the time to progression to acute myelogenous leukemia compared with best supportive care.3,4 Several studies have documented an increase in genome-wide promoter methylation in mononuclear cells and in CD34+ hematopoietic progenitor cells (HPCs) derived from the BM of MDS patients.5,6 However, drug-induced differences in genome-wide promoter methylation signatures and in gene-expression profiles have not been found to be correlated with the clinical response to hypomethylating agents.7 In addition, a correlation between changes in promoter methylation profiles and acquired resistance to 5AC or DAC has never been demonstrated8,9 Therefore, the role of DNA hypermethylation in the pathogenesis of MDS remains to be determined, as does the question of whether the therapeutic responses to 5AC and DAC result from the induction of gene-specific DNA hypomethylation as opposed to other sequelae such as the induction of a DNA-damage response, the activation of an immune response, or the induction of senescence.7
The nucleolus contains clusters of genes that encode the 45S pre-rRNA precursor of rRNA that is subsequently processed to generate the 18S, 5.8S, and 28S rRNA components.10,11 In human cells, there are approximately 400 copies of rDNA genes arranged as tandem repeats within nucleolar organizer regions located on chromosomes 13-15, 21, and 22, although not all of the genes are actively transcribed. Transcription of the rRNA genes depends on the chromatin structure of the promoter region.12-14 Promoters of active rRNA genes are devoid of CpG methylation and are associated with acetylated histones, whereas the reverse is found for silenced genes.15 DNA methyltransferase 1 (DNMT1) controls rDNA gene transcription16 and also regulates the nucleolar architecture by maintaining the heterochromatin state within intergenic repetitive sequences.17,18
Several recent observations have suggested that ribosomal protein synthesis is an essential component of normal hematopoietic stem cell (HSC) function. Haploinsufficiency of the ribosomal gene RPS14 in 5q− syndrome,19,20 the presence of RPS19 mutations in Diamond-Blackfan anemia,21 and the aberrant expression of multiple ribosomal proteins in MDS22-24 all suggest that altered ribosome biogenesis could play a major role in the pathogenesis of BM failure syndromes. To determine whether rRNA synthesis might be altered in HPCs from a broader spectrum of MDS patients, in the present study, we compared the level of rRNA expression and the extent of rDNA promoter methylation in CD34+ cells derived from BM of MDS patients with those in normal CD34+ cells. Our data demonstrate that rRNA expression is decreased in HPCs from MDS patients, whereas methylation within the rDNA upstream regulatory region is increased. We also show that rRNA expression and rDNA methylation are inversely correlated in combined data obtained from healthy controls and MDS patients. In addition, treatment of human myeloid leukemia cell lines with DAC resulted in both a decrease in promoter methylation and an increase in rRNA expression. We suggest that disruption of ribosomal biogenesis resulting from alterations in rRNA synthesis may also underlie HPC dysfunction in some MDS patients.
Methods
MDS patient samples
CD34+ mononuclear cells were enriched from freshly obtained BM from 22 MDS patients and 11 healthy donors by magnetic bead separation (Miltenyi Biotec). CD34+ purity was greater than 90% as assessed by flow cytometry. Cells were immediately snap-frozen and stored at −80°C until use. BM samples were obtained from patients after informed consent per the Declaration of Helsinki and according to an institutional review board–approved protocol.
Cell-culture experiments
The THP1, Mono Mac6, and ML-2 human myelomonocytic leukemia cell lines were incubated at 37°C and 5% CO2 in RPMI 1640 medium supplemented with 10% FBS (HyClone), 100 U/mL of penicillin, and 100 μg/mL of streptomycin (Invitrogen). Cells were treated with 0.1μM DAC (Sigma-Aldrich) for a maximum of 3 days and drug was added daily. Cells were harvested after 3 days of treatment and DNA and RNA were extracted according to the manufacturer's instructions (QIAGEN).
Quantitative RT-PCR
Total RNA was extracted from cell lines and BM-derived CD34+ cells using the RNeasy micro kit (QIAGEN). Total RNA was treated with DNAse (Ambion/Life Technologies) according to the manufacturer's instructions and reverse transcription was performed with random hexamer primers using the SUPERSCRIPT First-Strand Synthesis kit (Invitrogen). PCR primers were located within the 5′ externally transcribed sequence of the rDNA gene and pre-rRNA expression was measured by quantitative RT-PCR carried out in triplicate using SYBRGreen Master Mix (Applied Biosystems). cDNA was amplified using primers within the rDNA promoter and the absence of PCR product confirmed the lack of rDNA amplification. The RT-PCR primers used were: pre-rRNA: forward primer 5′-gaacggtggtgtgtcgttc-3′, reverse primer 5′-gcgtctcgtctcgtctcact-3′; GAPDH: forward primer 5′-ccccttcattgacctcaactacat-3′, reverse primer 5′-cgctcctggaagatggtga-3′.
Quantitative DNA methylation using pyrosequencing
Genomic DNA from CD34+ cells from 14 MDS samples and 11 normal samples (100-200 ng) and from myeloid cell lines (1 mg) was treated with sodium bisulfite using the EZ DNA methylation kit (Zymo Research). For 13 of 14 MDS samples and 8 of 11 control samples used for promoter methylation analysis, pre-rRNA expression was also studied by quantitative RT-PCR. Quantitative DNA methylation analysis was performed by pyrosequencing as described previously.25 Methylation of specific CpGs by pyrosequencing was evaluated using the published human rDNA repeat sequence (accession number U13369). A 247-bp rDNA promoter region was amplified using rDNA specific primers: forward: 5′-GTGTTTTTGGGTTGATTAGAGG-3′ and reverse: 5′-biotin CATCCCAAAACCCAACCTCTCC-3′. Three different primers (A1: 5′-GGTTGATTAGAGGGATT-3′; A2: 5′-TTTTGGGGATAGGTG-3′, and A3: 5′-TTYGGGGGAGGTATATTTT-3′) were annealed to the purified single-stranded 247-bp PCR product and pyrosequencing was performed using PyroMark Q24 (QIAGEN). Determining the ratio of cytosine to thymidine incorporation during pyrosequencing allows quantitation of the degree of methylation of target CpG sequences. Data were analyzed using PyroMark Q24 software. Of the 29 CpGs present within the 247-bp amplified PCR product, 6 were not included in the analysis because the ratio of observed to expected methylation (compared with in vitro methylated standard DNA) was not linear.
Sodium bisulfite sequencing
Sodium bisulfite–treated DNA from CD34+ normal and MDS samples were amplified using specific rDNA primers, as outlined in the previous paragraph. The 247-bp PCR products were purified from a 1.5% agarose gel using a gel-extraction kit (QIAGEN) and subcloned using the TOPO TA-Cloning kit (Invitrogen). Five clones were sequenced from each sample and methylation at 26 CpGs per clone was analyzed using SeqMan Pro software (DNASTAR).
Statistical analysis
The Mann-Whitney test with 2-sided α-level of 0.05 was used to compare the level of rRNA expression and the average percentage of promoter methylation between control and MDS samples. To examine the relationship between rRNA expression and promoter methylation at each individual CpG, Pearson correlation estimates were calculated with confidence intervals approximated using the Fisher Z transform. To examine the relationship between rRNA expression and the average percentage of promoter methylation, the Pearson correlation estimate was calculated and a linear model was fit, controlling for disease status. In multiple hypothesis testing, significance was determined by using the Bonferroni correction to control family-wise error rate at 0.05.
Results
Expression of pre-rRNA in MDS samples
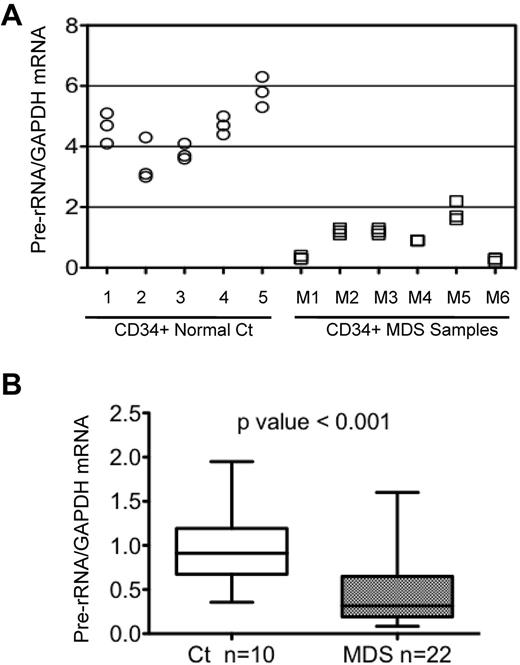
The level of expression of 48S precursor rRNA in CD34+ HPCs was determined by measuring the abundance of the transcript processed at externally transcribed regions, as described in “Methods.” Results from a representative experiment (Figure 1A) showed that pre-rRNA expression was significantly reduced (P < .01) in CD34+ HPCs from 6 MDS patients compared with that in cells from 5 healthy controls. Overall, pre-rRNA expression was studied in CD34+ HPCs from 22 MDS samples and 11 controls. The cumulative data from repetitive quantitative RT-PCR determinations revealed that pre-rRNA expression was significantly decreased (P < .001) in the 22 MDS samples compared with control samples (Figure 1B). These data demonstrate that rRNA synthesis is reduced in BM progenitor cells from the majority of MDS patients studied.
Pre-rRNA expression in CD34+ cells from healthy and MDS BM. (A) Levels of pre-rRNA expression were determined in CD34+ cells from 5 healthy controls (circles) and 6 MDS patients (squares) in a single representative experiment. GAPDH was used as the internal control and samples were run in triplicate. (B) Cumulative data showing levels of pre-rRNA in CD34+ cells from 10 healthy control and 22 MDS samples. GAPDH was used as the internal control. The ends of the whiskers represent minimum and maximum values and the bar indicates the median value (50th percentile). Significance was determined using the Mann-Whitney test.
Pre-rRNA expression in CD34+ cells from healthy and MDS BM. (A) Levels of pre-rRNA expression were determined in CD34+ cells from 5 healthy controls (circles) and 6 MDS patients (squares) in a single representative experiment. GAPDH was used as the internal control and samples were run in triplicate. (B) Cumulative data showing levels of pre-rRNA in CD34+ cells from 10 healthy control and 22 MDS samples. GAPDH was used as the internal control. The ends of the whiskers represent minimum and maximum values and the bar indicates the median value (50th percentile). Significance was determined using the Mann-Whitney test.
Methylation of the rDNA promoter in MDS CD34+ cells
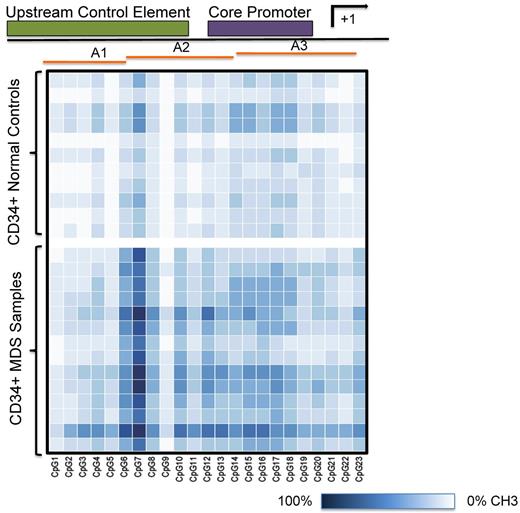
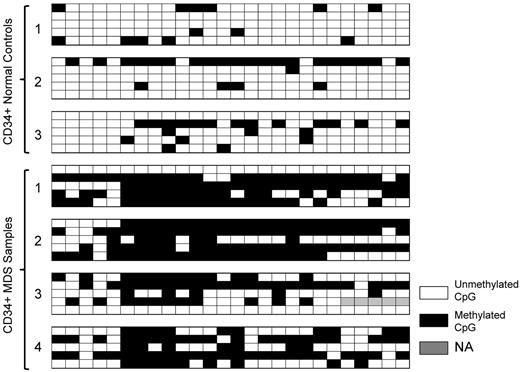
The rRNA gene promoter is highly enriched in CpG dinucleotides.26-28 We postulated that reduced rRNA levels in MDS could result from promoter hypermethylation. PCR amplification of sodium bisulfite–treated DNA and subsequent pyrosequencing of the PCR products was used to quantitate methylation at 23 of 29 CpG sites within the rDNA promoter. Six of the 29 CpGs were not included in the analysis because the ratios of observed to expected methylation found by in vitro methylated standard DNA was not linear. A 247-bp DNA segment was amplified using rRNA promoter–specific primers that overlapped the upstream core element and the core promoter, as illustrated in Figure 2. This amplicon served as a template for analyzing the degree of promoter methylation at 23 CpGs spanning the rDNA promoter region (−195 to +52) using the primers A1 to A3. The heat map shown is a schematic representation of the percentage DNA methylation at each of these 23 CpGs. The extent of methylation varied from 100% methylation (dark blue) to no methylation (white) in CD34+ HPCs from healthy controls (upper panel) and 14 MDS samples (lower panel). Densely methylated CpGs were present across the 247-bp rDNA promoter in MDS samples. The percentage of methylation at each CpG was significantly greater than that in control samples at 22 of 23 CpGs (P < .05; supplemental Figure 1A-D, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Sodium bisulfite sequencing was used to determine whether rDNA promoter methylation varied within individual PCR clones obtained from 3 healthy controls and 4 MDS CD34+ samples. As shown in Figure 3, sequencing of clones from control samples showed that the majority of clones were either unmethylated or methylated at low frequency. In contrast, the frequency of methylated CpGs per clone in MDS samples was high in the majority of clones analyzed. None of the clones obtained from patient samples was methylated at every CpG, and 2 clones were not methylated. These data further confirm the increased rDNA promoter methylation observed in patient samples obtained using pyrosequencing and also suggest that the number of transcriptionally active rDNA genes may be significantly fewer in MDS CD34+ cells.
Methylation of the rDNA promoter in MDS CD34+ cells using pyrosequencing. Heat map representation of the extent of rDNA gene promoter methylation at individual CpGs across the upstream core element and the core promoter region. A 247-bp DNA segment was amplified and the extent of methylation at 23 of 29 CpG sites (−195 to +52 bp) was determined by pyrosequencing using primers A1 to A3. Each square represents a single CpG and each row represents a sample. The extent of methylation is represented over the range of 0% (white) to 100% (dark blue). The heat map was constructed using R software.
Methylation of the rDNA promoter in MDS CD34+ cells using pyrosequencing. Heat map representation of the extent of rDNA gene promoter methylation at individual CpGs across the upstream core element and the core promoter region. A 247-bp DNA segment was amplified and the extent of methylation at 23 of 29 CpG sites (−195 to +52 bp) was determined by pyrosequencing using primers A1 to A3. Each square represents a single CpG and each row represents a sample. The extent of methylation is represented over the range of 0% (white) to 100% (dark blue). The heat map was constructed using R software.
rRNA promoter methylation by sodium bisulfite sequencing. DNA samples derived from CD34+ healthy subjects and MDS patients were treated with sodium bisulfite, PCR amplified, cloned, and sequenced. Each row represents an individual clone. The open squares represent unmethylated CpGs, closed squares represent methylated CpGs, and gray squares indicate that data were not obtained. One of the CpGs (CpG 6 in Figure 2) reported in the published rDNA sequence (accession number U13369) was found to be missing when individual clones were sequenced.
rRNA promoter methylation by sodium bisulfite sequencing. DNA samples derived from CD34+ healthy subjects and MDS patients were treated with sodium bisulfite, PCR amplified, cloned, and sequenced. Each row represents an individual clone. The open squares represent unmethylated CpGs, closed squares represent methylated CpGs, and gray squares indicate that data were not obtained. One of the CpGs (CpG 6 in Figure 2) reported in the published rDNA sequence (accession number U13369) was found to be missing when individual clones were sequenced.
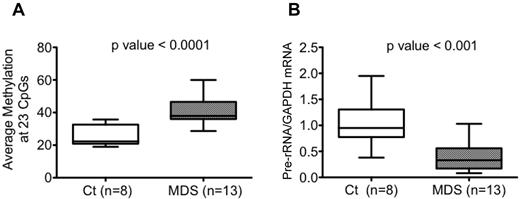
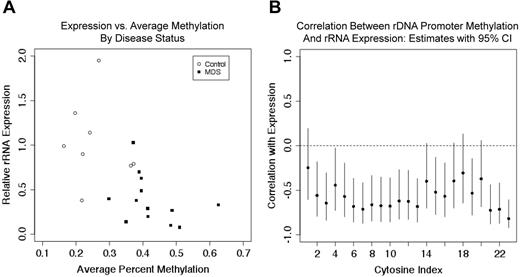
rDNA promoter methylation quantitated by pyrosequencing was then correlated with rRNA expression levels in 8 controls and 13 MDS samples (8 of 11 control and 13 of 22 MDS samples shown in Figure 1B). The average percentage of methylation across 23 CpGs in CD34+ HPC was significantly increased (P < .0001) in MDS samples, as shown in Figure 4A, and the pre-rRNA expression levels were significantly decreased (Figure 4B) in the same patient samples. When MDS and control samples were combined, the average percentage of methylation and expression was inversely correlated with a P < .01. The Pearson correlation coefficient estimate was −0.6, with a 95% confidence interval of −0.2 to −0.8. The relationship between the 3 variables (ie, average percent methylation, rRNA expression, and disease status) is shown in Figure 5A. The estimates of correlation, with 95% confidence intervals, between expression and methylation at each individual CpG (among the 13 MDS samples and 8 control samples) are shown in Figure 5B. All point estimates were negative, suggesting inverse correlation across the 23 CpGs, although we cannot declare significance at individual CpGs when correcting for multiple hypothesis testing. These data are concordant with studies that have shown an increase in overall methylation in MDS samples, but are the first to address specific methylation changes in the rDNA promoter region.
Hypermethylation of the rDNA promoter and decreased rRNA expression in MDS CD34+ cells. (A-B) The average percentage of DNA methylation across the 23 CpGs of the rDNA promoter was higher (P < .0001; A) and the pre-rRNA expression level was lower (P < .01; B) in the 13 MDS samples compared with the 8 control samples. The ends of the whiskers represent minimum and maximum values and the bar indicates the median value (50th percentile). Significance was determined using the Mann-Whitney test.
Hypermethylation of the rDNA promoter and decreased rRNA expression in MDS CD34+ cells. (A-B) The average percentage of DNA methylation across the 23 CpGs of the rDNA promoter was higher (P < .0001; A) and the pre-rRNA expression level was lower (P < .01; B) in the 13 MDS samples compared with the 8 control samples. The ends of the whiskers represent minimum and maximum values and the bar indicates the median value (50th percentile). Significance was determined using the Mann-Whitney test.
rRNA expression is correlated with the extent of rDNA promoter methylation. (A) As MDS samples had a higher average percentage of DNA methylation and lower rRNA expression, these variables were inversely correlated in all data (P < .01). (B) Pearson correlation estimates between pre-rRNA expression and rDNA promoter methylation at 23 CpGs in 8 control samples and 13 MDS samples are shown. Vertical lines represent 95% confidence intervals.
rRNA expression is correlated with the extent of rDNA promoter methylation. (A) As MDS samples had a higher average percentage of DNA methylation and lower rRNA expression, these variables were inversely correlated in all data (P < .01). (B) Pearson correlation estimates between pre-rRNA expression and rDNA promoter methylation at 23 CpGs in 8 control samples and 13 MDS samples are shown. Vertical lines represent 95% confidence intervals.
Effect of DAC treatment on pre-rRNA expression and promoter methylation
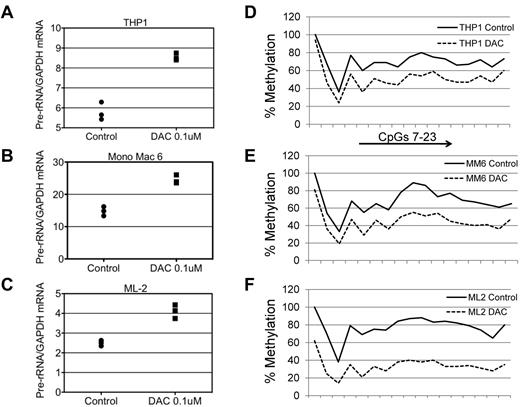
To address the question of whether DAC treatment would decrease rDNA promoter methylation and concordantly increase rRNA synthesis, we treated leukemic cell lines with the drug. Treatment with 0.1μΜ DAC for 5 days resulted in a gradual increase in pre-rRNA expression over days 2-5 compared with control cells treated with DMSO over the same interval. Increasing the DAC concentration from 0.1 to 0.5μM did not further increase rRNA expression (data not shown), suggesting that 0.1μM DAC is the optimal dose. We then treated 3 different myelomonocytic leukemia cell lines, THP1, Mono Mac 6, and ML2, with 0.1μM DAC for 3 days and observed an increase in pre-rRNA expression and a decrease in promoter methylation as measured by pyrosequencing at CpGs 7-23 in each cell line (Figure 6A-F). Because of limited number of cells obtained from the BM aspirates, we were unable to perform rRNA expression and promoter methylation analysis in CD34+–enriched populations from MDS patients.
Effect of DAC on rRNA expression and rDNA promoter methylation in myeloid leukemia cell lines. (A-C) THP1, Mono Mac 6, and ML2 cells were treated with DMSO or 0.1μM DAC for 3 days. The level of expression of pre-rRNA is shown relative to that in DMSO-treated cells. GAPDH was used as an internal control and the samples were run in triplicate. (D-F) The cells were treated with DMSO or 0.1μM DAC for 3 days and the extent of CpG methylation was determined by pyrosequencing at CpGs 7-23 spanning the rDNA upstream core element and the core promoter.
Effect of DAC on rRNA expression and rDNA promoter methylation in myeloid leukemia cell lines. (A-C) THP1, Mono Mac 6, and ML2 cells were treated with DMSO or 0.1μM DAC for 3 days. The level of expression of pre-rRNA is shown relative to that in DMSO-treated cells. GAPDH was used as an internal control and the samples were run in triplicate. (D-F) The cells were treated with DMSO or 0.1μM DAC for 3 days and the extent of CpG methylation was determined by pyrosequencing at CpGs 7-23 spanning the rDNA upstream core element and the core promoter.
Discussion
The requirement for ribosomal biogenesis reflects the overall demand for cellular protein synthesis. Formation of the mature 80S ribosome requires coordinated expression of 4 rRNAs and approximately 80 ribosomal proteins, and perturbation at any stage of ribosomal biogenesis or assembly would be expected to impair cellular proliferation.29 Our data showed decreased rRNA levels that were inversely correlated with rDNA promoter methylation in control and MDS CD34+ HPCs. The high-throughput array-based published reports for gene expression (Affymetrix gene-chip arrays)7,30,31 and promoter methylation (Nimble Gen oligonucleotide arrays or Illumina bead arrays)5,6 in MDS used arrays that lack probes for rRNA or rDNA, respectively. Therefore, this is the first report to specifically examine rRNA gene expression and rDNA promoter methylation in MDS. Our results suggest that the increase in methylation of this CpG-rich promoter is at least one of the mechanisms resulting in decreased rRNA gene expression, potentially altering the synthesis of ribosomal proteins32 and hampering ribosomal biogenesis.
Impairment of ribosome biogenesis characterizes several BMF syndromes.33 Multiple mouse models have demonstrated that mutation of genes, including those encoding Rps19, Rps14, and Rps6, result in defective hematopoiesis. Haploinsufficiency of Rps6 or of the Cd74-Nid67 region that contains 6 genes, including Rps14, results in macrocytic anemia, erythroid hypoplasia, megakaryocytic dysplasia, and a reduced number of HSCs/HPCs, all of which are characteristic features of 5q−syndrome.20,24 Similarly, conditional expression of mutant Rps19 or its haploinsufficiency in mice results in anemia, exhaustion of HSCs and BM failure.34,35 BM from these mice has a high level of p53 protein expression and increased apoptosis; breeding them with p53-deficient animals rescues the phenotype. Several studies have demonstrated that the release of specific ribosomal proteins, including RPL5 and RPL11, from the nucleolus increases p53 levels through an interaction with the MDM2 ubiquitin ligase, resulting in increased levels of p53.36,37 Decreased expression of RPS19 or RPS14 causes selective activation of the p53 pathway in erythroid progenitors, further establishing the sensitivity of hematopoietic precursors to disruptions in ribosome biogenesis.38 Therefore, a common mechanism underlying BM failure states appears to be the up-regulation of p53 through alterations in ribosome assembly.
Increased p53 stability also occurs when rRNA synthesis is decreased, either through down-regulation of the catalytic subunit of RNA polymerase I39 or through knock-out of TIF-1A, a protein required for rRNA transcription.40 The increase in p53 expression results in enhanced apoptosis and in disruption of nucleolar architecture. The chromatin status of rRNA genes maintains the appropriate ratio of expression among genes that are actively transcribed and those that are not, thereby regulating cellular proliferation.12 The nucleolar chromatin remodeling complex, an ATP-dependent chromatin remodeling complex that regulates rRNA gene silencing, interacts with both DNMT1 and with the histone deacetylase HDAC1.26,41 Our data raise the possibility that alterations in rRNA expression may underlie the up-regulation of p53 and apoptosis of BM progenitor cells in MDS patients. Further characterization of the epigenetic events regulating nucleolar rRNA gene expression, including both the extent of DNA methylation and the nature of histone modifications, may help to further our understanding of BM failure syndromes.
Reversal of DNA methylation by DNMT inhibitors has constituted a major rationale for treating MDS patients with these drugs. Although clinically significant responses occur in MDS patients treated with both 5AC and DAC,3,4,42,43 and although both drugs cause global DNA hypomethylation,31,44-46 specific methylation changes have not been identified that predict the clinical response.2,7-9 In addition, most patients ultimately develop resistance to 5AC or DAC.8,9 Our in vitro data with cell lines from the present study suggest that the response to hypomethylating agents could involve an increase in rRNA transcription, resulting in decreased apoptosis. This supposition is supported by recent data demonstrating that up-regulation of rRNA expression increases ribosome biogenesis and ubiquitination of p53, thereby reducing p53 levels.39 It will be of interest to obtain clinical data on the response of rRNA expression in CD34+ cells of patients receiving hypomethylating agents in relation to the durability of their response to this intervention.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a Leukemia & Lymphoma Society Specialized Centers of Research (SCOR) and translational grant to B.S.M.
Authorship
Contribution: A.R., P.L.G., and B.S.M. designed the experiments; A.R., K.J.S., and S.P. performed the experiments; and A.R., B.B.T., and B.S.M. analyzed the data and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beverly S. Mitchell, MD, George E. Becker Professor of Medicine, Stanford Cancer Institute, LLSC Research Bldg (SIM1), 265 Campus Dr, Rm G2167, Stanford, CA 94305-5458; e-mail: beverlym@stanford.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal