Abstract
We conducted a genome-wide association study to identify novel associations between genetic variants and circulating plasminogen activator inhibitor-1 (PAI-1) concentration, and examined functional implications of variants and genes that were discovered. A discovery meta-analysis was performed in 19 599 subjects, followed by replication analysis of genome-wide significant (P < 5 × 10−8) single nucleotide polymorphisms (SNPs) in 10 796 independent samples. We further examined associations with type 2 diabetes and coronary artery disease, assessed the functional significance of the SNPs for gene expression in human tissues, and conducted RNA-silencing experiments for one novel association. We confirmed the association of the 4G/5G proxy SNP rs2227631 in the promoter region of SERPINE1 (7q22.1) and discovered genome-wide significant associations at 3 additional loci: chromosome 7q22.1 close to SERPINE1 (rs6976053, discovery P = 3.4 × 10−10); chromosome 11p15.2 within ARNTL (rs6486122, discovery P = 3.0 × 10−8); and chromosome 3p25.2 within PPARG (rs11128603, discovery P = 2.9 × 10−8). Replication was achieved for the 7q22.1 and 11p15.2 loci. There was nominal association with type 2 diabetes and coronary artery disease at ARNTL (P < .05). Functional studies identified MUC3 as a candidate gene for the second association signal on 7q22.1. In summary, SNPs in SERPINE1 and ARNTL and an SNP associated with the expression of MUC3 were robustly associated with circulating levels of PAI-1.
Introduction
Plasminogen activator inhibitor type 1 (PAI-1) is a serine protease inhibitor protein encoded by the SERPINE1 gene. It is the principal inhibitor of tissue and urinary plasminogen activators, and therefore constitutes an important regulatory protein in fibrinolysis. PAI-1 is produced by vascular endothelium, liver, monocytes/macrophages, platelets, and adipose tissue. High plasma levels of PAI-1 (Ag or activity) are associated with increased risk of atherothrombotic diseases, particularly coronary artery disease (CAD) and myocardial infarction (MI),1-4 and PAI-1 accumulates in human atherosclerotic lesions.5,6 PAI-1–dependent mechanisms are also implicated in the pathogenesis of obesity, insulin resistance, and type-2 diabetes (T2D).7-9 The consistent association of PAI-1 with obesity and T2D contributes to a prevailing uncertainty regarding the role of PAI-1 as a causal factor in risk for cardiovascular disease.
Population-based case-control studies and family and twin studies have indicated that a major genetic component contributes to the variance of plasma PAI-1 concentration, with an estimated heritability of up to 0.83 in twin studies.10,11 Several common polymorphisms have been identified in SERPINE1, but only the 4G/5G insertion-deletion variant (rs1799889) located 675 bp from the transcriptional start site has been consistently associated with the plasma level of PAI-1. The proportion of the variation explained by this cis variant is modest (generally approximately 1%-3%), suggesting that as-yet-unknown variations in SERPINE1 or genes in other pathways or epistasis may also contribute.12-14 Further, some but not all systematic overviews have found that the homozygous 4G/4G genotype is associated with a modest increase in MI risk.15,16 Two high-coverage single nucleotide polymorphism (SNP) association studies on PAI-1, both with limited sample size, have been reported so far,13,17 neither of which identified loci showing genome-wide significance.
We hypothesized that there are multiple cis- and trans-acting loci, in addition to the 4G allele variant, that contribute to the variation in circulating PAI-1 level, and in the present study, we tested these hypotheses using data on 19 599 European-ancestry adults from 8 genome-wide association (GWA) studies, each with dense (approximately 2.5 million) imputed SNP genotypes. We then attempted replication of genome-wide significant findings in independent samples from 9 cohorts with a total of 10 796 subjects of European ancestry. SNPs found to be robustly associated with plasma PAI-1 levels in the discovery meta-analysis were examined for association with clinically apparent CAD and T2D in very large clinical GWA study meta-analyses.18,19 Finally, we performed expression analyses in human target tissues and silencing experiments in a relevant cell culture system to confirm the functional significance of the identified candidate genes.
Methods
Discovery and replication cohorts
Separate GWA studies of plasma PAI-1 Ag concentration were conducted in 8 independent cohorts of European-ancestry subjects. These cohorts included the Framingham Heart Study (FHS, n = 6634), the Precocious Coronary Artery Disease Study (PROCARDIS, n = 1922 cases and 2017 controls), Twins United Kingdom (n = 1294), MONICA/KORA (n = 1565), HealthABC (n = 1645), the Marseille Thrombosis Association study (MARTHA, n = 851), and the Prevention of Renal and Vascular End Stage Disease study (PREVEND, n = 3671). Detailed descriptions of the discovery cohorts and exclusion criteria adopted for the present study are given in section 1 of the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Sample sizes, mean age, and sex distribution of discovery study participants in each cohort at the time of the PAI-1 determination are summarized in supplemental Table 1. After a meta-analysis of a total of 19 599 subjects, genome-wide significant loci (P < 5.0 × 10−8) underwent replication in 10 796 independent samples from 9 cohorts of European ancestry (supplemental materials, section 2), characteristics of which are provided in supplemental Table 2. PAI-1 Ag levels (ng/mL) or PAI-1 activity (U/mL) were measured in plasma (EDTA or citrate) using ELISA and functional methods (supplemental Table 1 and supplemental materials, section 3).
All participating cohorts were granted approval by the appropriate research ethics committees for the research, and all participants provided written informed consent for the use of their DNA.
Genotyping and imputation
A description of the genotyping technologies used for the discovery cohorts is provided in supplemental Table 3, along with the quality control criteria for filtering and imputation methods. Briefly, approximately 2.5 M autosomal SNPs were imputed for every cohort using the HapMap II white sample from the Center d'Etude du Polymorphisme Humain as a reference panel.20,21 Before imputation, every cohort applied SNP exclusions by call rate (≤ 0.93-0.99 depending on the cohort), minor allele frequency (MAF) < 0.01, and deviation from Hardy-Weinberg equilibrium (P < 10−5-10−6). Details regarding genotyping, quality control, and imputation characteristics of the replication cohorts are provided in supplemental Table 4.
Statistical analysis of the discovery cohorts
PAI-1 values were natural-logarithm transformed because of skewness of the distribution. Genotype-phenotype association analyses were performed independently in each cohort according to a prespecified analysis plan. The association of measured and imputed SNPs with natural logarithm-transformed PAI-1 values was assessed by a linear model assuming additive genetic effects. Standard linear regression analysis was used in nonfamilial studies. All analyses were adjusted for age and sex and additionally on principal components or multidimensional scaling where appropriate. In PROCARDIS, analyses were further adjusted for antidiabetic medication in T2D. For family studies, FHS, and PROCARDIS, a linear mixed-effects model was also used to account for relatedness.22 A genomic control coefficient was computed for each discovery cohort estimating the GWA inflation coefficients and then applied individual SNP association test statistics to correct for cryptic relatedness. Specialized analysis tools and software packages are listed in the supplemental materials (section 4).
Meta-analysis of all discovery GWA studies using a total of 2 445 683 SNPs, was conducted using an inverse-variance weighted fixed-effects method as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html), and summary P values and β coefficients were calculated. The β value represents the per-allele effect on the natural log-transformed PAI-1 level. To ensure accuracy and reliability of results, the meta-analysis was conducted independently by 2 investigators (J.H. and M.S.-L.) at 2 separate sites. The prespecified threshold of genome-wide significance was set at the conventional level of P = 5.0 × 10−8.
Replication
Replication analysis was conducted for loci that contained at least one SNP passing the prespecified genome-wide significance threshold in the discovery meta-analysis. A total of 10 SNPs were selected for replication from the 4 loci (Table 1): the 4 lead SNPs (ie, the SNP with the lowest P value for each of the 4 genome-wide significant regions, also referred to as the index SNP), 2 nonsynonymous SNPs within 2 of these regions showing suggestive evidence of association (P < 5 × 10−7), and 4 additional SNPs selected to tag the main haplotypes in 2 of the candidate regions. Procedures for selecting tag SNPs are described in the supplemental materials (section 4).
Association results for the 10 selected SNPs in the discovery and replication phase and in secondary analyses
| Region, SNP . | Chr . | Closest gene . | Type . | Alleles . | MAF . | Discovery . | Replication . | Combined . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta . | P . | Beta . | P . | Beta . | P . | ||||||
| 3p25.2 | |||||||||||
| rs11128603 | 3 | PPARG | Index SNP | A → G | 0.102 | 0.086 | 2.9 × 10−8 | 0.031 | .1 | 0.066 | 9.4 × 10−8 |
| rs1801282 | 3 | PPARG | nsSNP | C → G | 0.127 | 0.069 | 1.4 × 10−7 | 0.026 | .1 | 0.052 | 3.1 × 10−7 |
| 7q22.1 | |||||||||||
| rs2227631 | 7 | SERPINE1 | Index SNP | A → G | 0.409 | 0.076 | 7.8 × 10−15 | 0.069 | 5.5 × 10−11 | 0.073 | 3.2 × 10−24 |
| 7q22.1 | |||||||||||
| rs6976053 | 7 | ACHE | Index SNP | T → C | 0.478 | 0.054 | 3.4 × 10−10 | 0.039 | 2.3 × 10−4 | 0.048 | 5.8 × 10−13 |
| rs2075756 | 7 | TRIP6 | nsSNP | A → G | 0.282 | 0.058 | 2.7 × 10−9 | 0.043 | 2.8 × 10−4 | 0.052 | 5.0 × 10−12 |
| rs314376 | 7 | SLC12A9 | Tag SNP | G → A | 0.483 | 0.052 | 2.4 × 10−9 | 0.041 | 1.4 × 10−4 | 0.048 | 2.0 × 10−12 |
| rs12672665 | 7 | SRRT | Tag SNP | G → A | 0.479 | 0.047 | 6.1 × 10−8 | 0.033 | 2.1 × 10−3 | 0.041 | 8.0 × 10−10 |
| rs3847067 | 7 | ACHE | Tag SNP | A → C | 0.289 | 0.059 | 4.1 × 10−10 | 0.038 | 1.1 × 10−3 | 0.051 | 4.6 × 10−12 |
| 11p15.2 | |||||||||||
| rs6486122 | 11 | ARNTL | index SNP | T → C | 0.311 | 0.051 | 3.0 × 10−8 | 0.038 | 9.8 × 10−4 | 0.046 | 1.7 × 10−10 |
| rs3816360 | 11 | ARNTL | Tag SNP | C → T | 0.333 | 0.048 | 9.9 × 10−8 | 0.031 | 5.1 × 10−3 | 0.041 | 3.9 × 10−9 |
| Region, SNP . | Chr . | Closest gene . | Type . | Alleles . | MAF . | Discovery . | Replication . | Combined . | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta . | P . | Beta . | P . | Beta . | P . | ||||||
| 3p25.2 | |||||||||||
| rs11128603 | 3 | PPARG | Index SNP | A → G | 0.102 | 0.086 | 2.9 × 10−8 | 0.031 | .1 | 0.066 | 9.4 × 10−8 |
| rs1801282 | 3 | PPARG | nsSNP | C → G | 0.127 | 0.069 | 1.4 × 10−7 | 0.026 | .1 | 0.052 | 3.1 × 10−7 |
| 7q22.1 | |||||||||||
| rs2227631 | 7 | SERPINE1 | Index SNP | A → G | 0.409 | 0.076 | 7.8 × 10−15 | 0.069 | 5.5 × 10−11 | 0.073 | 3.2 × 10−24 |
| 7q22.1 | |||||||||||
| rs6976053 | 7 | ACHE | Index SNP | T → C | 0.478 | 0.054 | 3.4 × 10−10 | 0.039 | 2.3 × 10−4 | 0.048 | 5.8 × 10−13 |
| rs2075756 | 7 | TRIP6 | nsSNP | A → G | 0.282 | 0.058 | 2.7 × 10−9 | 0.043 | 2.8 × 10−4 | 0.052 | 5.0 × 10−12 |
| rs314376 | 7 | SLC12A9 | Tag SNP | G → A | 0.483 | 0.052 | 2.4 × 10−9 | 0.041 | 1.4 × 10−4 | 0.048 | 2.0 × 10−12 |
| rs12672665 | 7 | SRRT | Tag SNP | G → A | 0.479 | 0.047 | 6.1 × 10−8 | 0.033 | 2.1 × 10−3 | 0.041 | 8.0 × 10−10 |
| rs3847067 | 7 | ACHE | Tag SNP | A → C | 0.289 | 0.059 | 4.1 × 10−10 | 0.038 | 1.1 × 10−3 | 0.051 | 4.6 × 10−12 |
| 11p15.2 | |||||||||||
| rs6486122 | 11 | ARNTL | index SNP | T → C | 0.311 | 0.051 | 3.0 × 10−8 | 0.038 | 9.8 × 10−4 | 0.046 | 1.7 × 10−10 |
| rs3816360 | 11 | ARNTL | Tag SNP | C → T | 0.333 | 0.048 | 9.9 × 10−8 | 0.031 | 5.1 × 10−3 | 0.041 | 3.9 × 10−9 |
The replication sample size was estimated to have 80% power to detect an explained variance of 0.5% at a MAF of 20.0%. The same statistical methods were used as for discovery. The significance threshold was set to P = .005 after Bonferroni correction for testing of 10 SNPs.
Conditional and secondary analysis
To confirm that 2 associated loci on the same chromosomal region (7q22.1) were independent, we conditioned on the sentinel SNP with the lowest P value (rs2227631) and repeated the association analysis between genotype and PAI-1 levels using the same model as was used in the discovery analysis. Given the adipose production of PAI-1 and the widely reported association of body mass index (BMI) with plasma PAI-1 levels, the genotype-phenotype association analyses were further adjusted for BMI.
Association with clinically apparent CAD and T2D
We explored associations of SNPs selected from the discovery meta-analysis with CAD and T2D to determine whether SNPs showing robust associations with PAI-1 levels may also be implicated in the pathogenesis of CAD or T2D, respectively. Genotype-phenotype association results for the selected SNPs were obtained from the Coronary Artery DIsease Genome-wide Replication And Meta-analysis (CARDIoGRAM) and the Coronary Artery Disease (C4D) Genetics consortia. CARDIoGRAM includes 22 233 cases of CAD and 64 762 controls,18 all of European ancestry, whereas C4D comprises 6424 CAD cases and 7268 controls of European ancestry.19 For T2D, genotype-phenotype association results for the selected SNPs were obtained from the GWAS meta-analysis of 8130 subjects with T2D and 38 987 controls in the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) consortium.23 SNP associations with CAD and T2D were assessed by logistical regression analysis under an additive model adjusting for age and sex. For the PROCARDIS component of C4D, a mixed-effects model was also used to account for relatedness. The significance threshold for disease association was set to P < .005 after Bonferroni correction for testing of 10 SNPs.
Association with gene expression in human tissues
Ex vivo studies of global gene-expression data from monocytes and macrophages were derived from the Cardiogenics study (http://www.cardiogenics.eu)24 and based on peripheral blood samples from both CAD patients and healthy controls. Global gene-expression data from liver, mammary artery intima media, aortic intima media, and aortic adventitia were obtained from the Advanced Study of Aortic Pathology (ASAP) study of patients undergoing aortic replacement and/or valve repair surgery.25 In silico lookup was also conducted on an eQTL transcriptome database of circulating monocytes (monocytes II) obtained from 1490 unrelated subjects.26
Details of the biobanks and the methods for gene-expression analysis, genotyping, and statistical analysis are provided in the supplemental materials (section 5).
Functional studies
The influence of MUC3 and TRIP6 on PAI-1 gene expression and protein secretion was studied in cultured HuH7 liver cells and the monocyte THP-1 cell line, respectively, using siRNA gene knockdown technology. Details on methods, cell culture, and verification experiments are provided in the supplemental materials (section 6).
Results
Discovery meta-analysis
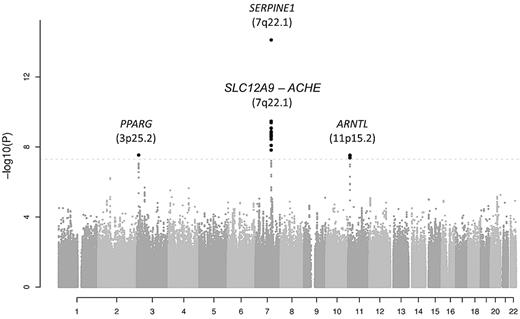
The results for the 2 445 683 meta-analyzed SNPs in the discovery cohorts are shown in Figure 1 organized by chromosome in a Manhattan plot. A quantile-quantile plot is also shown in supplemental Figure 1, with a λ value of 1.012. A total of 24 SNPs passed the genome-wide significance threshold of P = 5.0 × 10−8. All genome-wide significant SNPs were located in 3 chromosomal regions: 3p25.2, 7q22.1 (2 loci), and 11p15.2. The P values for the lead (index) SNPs in these regions are listed in Table 1, and the regional plots are shown in supplemental Figure 2. The MAFs of the top SNPs ranged from 0.10-0.48.
Manhattan plot showing the results for the 2 445 683 meta-analyzed SNPs in the discovery cohorts (a total of 19 683 subjects). SNPs are represented on the x-axis organized by chromosome. On the y-axis, statistical significance is expressed as −log10 of the P values. The horizontal line marks the P = 5.0 × 10−8 threshold of genome-wide significance.
Manhattan plot showing the results for the 2 445 683 meta-analyzed SNPs in the discovery cohorts (a total of 19 683 subjects). SNPs are represented on the x-axis organized by chromosome. On the y-axis, statistical significance is expressed as −log10 of the P values. The horizontal line marks the P = 5.0 × 10−8 threshold of genome-wide significance.
The lead SNP in the region on chromosome 3 (rs11128603, discovery P = 2.9 × 10−8, β = 0.086) is intronic to PPARG encoding peroxisome proliferator-activated receptor γ (supplemental Figure 2A).
The lead SNP on chromosome 7, rs2227631 (discovery P = 7.8 × 10−15, β = 0.076), is a proxy for the previously and consistently reported 4G/5G polymorphism in SERPINE1, the structural gene for PAI-1. A total of 19 SNPs located in a wide region of 266.6 kb between position 100 289 668 and 100 556 258 bp were found to surpass the genome-wide significance threshold. Analysis of the LD structure in this region showed that there are 2 independent significant signals on 7q22.1, one exclusively represented by SNP rs2227631 located in the promoter region of SERPINE1 gene, and another comprising a large LD block spanning 60 387 bp located 206 203 bp upstream of SNP rs2227631 (r2 < 0.10) (supplemental Figure 2B-C and supplemental Figure 3). The lead SNP in the second region on chromosome 7 is rs6976053 (P = 3.4 × 10−10, β = 0.054). The second region on chromosome 7 harbors several genes that contain SNPs with P values surpassing genome-wide significance: SLC12A9, TRIP6, SRRT, UFSP1, and ACHE (supplemental Figure 2C). All significant SNPs in this region are in strong LD with each other and are independent of rs2227631. This finding was confirmed by secondary analyses showing that the second region on chromosome 7 remained significant after conditional analysis on rs2227631, as shown in supplemental Table 5. Of a total of 120 SNPs analyzed in the 7q22.1 region, none had a significant interaction with rs2227631 (interaction P > .05); therefore, we did not find support for a potential effect of rare variants that might drive the 2 observed association signals.
Finally, the strongest association on chromosome 11 was found for rs6486122 (P = 3.0 × 10−8, β = 0.051). Four SNPs in this region showed genome-wide significant association with PAI-1 levels. These SNPs (rs6486122, rs10832027, rs2896635, and rs7947951) are in strong LD with each other and with the other SNPs exhibiting the lowest P values in this region (supplemental Figure 2D), and they are all intronic variants located in the aryl hydrocarbon receptor nuclear translocator-like (ARNTL) gene.
Association results for these 10 SNPs in each individual GWA study are detailed in supplemental Table 6. Secondary analyses with adjustment for BMI conducted in the discovery cohorts on the 10 SNPs selected for examination in the replication cohorts (supplemental Table 5) resulted in attenuation of the statistical significance for most of the genotype-phenotype associations encountered in all 4 PAI-1 loci. It is notable that the association with the 4G/5G proxy SNP rs2227631 was considerably strengthened by adjustment for BMI (from P = 7.8 × 10−15 to P = 3.0 × 10−22).
The total variation in PAI-1 levels in the discovery cohorts accounted for by the 10 SNPs, beyond that contributed by age and sex, ranged between 0.9% and 3.7%.
Replication
For all 10 SNPs followed up, the direction of the genetic effects was identical in the discovery and replication cohorts. The replication P value was less than the prespecified threshold of independent replication, P < .005, for 7 SNPs: rs2227631 (in SERPINE1), all 5 SNPs contained in the second locus on chromosome 7 harboring SLC12A9, TRIP6, SRRT, UFSP1, and ACHE, and rs6486122 (in the ARNTL region). The combined discovery and replication P value was less than 5.0 × 10−8 for all SNPs except for rs11128603 and rs1801282 in the PPARG region (Table 1).
Association with CAD and T2D
For CAD association, we meta-analyzed the 2 European-ancestry cohorts from C4D together with CARDIoGRAM to calculate a combined P value and effect size (Table 2). Two SNPs in ARNTL were nominally associated with CAD; the plasma PAI-1 raising alleles increased the risk of CAD (rs6486122, odds ratio [OR] = 1.04 and 95% confidence interval [CI], 1.01-1.07; rs3816360: OR = 1.03 and 95% CI, 1.01-1.06). These 2 SNPs also showed significant association with T2D in an in silico examination of the DIAGRAM results (rs6486122, OR = 1.06, 95% CI, 1.01-1.10; rs3816360, OR = 1.06, 95% CI, 1.02-1.11). The 2 SNPs in PPARG were also significantly associated with T2D, as has been reported previously, but not with CAD. For both of the T2D-associated loci, the alleles contributing to higher plasma PAI-1 levels increased the risk of T2D.
Association results for the 10 selected SNPs with CAD and T2D
| SNP . | Chr . | Closest Gene . | Type . | Alleles . | CAD* OR (95% CI) . | CAD P . | T2D† OR (95% CI) . | T2D P . |
|---|---|---|---|---|---|---|---|---|
| rs11128603 | 3 | PPARG | Index SNP | A → G | 0.99 (0.95-1.03) | .7 | 1.16 (1.09-1.24) | 1.3 × 10−5 |
| rs1801282 | 3 | PPARG | nsSNP | C → G | 1.00 (0.96-1.04) | 1.0 | 1.15 (1.08-1.22) | 8.0 × 10−6 |
| rs2227631 | 7 | SERPINE1 | Index SNP | A → G | 1.00 (0.97-1.03) | 1.0 | 1.02 (0.98-1.06) | .4 |
| rs6976053 | 7 | ACHE | Index SNP | T → C | 1.00 (0.98-1.03) | .7 | 0.99 (0.95-1.03) | .6 |
| rs2075756 | 7 | TRIP6 | nsSNP | A → G | 1.00 (0.97-1.03) | .8 | 0.98 (0.94-1.03) | .5 |
| rs314376 | 7 | SLC12A9 | Tag SNP | G → A | 1.01 (0.98-1.03) | .6 | 0.99 (0.95-1.03) | .7 |
| rs12672665 | 7 | SRRT | Tag SNP | G → A | 1.00 (0.97-1.02) | .7 | 0.99 (0.95-1.03) | .6 |
| rs3847067 | 7 | ACHE | Tag SNP | A → C | 0.99 (0.96-1.02) | .5 | 0.98 (0.94-1.03) | .5 |
| rs6486122 | 11 | ARNTL | index SNP | T → C | 1.04 (1.01-1.07) | 5.2 × 10−3 | 1.06 (1.01-1.10) | 1.0 × 10−2 |
| rs3816360 | 11 | ARNTL | Tag SNP | C → T | 1.03 (1.01-1.06) | 3.0 × 10−2 | 1.06 (1.02-1.11) | 3.9 × 10−3 |
| SNP . | Chr . | Closest Gene . | Type . | Alleles . | CAD* OR (95% CI) . | CAD P . | T2D† OR (95% CI) . | T2D P . |
|---|---|---|---|---|---|---|---|---|
| rs11128603 | 3 | PPARG | Index SNP | A → G | 0.99 (0.95-1.03) | .7 | 1.16 (1.09-1.24) | 1.3 × 10−5 |
| rs1801282 | 3 | PPARG | nsSNP | C → G | 1.00 (0.96-1.04) | 1.0 | 1.15 (1.08-1.22) | 8.0 × 10−6 |
| rs2227631 | 7 | SERPINE1 | Index SNP | A → G | 1.00 (0.97-1.03) | 1.0 | 1.02 (0.98-1.06) | .4 |
| rs6976053 | 7 | ACHE | Index SNP | T → C | 1.00 (0.98-1.03) | .7 | 0.99 (0.95-1.03) | .6 |
| rs2075756 | 7 | TRIP6 | nsSNP | A → G | 1.00 (0.97-1.03) | .8 | 0.98 (0.94-1.03) | .5 |
| rs314376 | 7 | SLC12A9 | Tag SNP | G → A | 1.01 (0.98-1.03) | .6 | 0.99 (0.95-1.03) | .7 |
| rs12672665 | 7 | SRRT | Tag SNP | G → A | 1.00 (0.97-1.02) | .7 | 0.99 (0.95-1.03) | .6 |
| rs3847067 | 7 | ACHE | Tag SNP | A → C | 0.99 (0.96-1.02) | .5 | 0.98 (0.94-1.03) | .5 |
| rs6486122 | 11 | ARNTL | index SNP | T → C | 1.04 (1.01-1.07) | 5.2 × 10−3 | 1.06 (1.01-1.10) | 1.0 × 10−2 |
| rs3816360 | 11 | ARNTL | Tag SNP | C → T | 1.03 (1.01-1.06) | 3.0 × 10−2 | 1.06 (1.02-1.11) | 3.9 × 10−3 |
Individual SNP results from the CARDIoGRAM and C4D consortia.
Individual SNP results from the DIAGRAM Consortium.
Association with expression in target tissues
Global gene-expression data from relevant tissues were used to explore the functional significance of candidate genes contained in the associated genomic regions. For each of the 10 identified PAI-1–associated SNPs, we investigated associations between genotype and expression levels of all genes located within ± 200 kb of the SNP. A total of 87 genes were contained within these regions. Table 3 reports all significant associations at P < .00408 (ie, false discovery rate = 0.05). In the candidate chromosome 3 region, the selected tag SNP rs1801282 showed a highly significant association with PPARG expression levels in macrophages (P = 1.7 × 10−50). In addition, monocyte expression of 3 genes (TRIP6, UFSP, and EPHB4) located in the chromosome 7 region were found to be significantly associated in 2 independent datasets with the SNPs selected from the meta-analysis of the GWA studies. Similar significant associations were found in macrophages for UFSP. However, these associations were not observed in the other studied cell/tissue types. Conversely, in liver, SNPs located on chromosome 7 showed significant associations with expression levels of MUC3 (P = 1.3 × 10−17) and acetylcholinesterase (ACHE, P = .00028). In the chromosome 11 candidate region, the selected tag SNP rs10832027 showed a significant association with the ARNTL expression levels in monocytes (P = 4.1 × 10−5).
Association between PAI-1–associated SNPs and proximal gene expression in human cells and tissues
| Chr . | SNP (alleles; β; P) . | Gene expression . | Monocytes (n = 849) . | Monocytes II (n = 1490) . | Macrophages (n = 614) . | Mammary artery intima-media (n = 88) . | Liver (n = 152) . | Aortic intima-media (n = 131) . | Aortic adventitia (n = 113) . |
|---|---|---|---|---|---|---|---|---|---|
| 3 | rs1801282 (C → G; 0.069; 1.4 × 10−7) | PPARG | 0.061.7 × 10−2 | NS | 0.38; 1.7 × 10−50 | NS | 1.11; 2.0 × 10−2 | NS | NS |
| 7 | rs314376 (G → A; 0.052; 2.4 × 10−9) | TRIP6 | −0.08; 4.7 × 10−15 | NA; 8.3 × 10−15 | −0.04; 6.2 × 10−3 | NS | NS | NS | NS |
| UFSP1 | −0.04; 3.5 × 10−9 | NA; 1.8 × 10−12 | −0.03; 6.7 × 10−7 | NS | 1.04; 4.0 × 10−2 | NS | NS | ||
| rs2075756 (A → G; 0.058; 2.7 × 10−9) | EPHB4 | −0.14; 2.9 × 10−24 | NA; 3.4 × 10−25 | NS | NS | NS | NS | 1.08; 5.0 × 10−2 | |
| rs6976053 (T → C; 0.054; 3.4 × 10−10) | ACHE | NS | NA | NS | NS | 1.06; 2.8 × 10−4 | NS | NS | |
| MUC3A | NA | NA | NA | NS | 1.16; 1.3 × 10−17 | NS | NS | ||
| 11 | rs10832027 (A → G; 0.051; 3.1 × 10−8) | ARNTL | −0.07; 4.1 × 10−5 | NA | −0.04; 9.2 × 10−3 | NS | NS | NS | NS |
| Chr . | SNP (alleles; β; P) . | Gene expression . | Monocytes (n = 849) . | Monocytes II (n = 1490) . | Macrophages (n = 614) . | Mammary artery intima-media (n = 88) . | Liver (n = 152) . | Aortic intima-media (n = 131) . | Aortic adventitia (n = 113) . |
|---|---|---|---|---|---|---|---|---|---|
| 3 | rs1801282 (C → G; 0.069; 1.4 × 10−7) | PPARG | 0.061.7 × 10−2 | NS | 0.38; 1.7 × 10−50 | NS | 1.11; 2.0 × 10−2 | NS | NS |
| 7 | rs314376 (G → A; 0.052; 2.4 × 10−9) | TRIP6 | −0.08; 4.7 × 10−15 | NA; 8.3 × 10−15 | −0.04; 6.2 × 10−3 | NS | NS | NS | NS |
| UFSP1 | −0.04; 3.5 × 10−9 | NA; 1.8 × 10−12 | −0.03; 6.7 × 10−7 | NS | 1.04; 4.0 × 10−2 | NS | NS | ||
| rs2075756 (A → G; 0.058; 2.7 × 10−9) | EPHB4 | −0.14; 2.9 × 10−24 | NA; 3.4 × 10−25 | NS | NS | NS | NS | 1.08; 5.0 × 10−2 | |
| rs6976053 (T → C; 0.054; 3.4 × 10−10) | ACHE | NS | NA | NS | NS | 1.06; 2.8 × 10−4 | NS | NS | |
| MUC3A | NA | NA | NA | NS | 1.16; 1.3 × 10−17 | NS | NS | ||
| 11 | rs10832027 (A → G; 0.051; 3.1 × 10−8) | ARNTL | −0.07; 4.1 × 10−5 | NA | −0.04; 9.2 × 10−3 | NS | NS | NS | NS |
Associations between 10 SNPs (see Table 2) and proximal genes (< 200 kb) were tested. For each cell and tissue type, β and P values for the association are shown calculated according to a linear additive model. Only SNP-gene combinations with significant associations are reported (FDR < 0.05, P < .00408).
NA indicates that the gene and/or SNP is not available in the dataset; and NS, P > .05 for the association.
Effects of silencing MUC3 and TRIP6 on gene expression and PAI-1 secretion
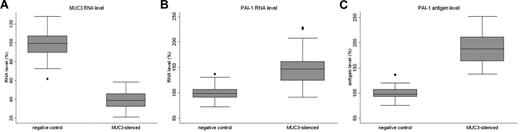
When prioritizing candidate genes for functional studies, we focused on genes with tissue expression levels in the top quartile of the overall gene-expression distribution that showed strong allele-specific associations with the lead-SNP genotype (false discovery rate < 0.05, P < .00408). Of the candidate genes contained in the chromosome 7 locus, only MUC3A expression in liver and TRIP6 expression in monocytes met these criteria. We therefore investigated MUC3 in liver cells and TRIP6 in monocytes using siRNA knock-down in in vitro experiments. In 15 independent experiments (with 3 replicates in each), the mean silencing of MUC3 was −61.0% (Figure 2A), which was accompanied by a highly statistically significant increase in SERPINE1 expression (+45.5%, P < .0001) when RNA was extracted 48 hours after transfection (Figure 2B). Verification experiments using 2 additional MUC3 silencers showed a consistent significant increase in SERPINE1 RNA levels (supplemental materials, section S6). Subsequent experiments (n = 10, with 3 replicates in each) demonstrated a highly significant increase in PAI-1 protein levels in the media collected from MUC3-silenced compared with control cells (increase of 88.8%, P < .0001, Figure 2C). In contrast, despite effective silencing of expression of TRIP6 in monocytes, we observed no significant changes in SERPINE1 expression or PAI-1 production (supplemental materials, section S6).
Box plots showing the differences in expression levels of MUC3 and PAI-1. Box plots (median, interquartile range and 95% CIs) show the differences in expression levels of MUC3 (A) and PAI-1 (B) comparing MUC3-silenced cells with control cells. (C) Differences in PAI-1 released into the media comparing MUC3-silenced cells with control cells. Gene-expression and protein levels are normalized to control (nonsilenced) cells (100%).
Box plots showing the differences in expression levels of MUC3 and PAI-1. Box plots (median, interquartile range and 95% CIs) show the differences in expression levels of MUC3 (A) and PAI-1 (B) comparing MUC3-silenced cells with control cells. (C) Differences in PAI-1 released into the media comparing MUC3-silenced cells with control cells. Gene-expression and protein levels are normalized to control (nonsilenced) cells (100%).
Discussion
This is the first meta-analysis of GWA studies to examine genetic determinants of plasma PAI-1. In our discovery meta-analysis, we identified 4 loci showing genome-wide significant associations with plasma PAI-1 Ag levels that were located in 3 regions on chromosomes 3p25.2, 7q22.1 (2 loci), and 11p15.2. Among the 4 loci, 3 contained obvious or plausible biologic candidate genes for the regulation of PAI-1 (SERPINE1, PPARG, and ARNTL), whereas one (harboring SLC12A9, TRIP6, SRRT, UFSP1, and ACHE) represents a greater mechanistic challenge. Independent replication was obtained for the first chromosome 7 locus encompassing SERPINE1; the second locus on chromosome 7 containing SLC12A9, TRIP6, SRRT, UFSP1, and ACHE; and the third locus on chromosome 11 containing ARNTL. The 3p25.2 locus, however, was not replicated (P = .1), which could be because of lack of power. Based on the observed MAF of 10% and the effect size of 0.031 for the lead SNP at 3p25.2 in the replication samples, our replication sample size of 10 796 subjects only had 35.2% power to replicate the PPARG finding made in the discovery cohorts. Evidence of genotype associations with risk of CAD and T2D was demonstrated for ARNTL by in silico examinations of databases generated by the CARDIoGRAM, C4D, and DIAGRAM consortia. Based on association studies between genotype and gene-expression levels in relevant tissues and gene silencing in liver cells, we identified MUC3 as a strong candidate gene for the second, independent association signal on chromosome 7.
Genes at chromosome 7q22.1
The strongest SNP association with plasma PAI-1 concentration was contributed by rs2227631, which is located in the structural gene for PAI-1 (SERPINE1). This SNP is in strong LD (D′ = 0.97, r2 = 0.78)12 with the widely reported 4G/5G polymorphism in the SERPINE1 promoter. The strong LD between the 2 SNPs renders it exceedingly difficult to distinguish which of the 2 SNPs is functionally responsible for the association with plasma PAI-1. However, functional studies of the 4G/5G polymorphism suggest that the 5G allele contains a binding site for a repressor protein that decreases PAI-1 transcription.27
Interestingly, we found another association signal on chromosome 7 located 206.2 kb upstream of SNP rs2227631, which had not been shown previously to be associated with PAI-1. Despite the proximity of this locus to SERPINE1, analysis of the LD structure in this region and secondary analyses conditional on rs2227631 confirmed that this locus is independent of known variation in SERPINE1. Among the genes contained in the associated region are SLC12A9, TRIP6, SRRT, UFSP1, and ACHE, none of which has been linked to PAI-1 regulation. To refine the association signal, we examined the relationship between PAI-1–associated SNPs and expression levels of genes in this region and nearby using human gene-expression data generated in relevant cells and tissues. Significant associations were found for expression levels of TRIP6, EPHB4, and UFSP1 in monocytes; for UFSP1 in macrophages; and for ACHE and MUC3 in liver. According to our ex vivo target tissue gene-expression analyses, MUC3 expression in the liver is strongly associated with the top PAI-1–associated SNPs in the second chromosome 7 locus (P = 1.3 × 10−17), constituting the strongest reported association in the liver. This finding suggests that MUC3 could have an important role in the regulation of PAI-1 synthesis in the liver. Moreover, our in vitro analyses in HuH7 liver cells showing that silencing of MUC3 increases SERPINE1 expression both at the RNA and protein levels are consistent with this hypothesis.
MUC3 belongs to a gene family encoding mucins, large, heavily glycosylated glycoproteins that exert important functions in the protection of the epithelium. They are also thought to be implicated in signal transduction, regulation of cell growth, cell-cell adhesion, and modulation of the immune system, and MUC3 has been implicated in common complex diseases such as rheumatoid arthritis28 and inflammatory bowel diseases.29 The mechanisms by which MUC3 is implicated in PAI-1 regulation remain to be elucidated. It is notable in this context that there are 2 transcripts from MUC3. Because of the high level of homology between MUC3A and MUC3B (the unique exonic sequences range from 94%-100% identity at the nucleotide level and the introns show approximately 95% identity), the probes used to detect and silence MUC3 expression in this study would recognize either MUC3A or MUC3B. For this reason, we chose to use the generic name for the gene, MUC3. However, it is worth mentioning that the tissue-expression patterns of MUC3A and MUC3B are different. Whereas expression of MUC3B has been detected exclusively in the small and large intestines, that of MUC3A has been observed in the intestines, heart, liver, thymus, prostate, and pancreas.29 Therefore, it is plausible to conclude that only MUC3A was detected and silenced in liver. It should also be emphasized that we cannot exclude that other genes contained in the second region on chromosome 7 are involved in the regulation of PAI-1, such as the ones showing allele-specific expression in monocytes/macrophages. However, we observed no significant changes in SERPINE1 expression when silencing TRIP6, the most likely candidate gene expressed in monocytes/macrophages.
Chromosome 11p15.2 locus containing ARNTL
The locus on chromosome 11 harbors the ARNTL gene, which encodes an important activator protein for circadian rhythm–associated genes. This is of major interest because plasma PAI-1 levels fluctuate in a circadian manner, resulting in reduced fibrinolytic activity during the early morning,30 with the peak in plasma PAI-1 concentration preceding an increased incidence of cardiovascular events.31 Furthermore, circadian variation in SERPINE1 expression in vitro is regulated by heterodimers formed from the circadian clock proteins CLOCK:ARNTL1 and CLOCK:ARNTL2, which bind to the SERPINE1 promoter and up-regulate its expression.32
In the present study, we have demonstrated highly significant associations of SNPs located in the ARNTL locus with both plasma PAI-1 concentrations and ARNTL mRNA levels in monocytes (P = 4.1 × 10−5). These findings confirm previous results obtained in vitro and show for the first time that ARNTL is implicated in PAI-1 regulation in vivo in humans. Furthermore, ARNTL influenced CAD risk and susceptibility to T2D. The CAD finding is in agreement with the previous observation that circadian variation in plasma PAI-1 levels is linked to variation in the incidence of MI31 and indicates that PAI-1 and/or pathways involving ARNTL may be causally related to the risk of CAD in some subjects.
Chromosome 3p25.2 locus containing PPARG
Although not significant in the replication studies and thus not demonstrated as being robustly associated with plasma PAI-1 level, the locus on chromosome 3 identified at a genome-wide significant level at the discovery stage is of considerable interest. It contains several SNPs located in the PPARG gene encoding peroxisome proliferator-activated receptor γ (PPARγ). PPARs are ligand-activated transcription factors that form functional heterodimers with retinoid X receptors and bind to specific peroxisome proliferator response elements in the promoter regions of their target genes. PPARγ is a regulator of adipocyte differentiation that has been implicated in the pathogenesis of obesity, T2D, atherosclerosis, and cancer.33 It is also important for inflammatory responses such as macrophage function and regulates PAI-1 expression in endothelial cells,34 adipose tissue,35 and the liver.36 Interestingly, secondary analysis with adjustment for BMI revealed that variation in BMI had only minor effects on the genotype-phenotype associations at the PPARG locus. In addition, expression analyses showed that the SNPs in the PPARG locus most strongly associated with plasma PAI-1 are also distinctly associated with expression levels of the PPARG transcript in macrophages (P = 1.70 × 10−50). Our results suggest that further studies should be conducted on PPARG as a potentially important regulator of PAI-1 expression independent of the degree of adiposity.
Relevance for cardiovascular disease
In addition to its important inhibitory role in fibrinolysis, PAI-1 is involved in the pathogenesis of obesity, insulin resistance, and T2D and appears to exert a wide range of pleiotropic functions in vascular remodeling, wound healing, fibrosis, tumor angiogenesis, bone remodeling, asthma, rheumatoid arthritis, glomerulonephritis, and sepsis.37 This wide array of functions has contributed to the prevailing uncertainty as to whether PAI-1 is a causal factor in risk for cardiovascular disease. It is notable that in this context, SERPINE1, the gene encoding PAI-1, was not associated with risk of CAD despite showing the strongest SNP association with circulating PAI-1 levels, a finding that is an argument against a causal involvement of PAI-1 in CAD. Against this background, the CAD association demonstrated for ARNTL may be of major significance and warrants further investigation to elucidate whether pathways involving ARNTL, rather than the PAI-1 molecule itself, are causally related to the risk for CAD.
Strengths and limitations
The strengths of this study include the large size (N = 19 599) of the sample used for the discovery meta-analysis, access to gene-expression data in target tissues from human subjects, and the inclusion of functional studies in cell culture systems as a complementary approach to gene identification. Conversely, it should be emphasized that the GWA study strategy based on the SNP arrays used here primarily identifies marginal to small effects of common genetic variations (MAF > 0.10). Low-frequency and rare variants that may influence plasma PAI-1 levels are likely to have escaped detection in the present study and may contribute to the missing heritability, and additional common variants with real effects on plasma PAI-1 levels but smaller effect sizes did not attain the conservative threshold for genome-wide significance. Furthermore, gene-gene and gene-environment interactions, which were not examined, may play an important role in the regulation of PAI-1. It should also be emphasized that the results were generated in European ancestry populations, so additional studies are needed in other ethno-geographic groups. Two of our discovery cohorts (KORA and MARTHA) measured PAI-1 activity and the other cohorts measured PAI-1 Ag. These 2 cohorts comprised only approximately 10% of the total discovery samples, and the effect sizes for all 10 SNPs were consistent and comparable across all 8 discovery cohorts (heterogeneity P > .05). Finally, very few clinical or population-based studies with adequate numbers of prevalent or incident CVD/T2D events have included measurements of plasma PAI-1 levels. Accordingly, the power of a conventional Mendelian randomization study of PAI-1 would unfortunately be low. The best substitute available are PAI-1–associated SNPs in adequately powered genetic case-control studies, such as C4D and Cardiogram, which was conducted in our study.
Conclusions
Using genome-wide, high-coverage genotype data from a total of 19 599 subjects of European ancestry, we identified 4 loci showing genome-wide significant associations with plasma PAI-1 Ag levels and successfully replicated findings for 3 of the loci. Among the 4, 3 loci contained obvious or plausible biologic candidate genes (SERPINE1, PPARG, and ARNTL), whereas one, for which MUC3 was identified as a strong candidate gene in functional studies, represents an entirely new mechanism. Although only a fairly small sample was available for formal replication of the discovery GWA meta-analysis findings, independent replication was obtained for SERPINE1, ARNTL, and the locus on chromosome 7 for which MUC3 was identified as a candidate gene. Evidence of genotype associations with risk of CAD was demonstrated for ARNTL, but not for SERPINE1 and MUC3, indicating that pathways involving ARNTL, rather than the PAI-1 molecule itself, may be causally related to the risk for CAD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
For acknowledgments and financial support, please see the supplemental materials (sections 7 and 8).
National Institutes of Health
Authorship
Contribution: A.H and C.O.J designed the study and organized the project; J.H. and M.S.-L. collected the data and ran the meta-analysis; M.S.-L. conducted the in vitro cell study. J.H., M.S.-L., F.W.A., D.T., A.D.J., N.L.S., S.M.W., W.T., P-E.M., P.v.d.H., Y.L, C.J.O., and A.H. wrote the manuscript; and all authors were involved in individual GWAS analysis and reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anders Hamsten, MD, FRCP, Atherosclerosis Research Unit, Center for Molecular Medicine, Bldg L8:03, Karolinska University Hospital Solna, S-171 76 Stockholm, Sweden; e-mail: anders.hamsten@ki.se; or Christopher J. O'Donnell, MD, MPH, National Heart, Lung and Blood Institute's Framingham Heart Study, 73 Mt Wayte St, Suite 2, Framingham, MA 01702; e-mail: odonnellc@nhlbi.nih.gov.
References
Author notes
J.H. and M.S.-L. contributed equally to this work.
C.J.O. and A.H. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal