Abstract
We recently identified 68 genomic loci where common sequence variants are associated with platelet count and volume. Platelets are formed in the bone marrow by megakaryocytes, which are derived from hematopoietic stem cells by a process mainly controlled by transcription factors. The homeobox transcription factor MEIS1 is uniquely transcribed in megakaryocytes and not in the other lineage-committed blood cells. By ChIP-seq, we show that 5 of the 68 loci pinpoint a MEIS1 binding event within a group of 252 MK-overexpressed genes. In one such locus in DNM3, regulating platelet volume, the MEIS1 binding site falls within a region acting as an alternative promoter that is solely used in megakaryocytes, where allelic variation dictates different levels of a shorter transcript. The importance of dynamin activity to the latter stages of thrombopoiesis was confirmed by the observation that the inhibitor Dynasore reduced murine proplatelet for-mation in vitro.
Introduction
A genome-wide association meta-analysis study (GWAS) in nearly 67 000 persons identified 68 independent loci associated with the mean platelet volume (MPV) and the count of platelets (PLT) at genome-wide (GW) significance, representing an ideal resource to make discoveries in megakaryocyte (MK) and PLT biology.1 Although such GWASs unequivocally identified loci that are implicated in the formation and survival of PLTs, to confirm which genetic variants are responsible for the observed associations and which genes mediate their effects remains a challenge. Strategies to prioritize functional follow-up studies are therefore highly desired. Fifteen of the 68 loci are likely to act through nonsynonymous variants. The remaining 53 loci have potential roles in altering transcriptional regulation. Indeed, at one of the MPV loci at chromosome 7q22.3, we recently showed that the minor allele of the noncoding variant rs342293 disrupts a binding site of the transcription factor EVI1/MECOM, leading to a higher transcript level of the downstream gene PIK3CG.2
Here we present a strategy to prioritize the functional follow-up of GWAS hits whereby sentinel single nucleotide polymorphisms (SNPs; those with the lowest P value at an associated locus) and those in strong linkage disequilibrium (r2 > 0.8 in Europeans; proxy SNPs hereafter) are overlapped with the GW DNA binding patterns of a lineage restricted transcription factor as determined experimentally by ChIP combined with deep sequencing (ChIP-seq). This strategy has the advantage of providing hypotheses about the underlying genetic mechanisms behind the observed associations that can be readily followed. We selected the transcription factor MEIS1 for several reasons. First, this 3 amino acid loop extension homeodomain protein was identified as a common viral integration site in myeloid leukemias of BXH-2 mice3 and has since been established as a strong regulator of hematopoiesis and myeloid leukemogenesis.4-7 Second, MEIS1 is exclusively transcribed in hematopoietic stem/progenitor cells and in the MK lineage; therefore, MEIS1 binding events are highly likely to be critical for the identity of this lineage.8,9 Third, gene knockout in mice results in early embryonic lethality with a complete lack of megakaryopoiesis,10,11 and knockdown studies of meis1 in zebrafish showed major effects on both hematopoiesis and vasculogenesis, confirming the abrogation of the formation of thrombocytes, the fish equivalent of MKs and PLTs.12,13 Finally, little is known about how MEIS1 regulates megakaryopoiesis, resulting in these striking phenotypes.
In this study, we report on the successful identification of a set of MEIS1-regulated genes by ChIP-seq in megakaryocytic cells. The set is significantly enriched for genes implicated in megakaryopoiesis and PLT function and contains, as postulated, more PLT GWAS loci than to be expected by chance. Detailed functional studies of a dynamin isoform DNM3, one of the identified GWAS loci that colocalizes with a binding event, revealed that MEIS1 binds a novel promoter that is uniquely used in the megakaryocytic lineage and where the promoter activity displays differential allelic effects at the putative functional variant. We show that the formation of pro-PLTs from murine in vitro–derived MK was inhibited by the dynamin inhibitor Dynasore, confirming the importance of dynamin activity to the late stages of megakaryopoiesis.
Methods
Cell culture
CHRF 288-11 cells14 (hereafter CHRF cells) were cultured in RPMI 1640 (Sigma-Aldrich), 10% FBS (Invitrogen), and 200μM l-glutamine, 100 U penicillin, and 100 μg/mL streptomycin (Sigma-Aldrich). The 1603MED cells15 were cultured in DMEM, high glucose, with sodium pyruvate, without L-glutamine (PAA Laboratories), and supplemented with 10% FBS, 1% nonessential amino acids (HyClone, Thermo Scientific), and 200μM l-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin.
MK culture
MKs were obtained by culture of CD34+ hematopoietic progenitor cells as previously reported.16 In short, cord blood was collected with informed consent, and CD34+ cells were prepared by magnetic bead selection. Purified cells were cultured (1 × 105 cells/mL) for up to 12 days in serum-free medium supplemented with human thrombopoietin and IL-1β to differentiate into MKs. Cells were also harvested at days 3, 5, 7, 9, 10, and 12 and characterized by immunophenotyping with monoclonal antibodies against relevant CD markers and by analysis of RNA on Illumina whole genome expression arrays.1 By day 10, > 90% of cells stained positive for CD41 and negative for CD34.
ChIP and massive parallel sequencing
Twenty million CHRF cells per condition were crosslinked for 10 minutes with 1% formaldehyde. Cells were lysed for 10 minutes on ice (10mM Tris, pH 8.0, 10mM NaCl, 0.2% NP-40). Nuclei were then lysed for 10 minutes on ice (50mM Tris, pH 8.1, 10mM EDTA, 1% SDS). Crosslinked chromatin was sheared for 2 × 5 minutes by sonication using a Bioruptor (Diagenode) and precleared with 100 μg anti–rabbit IgG preimmune serum (Sigma-Aldrich) using protein G-Sepharose beads (Roche Diagnostics). ChIP was performed overnight in IP dilution buffer (20mM Tris, pH 8.1, 2mM EDTA, 150mM NaCl, 1% Triton X-100, 0.01% SDS), using 14 μg anti-MEIS1 (Abcam, ab19867, lot no. 153899) or anti–rabbit IgG (Sigma-Aldrich I5006, lot no. 115K7551), respectively. Beads were washed twice with buffer 1 (20mM Tris, pH 8.1, 2mM EDTA, 50mM NaCl, 1% Triton X100, 0.1% SDS), once with buffer 2 (10mM Tris, pH 8.1, 1mM EDTA, 250mM LiCl, 1% NP40, 1% sodium deoxycholate monohydrate), and twice with TE (10mM Tris, 1mM EDTA, pH 8.0) buffer. Bound chromatin was eluted from the beads twice with 100mM NaHCO3 containing 1% SDS. After reverse crosslinking and RNaseA and proteinase K digestion, chromatin was cleaned up using the QIAGEN PCR purification kit (QIAGEN). ChIP-seq library generation, cluster formation, and sequencing were performed at the Michael Smith Genome Sciences Center (Vancouver, BC) on an Illumina GAII analyzer. The resulting 9 513 449 single-end reads were mapped to the human genome (hg19) using stampy 1.0.11_(r880).17 Of the reads, 7 323 093 were of high mapping quality (> 30). Peaks were called using MACS18 and PICS.19 Peak overlap was generated by requiring a minimum overlap of 150 nucleotides. Results were deposited at ArrayExpress (www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-859.
Overlap with GWAS loci and significance analysis
For each of the 68 associated loci, candidate functional SNPs were selected by identifying all single nucleotide variants with an r2 > 0.8 and within 100 kb of the sentinel SNP in the European samples of the 1000 Genomes project (June 2011 release). To establish whether the association of a locus could potentially be of regulatory origin, we determined whether at least one candidate functional SNP overlapped with a ChIP-seq peak. Because the chance of identifying an overlap between a sentinel SNP and a MEIS1 binding peak is sensitive to the number of peaks considered, the overlap analysis was carried by successively increasing the number of peaks by including peaks with decreasing peak height. We estimated the significance of observing a given number of overlaps by resampling. A total of 50 000 sets of 66 loci (2 GWAS SNPs in the HLA locus were removed because of its complicated haplotype structure) were drawn from the same SNPs onto which the GWAS data were imputed (∼ 2.5 million SNPs from HapMap2)20 while preserving the distribution of allele frequencies. All analyses were carried out in the R/Bioconductor environment.
Functional analysis of MEIS1 binding sites
To identify annotated protein coding genes enriched in the proximity of MEIS1 binding sites, the set of 13 842 peaks jointly called by 2 algorithms was analyzed using the GREAT software (Version 1.8.1).21 Enrichment was tested against the whole human genome (hg19) using standard parameters. Significance was measured by binomial P values and the false discovery rate q values. In addition, a set of 1285 protein coding genes with MEIS1 binding sites located within an annotated gene body, including 5 kb each upstream and downstream, was tested for over-representation of Gene Ontology terms using FatiGO as part of the Babelomics Version 4.2 environment.22 Enrichment was tested compared with the rest of the human genome. Adjusted P values stem from the Fisher exact test, after correcting for multiple testing, using the false discovery rate procedure of Benjamini and Hochberg.23
RNA sequencing
Total RNA from 10 day cultured MKs was obtained by the Trizol method as described.16 Polyadenylated RNA was purified from this by 2 rounds of magnetic Oligo dT25 selection (Dynabeads Oligo (dT)25, Invitrogen) in the presence of RNase inhibitor (New England Biolabs), combined with DNase treatment (Ambion, Invitrogen). The mRNA was fragmented using RNA fragmentation reagent (Ambion, Invitrogen) and precipitated with added glycogen (Ambion), then cDNA produced by Superscript Double Stranded cDNA synthesis kit (Invitrogen), replacing oligo-dT primer with random primer. Thereafter, library preparation was as per mRNA-sequencing kit (Illumina), starting with end repair. Bands of ∼ 300-450 bp were gel extracted for paired-end sequencing by Illumina GAII analyzer with a read length of 76 bp. The RNA-seq results are deposited at ArrayExpress under accession number E-MTAB-918.
FAIRE
Formaldehyde-assisted isolation of regulatory elements (FAIRE) experiments were carried out as previously described.24 In short, 20 million cells were cross-linked with 1% formaldehyde for 12 minutes and subjected to 12 sonication cycles using the Bioruptor UCD-200. The sample was cleaned-up using the MinElute PCR Purification Kit (QIAGEN). FAIRE DNA was processed following the Illumina paired-end library generation protocol. The genomic libraries were sequenced at 54-bp paired-end reads on an Illumina GAII analyzer. Sequence reads were aligned to the human genome (hg19) using stampy 1.0.11_(r880) with default parameters. The FAIRE-seq data are deposited at ArrayExpress under accession number E-MTAB-858.
MPV
EDTA anticoagulated blood from volunteers of the Cambridge BioResource (http://www.cambridgebioresource.org.uk) was obtained with informed consent under the Cardiosome Project: Genes and Mechanisms in Cardiovascular Disease. Ethical approval for this study, conducted in accordance with the Declaration of Helsinki, was given by the Cambridgeshire 1 Research Ethics Committee. Full blood counts, including the measurement of MPVs, were obtained from blood samples within 2 hours after collection using a Coulter LH500 hematology analyzer (Beckman-Coulter).
Platelet isolation
Citrate anticoagulated samples of blood were obtained from Cambridge BioResource volunteers and centrifuged for 20 minutes at 150g to obtain PLT-rich plasma (PRP). The PRP was centrifuged twice more, each time retaining the supernatant and discarding the leukocyte-rich pellet together with the 0.5-mL PRP layer immediately above it. The PRP was further leukocyte-depleted by mixing with anti-CD45 magnetic beads (Dynabeads CD45, Invitrogen; 33 μL beads/mL of PRP) and rotating at room temperature for 20 minutes. The beads were removed by Dynal MPC-L magnet, using 2 cycles of 2-minute magnetization steps and transferring the PRP to fresh tubes after each step. The leukocyte-depleted PRP was centrifuged for 10 minutes at 1500g. The supernatant was discarded and the PLT pellet resuspended in 2 mL Trizol (Invitrogen) until a particle-free solution was obtained; 1-mL aliquots were frozen on dry ice and stored at −80°C. Starting PRP volumes ranged from 5 to 15 mL, and corresponding yields ranged from 0.5 to 2.5 × 109 PLTs.
RNA preparation and cDNA synthesis
RNA from cultured cells and PLTs was prepared using Trizol essentially according to the manufacturer's instructions, except that 2-mL Phase Lock Gel tubes (5′) were used for the phase separation. RNA pellets were air dried for no more than 7 minutes and then resuspended in 15 μL nuclease-free water (Ambion, Invitrogen). RNA yields averaged 940 ng/109 PLTs with A260/280 ratios between 1.33 and 1.66. The 25-ng PLT RNA was processed by the ABI TaqMan Reverse Transcription Reagents (Applied Biosystems) in a 25-μL volume, according to the manufacturer's protocol. At the end of the reaction, RNA was diluted to 250 μL with nuclease-free water and stored at −20°C. In addition, total RNA from human brain (P/N #54005, 54007, and 540117) was obtained from Stratagene (Agilent Technologies).
Gene expression quantitative real-time PCR
Absolute quantification of DNM3 transcript abundance in RNA samples from human PLTs and other cells was carried out on a ABI Prism 7900HT Sequence Detection System (Applied Biosystems) using the following protocol: 50°C 2 minutes, 95°C 10 minutes, 40 × (95°C 15 seconds, 60°C 1 minute). Product numbers of the used TaqMan gene expression assays (Applied Biosystems) can be found in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). To test for abundance of the novel DNM3 transcript variant, a custom TaqMan assay was designed with the probe spanning the boundary between the novel exon 2B and exon 3. Transcript levels were normalized to GAPDH. Reactions were measured in triplicate and primer efficiencies obtained through cDNA standard dilution series.
RACE
5′-Rapid amplification of cDNA ends (RACE) on RNA samples from CHRF and MK cells and PLTs was performed using the 5′/3′ RACE Kit, 2nd Generation (Roche Diagnostics) following the manufacturer's protocol. For the PCR amplification steps, recombinant Taq polymerase and dNTPs from Fermentas were used (Fermentas). DNA fragments were subcloned into pGem T easy (Promega) and sequenced. Primer and clone sequences can be found in supplemental Table 2.
Dual Luciferase reporter assays
DNA fragments were cloned into the firefly Luciferase vector pGl4.10 (Promega). Ten million cells were transfected with 10 μg firefly Luciferase vector and 500 ng Renilla Luciferase vector pGl4.74 (Promega). Transfection was performed via electroporation using a Bio-Rad GenePulser Xcell (exponential curve, 220 V, 900 μF, resistance ∞, 4-mm cuvettes). Luciferase activity was measured in a LUMIstar Optima luminometer (BMG Labtech) using the Dual-Luciferase Reporter Assay kit (Promega). Random DNA sequences for use as negative controls were generated by the Random DNA Sequence Generator (http://www.faculty.ucr.edu/∼mmaduro/random.htm) and produced by artificial gene synthesis (GeneArt). For sequences of cloning primers and construct inserts, see supplemental Table 3.
Electrophoretic mobility shift assay
See supplemental Methods.
MK suspension cultures
Mouse fetal liver cells were collected from WT CD1 mice (Charles River Laboratories) on day E13.5 and cultured at 37°C and 5% CO2 in the presence of 0.1 μg/mL purified recombinant mouse c-Mpl ligand for 5 days. Fetal liver cell cultures were layered on a single-step gradient (1.5%-3.0% BSA) on culture day 4, and MKs were allowed to sediment for 30 minutes.25 The MK pellet was then resuspended in fresh media and cultured for an additional 24 hours alone or in the presence of either 0.3% DMSO or 100μM DynaSore in DMSO. MK cultures were examined on day 5 by phase-contrast microscopy using a Nikon eclipse TS100 benchtop microscope (Nikon) at 20× magnification; digital images were collected on a Hamamatsu C2400 CCD camera and analyzed using ImageJ. Mature MKs were identified by size (> 10μm diameter) and distinguished from pro-PLT-producing MKs because of the presence or absence of long pro-PLT elongations (circularity ≥ 0.7). Samples were examined in triplicate, and at least 78 cells were counted for each condition tested. All studies complied with institutional guidelines approved by the Children's Hospital animal care and use committee, and the Institutional Animal Care and Use Committee.
Statistical analysis
Results are expressed as mean ± SD with number of experiments. Statistical comparisons between groups were performed by 2-tailed t test using Prism unless stated otherwise. Statistical comparisons of Luciferase data were performed on log transformed signal intensities using mixed linear models to account for the hierarchical nature of the data.
Results
MEIS1 putatively regulated genes are important determinants of the PLT lineage
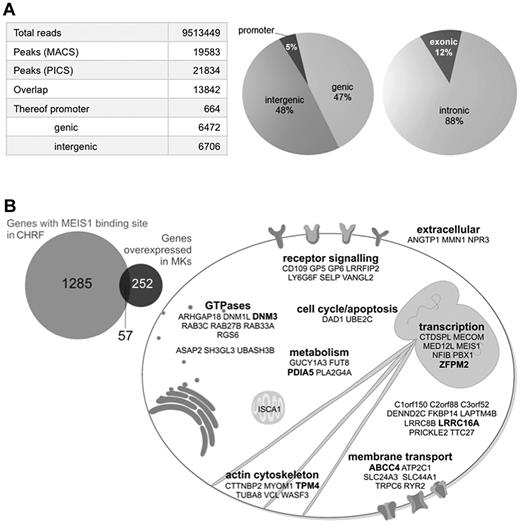
The expression of MEIS1 is restricted to the megakaryocytic lineage, among differentiated blood cells (supplemental Figure 1).8,9 To map MEIS1 binding events, we performed ChIP-seq in the human megakaryocytic cell line CHRF, a close model for MKs based on its transcriptional profile (supplemental Figure 2). MEIS1 peaks were called using magnetic-activated cell sorting18 and PICS19 from which a high-confidence dataset of 13 842 binding events detected by both algorithms was generated (Figure 1A). Genes nearby MEIS1 binding events were analyzed with GREAT21 (Genomic Regions Enrichment of Annotations Tool), revealing that putatively regulated genes are significantly over-represented in megakaryopoiesis and PLT biology categories (Table 1; supplemental Figure 3). To increase stringency in assigning putatively regulated genes, a list of 1285 potentially MEIS1-regulated genes was generated by filtering for peaks with a minimum height of 30 reads (80th percentile of peak height) and within ± 5 kb of protein coding genes (supplemental Methods). Analysis of their expression patterns showed a significant over-representation of 57 genes of a set of 252 that were shown to be strongly overexpressed in MKs relative to the other 7 mature blood cell types (P < 5 × 10−19; Figure 1B) in the HaemAtlas compendium of blood cell transcripts.8 These observations are consistent with our hypothesis that MEIS1, because of its characteristic lineage restriction, regulates many genes critical to the identity of MKs and PLTs. Indeed, these 57 genes contain many well-known regulators of their function, such as the collagen signaling receptor GlycoProtein (GP) VI (GP6), 1 of the 4 subunits of the PLT receptor for von Willebrand Factor, GPV (GP5), and the α granule membrane protein P-selectin (SELP).
MEIS1 ChIP-seq identifies genes regulating MK and PLT biology. (A) MEIS1 ChIP-seq in CHRF 288-11 cells identifies mainly intronic and intergenic binding sites. (B) Cellular functions of 57 genes with MEIS1 binding events and overexpressed in MKs. Bold represents 6 loci containing sentinel SNPs associated with MPV or PLT count.
MEIS1 ChIP-seq identifies genes regulating MK and PLT biology. (A) MEIS1 ChIP-seq in CHRF 288-11 cells identifies mainly intronic and intergenic binding sites. (B) Cellular functions of 57 genes with MEIS1 binding events and overexpressed in MKs. Bold represents 6 loci containing sentinel SNPs associated with MPV or PLT count.
Functional assessment of MEIS1 binding sites by GREAT shows enrichment of megakaryocyte and platelet phenotypes
| Enriched term . | Noncorrected P . | FDR adjusted q . |
|---|---|---|
| Mouse phenotype | ||
| Abnormal platelet physiology | 6.5 × 10−30 | 7.3 × 10−28 |
| Abnormal platelet activation | 4.3 × 10−22 | 3.3 × 10−20 |
| Abnormal platelet aggregation | 1.1 × 10−21 | 7.8 × 10−20 |
| Decreased platelet aggregation | 6.6 × 10−20 | 4.1 × 10−18 |
| Abnormal vascular branching morphogenesis | 1.0 × 10−19 | 6.0 × 10−18 |
| Increased bleeding time | 7.3 × 10−19 | 4.0 × 10−17 |
| Decreased common myeloid progenitor cell number | 3.9 × 10−14 | 1.4 × 10−12 |
| Abnormal bone marrow development | 6.4 × 10−8 | 1.2 × 10−6 |
| MSigDB pathway | ||
| Pertussis toxin-insensitive CCR5 signaling in macrophage | 4.6 × 10−13 | 5.1 × 10−11 |
| TPO signaling pathway | 1.5 × 10−9 | 4.5 × 10−8 |
| MSigDB perturbation | ||
| Genes essential to the development of megakaryocytes, as expressed in normal cells and essential thrombocythemic cells | 1.7 × 10−22 | 2.4 × 10−20 |
| Enriched term . | Noncorrected P . | FDR adjusted q . |
|---|---|---|
| Mouse phenotype | ||
| Abnormal platelet physiology | 6.5 × 10−30 | 7.3 × 10−28 |
| Abnormal platelet activation | 4.3 × 10−22 | 3.3 × 10−20 |
| Abnormal platelet aggregation | 1.1 × 10−21 | 7.8 × 10−20 |
| Decreased platelet aggregation | 6.6 × 10−20 | 4.1 × 10−18 |
| Abnormal vascular branching morphogenesis | 1.0 × 10−19 | 6.0 × 10−18 |
| Increased bleeding time | 7.3 × 10−19 | 4.0 × 10−17 |
| Decreased common myeloid progenitor cell number | 3.9 × 10−14 | 1.4 × 10−12 |
| Abnormal bone marrow development | 6.4 × 10−8 | 1.2 × 10−6 |
| MSigDB pathway | ||
| Pertussis toxin-insensitive CCR5 signaling in macrophage | 4.6 × 10−13 | 5.1 × 10−11 |
| TPO signaling pathway | 1.5 × 10−9 | 4.5 × 10−8 |
| MSigDB perturbation | ||
| Genes essential to the development of megakaryocytes, as expressed in normal cells and essential thrombocythemic cells | 1.7 × 10−22 | 2.4 × 10−20 |
FDR indicates false discovery rate; MSigDB, molecular signatures database; and TPO, thrombopoietin (the principal growth factor for megakaryocytes).
MEIS1 binding profile aids in the prioritization of functional studies of GWAS loci
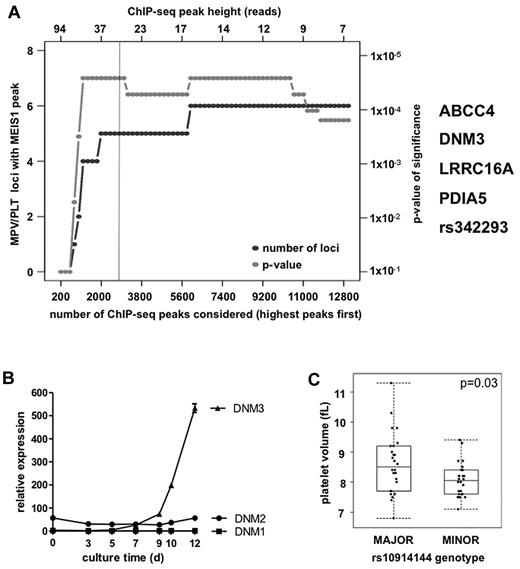
Six of the 57 MK overexpressed genes with a MEIS1 binding site (Figure 1B genes labeled in bold) harbored a sentinel GWAS SNP. Importantly, a GW analysis of the spatial co-occurrence of MEIS1 binding peaks with 66 sentinel SNPs (2 GWAS SNPs in the HLA locus were removed because of its complicated haplotype structure) for PLT volume and count and their proxy SNPs robustly identified at least 5 sites of colocalization when only the top 20% of MEIS1 peaks was considered (Figure 2A). Such degree of overlap was significantly greater than expected by chance (P < 1 × 10−5).
DNM3 harbors MEIS1 binding sites and is associated with differences in MPV. (A) Co-occurrence of MPV and PLT count association SNPs with MEIS1 ChIP-seq peaks. (B) Transcript levels of DNM genes during in vitro differentiation of MKs from human cord blood CD34+ cells, as measured by quantitative real-time RT-PCR (n = 3). Error bars represent SD. (C) MPV of the PLTs of persons according to genotype for sentinel SNP rs10914144 (n = 26 of 26). Mann-Whitney test: P = .03.
DNM3 harbors MEIS1 binding sites and is associated with differences in MPV. (A) Co-occurrence of MPV and PLT count association SNPs with MEIS1 ChIP-seq peaks. (B) Transcript levels of DNM genes during in vitro differentiation of MKs from human cord blood CD34+ cells, as measured by quantitative real-time RT-PCR (n = 3). Error bars represent SD. (C) MPV of the PLTs of persons according to genotype for sentinel SNP rs10914144 (n = 26 of 26). Mann-Whitney test: P = .03.
To investigate the consequences of the observed colocalization, we selected the DNM3 locus because the locus is strongly associated with MPV (sentinel SNP rs10914144 P = 1.11 × 10−24), and there are no other genes localized in the recombination interval that harbors this variant. The sentinel SNP (minor allele frequency in Europeans 0.173; 2.99% of persons are homozygous for the minor allele) is noncoding, and no coding proxy SNPs could be identified in the 1000 Genomes project catalog.26 Moreover, whereas DNM2 is ubiquitously expressed, DNM1 and DNM3 show tissue-specific expression profiles,27,28 whereby in hematopoiesis, DNM3 expression is restricted to the megakaryocytic lineage (Figure 2B; supplemental Figure 4). In other tissues, DNM3 transcript is also found in brain, lung, heart, and testis (supplemental Figure 5A-B).8,9,29,30
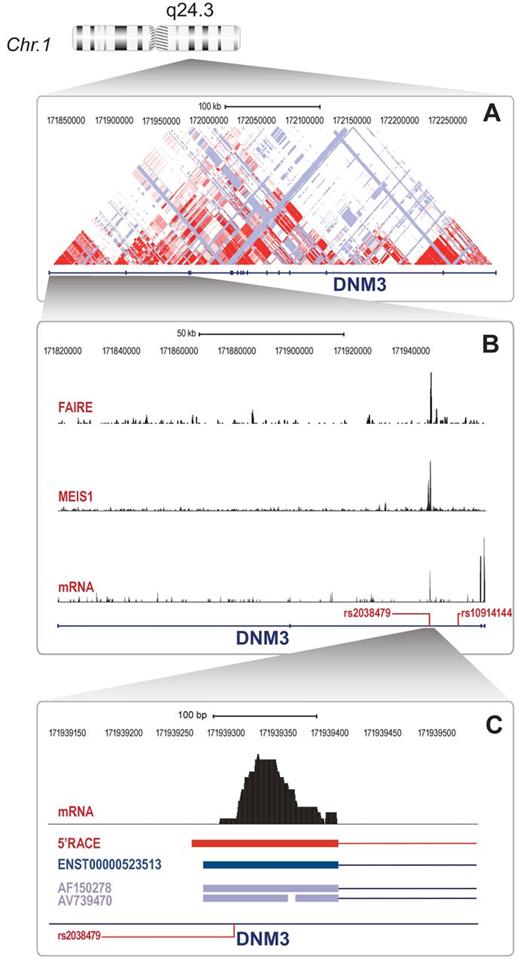
MEIS1 and variant rs2038479 co-occur in the proximity of a MK specific alternative DNM3 transcription start site
In the DNM3 gene locus, the MEIS1 binding event occurs very close to, although does not contain, variant rs2038479 (90 bp from MEIS1 peak maximum). This variant is 10 460 bp upstream and in strong linkage disequilibrium with the GWAS sentinel SNP rs10914144 (r2 = 0.991 in Europeans; 1000 Genomes, June 2011 release). Both the MEIS1 binding site and variant rs2038479 lie at a putative promoter of an alternative transcript ENST00000523513 (termed alternative transcript hereafter) that is so far solely based on EST clones derived from CD34+ cells obtained from cord blood (AF150278 and AV739470). RNA-seq data for cultured MK revealed transcription surrounding rs2038479 (Figure 3B), suggesting a novel DNM3 exon of ∼ 133 bp (exon 2B hereafter). 5′ RACE with RNA from cultured MKs, CHRF cells, and PLTs (Figure 3C) confirmed the expression of exon 2B and the presence of an alternative transcription start site. Furthermore, RACE analysis revealed that exons 1 and 2 are not part of the predominant DNM3 transcript in MKs (see supplemental Table 2 for RACE clone sequences). Using primers in exon 2B and the annotated 3′-untranslated region, a PCR product was obtained from MK cDNA that contained an open reading frame extending from exon 2B to the annotated stop codon after Asp859 (data not shown).
The DNM3 locus harbors a regulatory element and an alternative exon in annotated intron 2. (A) DNM3 gene locus showing CEU LOD plot (HapMap release 22). (B) Close-up of gene region, including exons 1-4 with regional plots from FAIRE-seq, MEIS1 ChIP-seq, and RNA-seq data. Also depicted are key SNPs rs10914144 and rs2038479. (C) Further close-up of the region of the MEIS1 peak in intron 2 showing RNA-seq peak, sequence location of the longest 5′RACE clone, alternative transcript ENST00000523513, and 2 EST clones as well as position of variant rs2038479.
The DNM3 locus harbors a regulatory element and an alternative exon in annotated intron 2. (A) DNM3 gene locus showing CEU LOD plot (HapMap release 22). (B) Close-up of gene region, including exons 1-4 with regional plots from FAIRE-seq, MEIS1 ChIP-seq, and RNA-seq data. Also depicted are key SNPs rs10914144 and rs2038479. (C) Further close-up of the region of the MEIS1 peak in intron 2 showing RNA-seq peak, sequence location of the longest 5′RACE clone, alternative transcript ENST00000523513, and 2 EST clones as well as position of variant rs2038479.
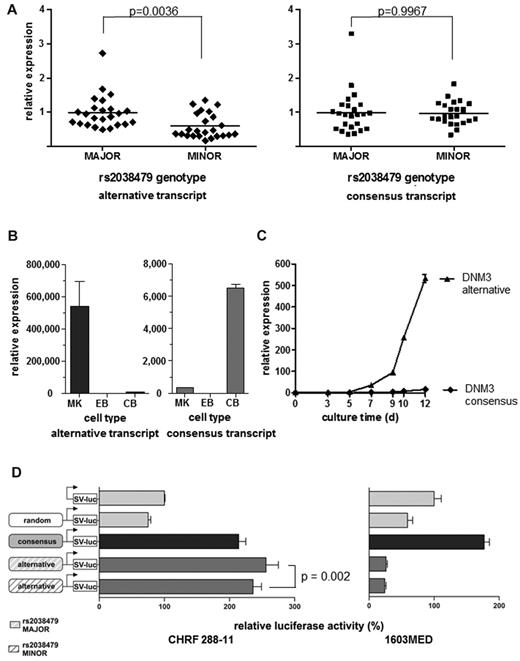
Because the annotated DNM3 consensus transcript is derived from human brain tissue, we quantified expression levels of both transcripts in cerebellum and MKs. This revealed a striking tissue specific expression pattern, with the consensus transcript being used in the former and the alternative one in the latter (Figure 4B). Indeed, expression of the alternative transcript appears restricted to the megakaryocytic lineage (supplemental Figures 5 and 6A-C), with the expression of this isoform substantially increasing during megakaryopoiesis (Figure 4C; supplemental Figure 6D).
Identification of functional SNP rs2038479 in a novel, lineage-specific promoter. (A) Relative DNM3 transcript levels in PLTs from persons homozygous for the major or minor allele of variant rs2038479. Quantitative real-time RT-PCR results with a probe specific for the novel exon 2B-exon 3 boundary (left) versus the annotated exon 2-exon 3 boundary (right) (n = 24/23, unpaired t test, including Welch correction for unequal variances). (B) Relative transcript abundance of tissues as measured by quantitative PCR. MK indicates MK; EB, erythroblast; and CB, cerebellum. n = 3. Error bars represent SD, relative to EB = 1. (C) Quantitative PCR comparison of 2 major DNM3 transcripts during MK maturation (n = 3). Error bars represent SD, relative to annotated transcript day 0 = 1. (D) In vitro dual Luciferase reporter assay of promoter activity of the region 5′ of alternative exon 2B, including variant rs2038479 compared with the consensus promoter 5′ of exon 1. Left: Megakaryocytic CHRF cells. Right: Medulloblastoma cells 1603MED, relative to empty firefly vector = 100%. Error bars represent SD. The significance of the effect of the variant at the alternative promoter (P = .002) was determined using a linear mixed model based on 3 independent experiments with 3 replicates per experiment.
Identification of functional SNP rs2038479 in a novel, lineage-specific promoter. (A) Relative DNM3 transcript levels in PLTs from persons homozygous for the major or minor allele of variant rs2038479. Quantitative real-time RT-PCR results with a probe specific for the novel exon 2B-exon 3 boundary (left) versus the annotated exon 2-exon 3 boundary (right) (n = 24/23, unpaired t test, including Welch correction for unequal variances). (B) Relative transcript abundance of tissues as measured by quantitative PCR. MK indicates MK; EB, erythroblast; and CB, cerebellum. n = 3. Error bars represent SD, relative to EB = 1. (C) Quantitative PCR comparison of 2 major DNM3 transcripts during MK maturation (n = 3). Error bars represent SD, relative to annotated transcript day 0 = 1. (D) In vitro dual Luciferase reporter assay of promoter activity of the region 5′ of alternative exon 2B, including variant rs2038479 compared with the consensus promoter 5′ of exon 1. Left: Megakaryocytic CHRF cells. Right: Medulloblastoma cells 1603MED, relative to empty firefly vector = 100%. Error bars represent SD. The significance of the effect of the variant at the alternative promoter (P = .002) was determined using a linear mixed model based on 3 independent experiments with 3 replicates per experiment.
Variant rs2038479 displays allelic differences in the promoter activity of the MK specific DNM3 transcript
To test whether the variant affected DNM3 expression, we typed the sentinel SNP rs10914144 in 5034 healthy volunteers from the Cambridge BioResource and recalled 2 groups of 26 persons homozygous for each allele. Typing of the samples for rs2038479 showed complete concordance in all but 1 sample from a person homozygous for the minor allele of rs10914144, which tested heterozygous for rs2038479. MPV and PLT DNM3 RNA levels were compared between groups and, as expected, the minor allele was associated with lower MPV, consistent with the GWAS results derived from nearly 67 000 persons (P = .03; Figure 2C). Comparison of the PLT DNM3 transcript levels with a probe specific for the alternative exon 2B revealed a statistically significant difference between groups, with lower levels in persons homozygous for the minor allele (P = .0036; Figure 4A). This difference was not observed with a probe specific for the consensus transcript (P = .9967).
Luciferase reporter constructs for the consensus and alternative promoters confirmed the quantitative PCR data. Whereas the consensus promoter was active in both CHRF and 1603MED (a human medulloblastoma cell line) cells, the alternative one was exclusively active in CHRF cells (Figure 4D). We then tested the effect of the rs2038479 variant on the activity of the alternative promoter and observed reduced activity in CHRF cells with the minor versus the major allele (Figure 4D; P = .002), which is consistent with the allelic differences in PLT DNM3 transcript levels between the 2 genotype groups. Electrophoretic mobility shift assay with probes harboring alternative forms of rs2038479 incubated with CHRF nuclear lysates displayed a striking difference between the 2 alleles with strong band shifting for the minor allele, thus revealing preferential binding of nuclear factors to this allele (supplemental Figure 7). Taken together, these data strongly suggest that rs2038479 is the more likely causative variant underlying the observed association with MPV, resulting in increased binding of a repressor complex at this locus.
Loss of Dynamin activity results in impaired pro-PLT formation
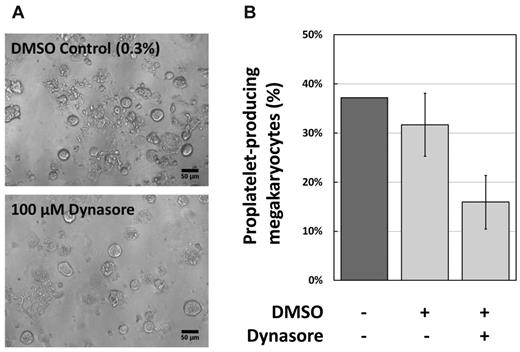
We reasoned, because of the observed positive correlation between PLT DNM3 transcript levels and MPV (Figures 2C and 4A), that inhibition of the DNM proteins may exert an effect on the processes of megakaryopoiesis and pro-PLT formation. To test this, MKs were generated by culture of murine fetal liver cells in the presence and absence of Dynasore, an inhibitor of DNM GTPase activity.31 The proportion of MKs that formed pro-PLT processes was recorded (Figure 5A) and compared between the treated and control cultures. A significant reduction was observed (Figure 5B), providing additional evidence for the importance of DNM activity in this process.
Inhibition of dynamin in MK cultures. (A) Representative phase-contrast photomicrographs from the 2 treatments shown. DMSO was used at 0.3% final, DynaSore at 100μM final (taken with a 20× objective). Bar represents 50 μm. (B) The proportion of murine MK developing pro-PLTs after 4 days culture of fetal liver cells, quantified as stated in “Methods.”
Inhibition of dynamin in MK cultures. (A) Representative phase-contrast photomicrographs from the 2 treatments shown. DMSO was used at 0.3% final, DynaSore at 100μM final (taken with a 20× objective). Bar represents 50 μm. (B) The proportion of murine MK developing pro-PLTs after 4 days culture of fetal liver cells, quantified as stated in “Methods.”
Discussion
The volume and count of PLTs vary widely in the healthy population but are tightly regulated within narrow ranges at the individual level. The population variation in MPV and PLT is for a large extent genetically controlled. In a GWAS in nearly 67 000 persons of European ancestry, we identified 68 independent genetic loci associated with either or both PLT traits at GW significance.1 The sentinel SNP of 54 of the loci mapped within 10 kb of a gene providing probable biologic candidate proteins. At 15 of the 54 loci, the sentinel SNP or a proxy SNP in high linkage disequilibrium (r2 > 0.8) altered the amino acid sequence of the implicated protein, thus providing a plausible functional explanation for the observed associations. The sentinel SNPs at the 53 remaining loci are synonymous or noncoding ones, generally localized in introns or sometimes in intergenic regions and therefore are likely to exert their effect on megakaryopoiesis and the formation and survival of PLTs by regulating gene transcription, possibly through lineage-specific regulatory elements.2
Here we have presented a prioritization strategy to select genes and variants for functional follow-up studies to investigate their role in the megakaryocytic lineage based on only considering associated variants that are also located at MEIS1 binding sites. This has resulted in a small list of readily tractable loci of which we selected DNM3 for in-depth analysis.
A similar overlapping strategy has been adopted previously using regions of open chromatin as markers for regulatory elements as determined by FAIRE. For example, in Paul et al,2 we used FAIRE-ChIP to identify potential regulatory variants and showed that the major (C) allele of the MPV variant rs342293 at 7q22.3 is bound by the transcription factor EVI1/MECOM more firmly than the minor (G) allele. Binding of EVI1/MECOM to the major allele at this position is likely to be associated with repression of transcription and results in a lower PIK3CG transcript level in persons homozygous for the major allele compared with those homozygous for the minor allele. Although the selection of EVI1/MECOM as likely candidate was based on computational prediction followed by experimental validation, the present study used in vivo transcription factor binding to prioritize potential regulatory variants.
To explore the mechanisms that underlie the observed associations between noncoding sequence variants and the PLT traits of volume and count, we chose to focus on the transcription factor MEIS1. In lineage-committed blood cells MEIS1 is uniquely transcribed in megakaryocytic cells, and not in the other 7 major mature blood cell types (supplemental Figure 1).8 The functional role of MEIS1 in hematopoiesis and leukemogenesis has been studied extensively,32-34 but its role in megakaryopoiesis and PLT formation has not yet been fully appreciated. In this study, we postulated that a fraction of the association SNPs identified by GWAS or their proxy SNPs may modify target gene transcription via the transcription factor MEIS1 or one of its binding partners.
The first step in exploring this notion was to establish a comprehensive catalog of genes regulated by MEIS1. For this, we performed a ChIP-seq experiment in the megakaryocytic cell line CHRF. This revealed MEIS1 binding events in a set of 1285 genes (Figure 1A-B), and this gene set showed a number of highly significant and interesting features. First, the set was strongly enriched for the terms relevant to megakaryopoiesis and PLT biology (Table 1). Second, there was an excess of genes, which are relatively overexpressed in MKs compared with the other 7 mature blood cell elements (Figure 1B; P < 5 × 10−19). Furthermore, 6 of the 57 MK overexpressed genes (ABCC4, DNM3, LRRC16A, PDIA5, TPM4, and ZFPM2) harbored a sentinel GWAS SNPs (Figure 1B). Finally, the GW prioritization through co-occurrence of MEIS1 binding events with association SNPs identified 4 of the latter 6 genes plus SNP rs342293 at 7q22.3. Thus, not only did GW profiling of MEIS1 binding sites support the assumption that MEIS1 has an important role in conferring the PLT its cellular identity, but it also supported our assumption that allelic alteration of gene regulation by MEIS1 or its binding partners is possibly responsible for the observed associations with MPV and PLT at 5 GWAS loci.
We selected the DNM3 gene on chromosome 1 of these 5 GWAS loci for further functional studies. The gene spans 571 kb, has 21 exons, and is the single gene in the recombination interval that harbored the PLT-volume sentinel SNP rs10914144. We therefore reasoned that either the sentinel SNP or one of its proxy SNPs was highly likely to regulate the transcription of the DNM3 gene itself and not one of the other more distantly removed cis-positioned genes. Indeed, the analysis of 3 new GW datasets (Figure 3B-C) confirmed this assumption. It showed that SNP rs2038479, one of the proxy SNPs of the sentinel SNP, co-occurs with the MEIS1 binding event in the second intron of the DNM3 locus. The FAIRE analysis showed that this SNP is at a position of open chromatin in CHRF cells (Figure 3B), which is absent in the erythroid K562 cell line (data not shown). The lineage-restricted open chromatin signature around SNP rs2038479 together with the presence of a MEIS1 binding event and already published ChIP-seq datasets from MKs35 is strongly suggestive of a regulatory element specific for the MK lineage at this position (see supplemental Figure 8). Indeed, the sequencing of RNA from MKs showed that the same region is transcribed (Figure 3B), and suggested that the first 2 exons were transcribed at much lower levels. The presence of an alternative transcription start site and DNM3 transcript with exon 2B as a first exon was confirmed by 5′RACE analysis and by quantitative PCR tests with transcript specific probes (Figure 4B; supplemental Figures 5 and 6). Moreover, we showed that the levels of the alternative transcript increase sharply during megakaryopoiesis (Figure 4C).
A recall study of persons from the Cambridge BioResource was used to compare the levels of the alternative DNM3 transcript in PLTs between 2 groups of volunteers each, homozygous for the major (n = 24) and minor alleles (n = 23) of rs10914144 and rs2038479, respectively. Both the MPV and RNA levels between the groups differed significantly (Figures 2C and 4A), indicating that the minor allele dictates lower levels of the DNM3 transcript and these reduced levels are possibly causative of the formation of smaller PLTs. This in vivo observation was corroborated by the results of Luciferase-reporting assays, which revealed a significant lower signal in CHRF cells for the reporter construct containing the minor allele compared with the signal produced by the same construct harboring the major allele (Figure 4D). If rs2038479 is indeed the causative variant, this variant would have been expected to have a lower P value in the GWAS. Closer inspection of the meta-analysis revealed that rs2038479 (P = 2.6 × 10−22 for association with MPV) was genotyped or imputed in a smaller number of samples than rs10914144, consequently resulting in a higher P value.
DNM3 is a member of the dynamin family of large GTPases that each contain an amino-terminal GTPase domain, 3 central domains (middle, pleckstrin homology, and coiled coil) involved in self-assembly and membrane binding, and a carboxy-terminal proline-rich domain that links to SH3 domain-containing signaling/cytoskeletal partner proteins.36 The dynamins have been shown to mediate endocytosis of clathrin-coated pits, vesicle budding, and pseudopodia formation.37-40 The DNM3 protein has been observed immunochemically in human MK and murine pro-PLTs.30 Interestingly, silencing of the DNM3 gene in human CD34+ hematopoietic precursor cells seems to impair the formation of MKs, although this is possibly caused by the absence of the consensus DNM3 protein in precursor cells.41 Here we provide evidence that pharmacologic inhibition of dynamin GTPase activity inhibits pro-PLT formation (Figure 5).
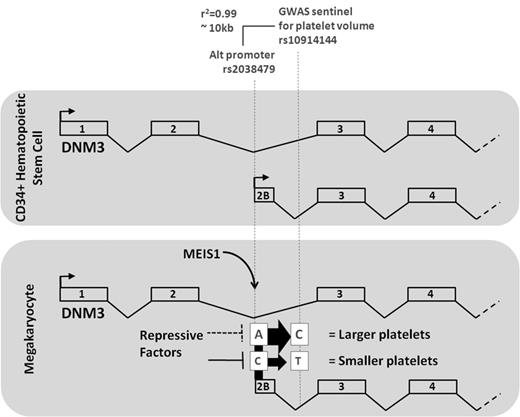
In conclusion, we have obtained evidence that 5 of the 68 GWAS SNPs for PLT count and volume are colocalized with a MEIS1 binding event. By focusing on one of these sites, we provide strong evidence that variant rs2038479 in the DNM3 locus is the functionally causative allele, which underlies the observed association with PLT volume, controlling a MK-specific alternative promoter of the DNM3 gene involved in PLT formation. Importantly, expression levels and promoter activity of the MK transcript are significantly and concordantly associated with the genotype of the original GWAS association SNP (rs10914144) and its proxy variant rs2038479, which resides at a MEIS1 binding site and marks the position of the alternative promoter. A scheme showing the 2 SNPs and their relationship to DNM3 gene transcription and PLT volume is shown in Figure 6. Further studies are required to define the functional role of the alternative DNM3 transcript in megakaryopoiesis and PLT formation.
From a PLT volume, GWAS variant at rs10914144 in the DNM3 locus to the discovery of the functional variant rs2038479 at a MEIS1/RUNX1 binding site. A schema depicting the transcription of the DNM3 gene at 1q24.3 in CD34+ hematopoietic stem cells (top panel) and in MKs (bottom panel), respectively. A GWAS identified variant rs10914144 in intron 2 as being associated with differences in MPV. The MPV of persons homozygous for the major (C) allele of the variant is higher compared with the values observed in persons who are homozygous for the minor (T) allele. Functional annotation of the genome of megakaryocytic cells identified binding sites for the transcription factor MEIS1 in intron 2 at a position of an alternative transcription start site, which is uniquely used in MKs. The same element harbors SNP rs2038479, which is in high linkage disequilibrium with the GWAS sentinel rs10914144. DNM3 transcript levels (represented by the sizes of arrows at the consensus and alternative promoters) in PLTs from persons homozygous major (T) for rs2038479 were significantly lower than in PLTs from homozygous minor ones (C).
From a PLT volume, GWAS variant at rs10914144 in the DNM3 locus to the discovery of the functional variant rs2038479 at a MEIS1/RUNX1 binding site. A schema depicting the transcription of the DNM3 gene at 1q24.3 in CD34+ hematopoietic stem cells (top panel) and in MKs (bottom panel), respectively. A GWAS identified variant rs10914144 in intron 2 as being associated with differences in MPV. The MPV of persons homozygous for the major (C) allele of the variant is higher compared with the values observed in persons who are homozygous for the minor (T) allele. Functional annotation of the genome of megakaryocytic cells identified binding sites for the transcription factor MEIS1 in intron 2 at a position of an alternative transcription start site, which is uniquely used in MKs. The same element harbors SNP rs2038479, which is in high linkage disequilibrium with the GWAS sentinel rs10914144. DNM3 transcript levels (represented by the sizes of arrows at the consensus and alternative promoters) in PLTs from persons homozygous major (T) for rs2038479 were significantly lower than in PLTs from homozygous minor ones (C).
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the volunteers of the Cambridge BioResource.
This work was supported in part by the National Institute for Health Research (program grant RP-PG-0310-1002; S.T.N., P.A.S., and W.H.O.), the British Heart Foundation (RG/09/12/28096; A.R.), and the Wellcome Trust (project grant WT-084183/2/07/2; J.G.S.). The Cambridge BioResource, a local resource for genotype-phenotype association studies, is supported by the National Institute for Health Research to the Cambridge Biomedical Research Center (J.G.S. and H.L.-J.). D.S.P. and K.V. were supported by the Marie-Curie NetSim ITN (grant EC-215820). M.R.T. was supported by a Marie-Curie Intra-European Fellowship (237296). The mouse study was supported in part by the National Institutes of Health (grant HL68130; J.E.I.). J.E.I. is an American Society of Hematology Junior Faculty Scholar. J.N.T. is an American Society of Hematology Scholar.
National Institutes of Health
Authorship
Contribution: D.S.P., J.E.I., J.N.T., P.A.S., and S.T.N. designed and performed experiments and analyzed the data; A.R. analyzed the data; K.V., M.R.T., H.L.-J., and J.G.S. provided samples; N.S. provided data; P.D. and B.G. supervised experiments; W.H.O. supervised the project; and A.R., P.A.S., S.T.N., and W.H.O. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A list of the HaemGen Consortium members is provided in the online supplemental Appendix.
Correspondence: Willem H. Ouwehand, Department of Haematology, University of Cambridge, Cambridge, United Kingdom; e-mail: who1000@cam.ac.uk.
References
Author notes
S.T.N. and A.R. contributed equally to this study.