Abstract
The P2RY8-CRLF2 fusion defines a particular relapse-prone subset of childhood acute lymphoblastic leukemia (ALL) in Italian Association of Pediatric Hematology and Oncology Berlin-Frankfurt-Münster (AIEOP-BFM) 2000 protocols. To investigate whether and to what extent different clone sizes influence disease and relapse development, we quantified the genomic P2RY8-CRLF2 fusion product and correlated it with the corresponding CRLF2 expression levels in patients enrolled in the BFM-ALL 2000 protocol in Austria. Of 268 cases without recurrent chromosomal translocations and high hyperdiploidy, representing approximately 50% of all cases, 67 (25%) were P2RY8-CRLF2 positive. The respective clone sizes were ≥ 20% in 27% and < 20% in 73% of them. The cumulative incidence of relapse of the entire fusion-positive group was clone size independent and significantly higher than that of the fusion-negative group (35% ± 8% vs 13% ± 3%, P = .008) and primarily confined to the non–high-risk group. Of 22 P2RY8-CRLF2–positive diagnosis/relapse pairs, only 4/8 had the fusion-positive dominant clone conserved at relapse, whereas none of the original 14 fusion-positive small clones reappeared as the dominant relapse clone. We conclude that the majority of P2RY8-CRLF2–positive clones are small at diagnosis and virtually never generate a dominant relapse clone. Our findings therefore suggest that P2RY8-CRLF2–positive clones do not have the necessary proliferative or selective advantage to evolve into a disease-relevant relapse clone.
Introduction
B-cell precursor acute lymphoblastic leukemias (BCP-ALLs) with either a genomic rearrangement or deregulated expression of the cytokine receptor–like factor 2 (CRLF2), or both, practically never concur with ETV6-RUNX1, MLL, TCF3, or BCR-ABL fusions.1-6 They are primarily classified as standard (SR) or high risk (HR) in Children's Oncology Group (COG) protocols and as corresponding SR and intermediate risk (IR) in Italian Association of Pediatric Hematology and Oncology Berlin-Frankfurt-Münster (AIEOP-BFM) protocols, with a significantly increased risk of relapse in non–Down syndrome (DS) patients.1,2,6-9
The 2 most prominent genomic rearrangements are those that either juxtapose the first noncoding exon of P2RY8 to the CRLF2 coding region or put the latter under the control of the IGH@ gene enhancer.1-3,10,11 The P2RY8-CRLF2 fusion in particular results from an approximately 320-kb large interstitial deletion within the pseudoautosomal region 1 (PAR1) that is located on the short arm of both sex chromosomes. Apart from rare CRLF2-activating mutations and additional copies of the respective gene-containing chromosomal region, no other consistent genetic factors are currently known to be associated with a CRLF2 overexpression.1,4-6,8,12 With an incidence of 5% to 7%, cases with a P2RY8-CRLF2 fusion are 2 to 5 times as common as those with an IGH@-CRLF2 translocation and at > 50% they are particularly frequent in DS ALL patients.1-3,10,11
Of note, in AIEOP-BFM studies apparently only half of the P2RY8-CRLF2 fusion-positive cases overexpress CRLF2.8 Conversely, only half of the overexpressing cases in these studies, and also those from COG focusing on HR ALL cases, carry a CRLF2 genomic lesion.1,4-8 This intriguing discrepancy raised the question of why not all CRLF2 fusions cause a similar CRLF2 overexpression, and consequently, whether the fusion or the overexpression might be the preferred predictive factor. The discussion was fueled by the heterogeneous results that were obtained by the various study groups, because they used different patient cohorts and numbers, protocols and observation periods, and screening approaches.1-8,10,11 P2RY8-CRLF2–positive cases, for example, can be ascertained at the genomic level by either fluorescence in situ hybridization (FISH), genomic PCR, or multiplex ligation-dependent probe amplification, and on the RNA level with RT-PCR. CRLF2 expression levels, in contrast, can be determined by various absolute and relative measurement methods with arbitrary cut-off levels for overexpression.
The BFM group originally screened for high CRLF2 expression and found that especially patients with a concurring P2RY8-CRLF2 fusion in the non-HR group had a significantly increased relapse risk.1 These findings were recently corroborated by the AIEOP-BFM 2000 study that concluded that the P2RY8-CRLF2 fusion is the relevant factor.8 Thus, at least for the non-HR groups in the AIEOP-BFM protocols, the clinically most crucial problem in this context seems to have been answered for the time being. Nevertheless, it remains difficult to understand why and how such genomic lesions, even without the appropriate simultaneous evidence of a corresponding functional correlate in form of CRLF2 overexpression, should be able to increase the relapse risk. The most likely explanation is that the extent of the overall expression levels merely reflects or correlates with fluctuating proportions of the fusion-positive clones. Along these lines, one also would expect that fusion-positive clones should have either a selective or proliferative advantage, or both, to be able to directly contribute to disease progression and relapse development.
We addressed these questions with the help of unbiased and highly sensitive qualitative and quantitative PCR analyses to determine the respective clone sizes in the diagnostic and relapse samples, correlated the ensuing results with the corresponding CRLF2 expression levels, and we also checked whether P2RY8-CRLF2 fusions were already present in Guthrie card blood samples that had been taken at birth.
Methods
Patient samples
Frozen viable cells or DNA and RNA were obtained from the diagnostic, remission, and relapse bone marrow samples of children and adolescents with BCP-ALL that had been consecutively enrolled in the BFM ALL 2000 study in Austria over a 10-year period (2000-2010). Leukemia samples (with at least 85% blasts) were obtained from the Austrian Study center on institutional review approval and with approval of the ethics committee. Informed consent for tissue banking and research studies was obtained from patients, their parents, legal guardians, or a combination in accordance with the Declaration of Helsinki. Cytogenetic and genetic characterization of all samples was performed as a routine diagnostic workup. Final risk assignment was performed according to published clinical and minimal residual disease criteria.13-15
Detection of P2RY8-CRLF2
Genomic PCR.
Genomic PCR for the PAR1 deletion that results in a P2RY8-CRLF2 fusion was performed as described previously.2 In brief, 100 ng of genomic DNA was amplified using Hot-start Phusion High-Fidelity DNA polymerase II (Biozym) on a C1000 thermal cycler (Bio-Rad Laboratories). Primer sequences and PCR conditions can be found in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). PCR products were separated on agarose gels, purified (QIAquick PCR purification kit; QIAGEN), and sequenced from both sides (Eurofins MWG Operon). Sequences were confirmed from a second independent PCR amplification.
Long-distance multiplex PCR.
A set of 21 primers, covering intron 1 of P2RY8 and positioned at a distance of 1500 to 2000 bp apart from each other, and a second set of 30 primers, distributed over a 50-kb region upstream of CRLF2 exon 1, were designed (sequences and PCR conditions are available on request). Genomic DNA from 6 transcript-positive cases without the common P2RY8-CRLF2 fusion was amplified with various primer mixes containing 10 forward (or reverse) primers each and 1 of the reverse (or forward) primers. PCR products were visualized on agarose gels and sequenced. Sequences were verified from a second independent PCR amplification.
Quantification of genomic P2RY8-CRLF2.
The amounts of genomic fusion were determined by TaqMan PCR with a 7500 real-time PCR system (Applied Biosystems) using the following primer-probe combination: P2RY8_q_fw, CRLF2_q_rv, and P-C_q_pr (corresponding primer sequences are shown in supplemental Table 1). A standard curve with 10-log dilution steps from 1 × 10-1 to 1 × 10-5 from 1 case with one copy of the P2RY8-CRLF2 in virtually all leukemia cells was used for quantification of the genomic fusion in test samples. Samples were run in triplicates. In the 2 relapse samples with a blast count of < 85%, the genomic fusion levels were related to the respective blast counts.
PCR for P2RY8-CRLF2 transcripts.
Total leukemic cell RNA was extracted using the RNeasy mini kit (QIAGEN). We reverse transcribed 2 μg of RNA with Moloney murine leukemia virus reverse transcriptase (Promega) and amplified using Hot-start Phusion HF polymerase II using published primers.2 Cycling conditions were optimized to detect low abundant transcripts (provided in supplemental Table 1).
Quantitative real-time PCR for CRLF2 expression.
CRLF2 and endogenous control ALAS1 gene expression levels were quantified by quantitative (q)RT-PCR using a 7500 real-time PCR system (Applied Biosystems) and the following primer and probe combinations: CRLF2_q_fw, CRLF2_q_rv and CRLF2_q_pr, as well as ALAS1_q_fw, ALAS1_q_rv, and ALAS1_q_pr (primer sequences are shown in supplemental Table 1). Samples were run in triplicates, and transcript levels are indicated as fold changes relative to an arbitrary calibrator (mean CRLF2 expression of peripheral blood mononuclear cells [MNCs] from 8 healthy controls) using comparative Ct analysis (2−ΔΔCt) after normalization to endogenous ALAS1 expression. The standard curve was obtained by dilutional series of RNA/cDNA extracted from the erythropoietin-stimulated cell line TF1 (DSMZ).
To compare the fold changes obtained from this qRT-PCR, quantification of CRLF2 expression also was performed using inventoried assays for CRLF2 (Hs00845692_m1) and EEF2 (Hs00157330_m1; both from Applied Biosystems). The assays were run at the default conditions recommended by the manufacturer. Samples were run in triplicates, and transcript levels were calculated as ΔCt values, essentially as described by Chen et al.6 The same cut-off of ≤ 8 ΔCt for CRLF2 overexpression was applied.
Detection of the P2RY8-CRLF2 fusion in Guthrie cards.
DNA was extracted from one-third of a neonatal blood spot (Guthrie card), essentially as described previously with minor variations (see supplemental Figure 5).16-18 From cases with the previously published breakpoints, the genomic fusion was determined in whole genome–amplified Guthrie card DNA by TaqMan qRT-PCR as described in “Quantification of genomic P2RY8-CRLF2.” For the case with the newly identified breakpoint, a patient-specific nested 2-round PCR was established using a standard curve (10-log dilutions) from the respective diagnostic leukemia DNA. P2RY8_fw1 and CRLF2_rv10 were used for the first round and P2RY8_fw10 was used with a patient-specific reverse primer for the second-round PCR (primer sequences in supplemental Table 1). The patient-specific primer was placed over the breakpoint. PCR products were size separated on agarose gels, purified, and sequenced.
Statistical analyses
Associations between categorical variables were examined using Fisher exact test. Event-free survival (EFS) and overall survival (OS) were analyzed according to the Kaplan-Meier method and compared by the log-rank test.19 The cumulative incidence of relapse (CIR) functions were calculated by the method of Kalbfleish and Prentice and compared using the Gray test.20,21
Spearman rank correlation was used to study the correlation between genomic P2RY8-CRLF2 amounts and CRLF2 expression and between the corresponding CRLF2 expression levels determined by 2 different methods.
Results
Screening for genomic P2RY8-CRLF2 breakpoints and identification of novel deletion breakpoints upstream of CRLF2
We restricted our screening to those 277 BCP-ALL cases in the Austrian ALL BFM 2000 treatment protocol (n = 555, including 25 individuals with DS) that neither had an HD karyotype nor an MLL, BCR-ABL, or ETV6-RUNX1 gene rearrangement. Moreover, we excluded all 8 cases with an IGH@-CRLF2 translocation from these analyses. DNA was available from 268 (97%) of these cases, including 20 with DS. The clinical and biologic data of these cases are summarized in Table 1.
Patient characteristics and response to treatment according to P2RY8-CRLF2 fusion in 268 patients with childhood BCP-ALL
| . | P2RY8-CRLF2 . | P2RY8-CRLF2 + . | ||||
|---|---|---|---|---|---|---|
| Positive . | Negative . | P . | Major sub/clone* . | Minor subclone* . | P . | |
| No. of patients (%) | 67 (25.0) | 201 (75.0) | 18 (6.7) | 49 (18.3) | ||
| Sex (%) | .724 | .974 | ||||
| Male | 37 (55.2) | 106 (52.7) | 10 (55.6) | 27 (55.1) | ||
| Female | 30 (44.8) | 95 (47.3) | 8 (44.4) | 22 (44.9) | ||
| Age, y (%) | .003 | .11 | ||||
| 1-10 | 55 (82.1) | 126 (62.7) | 17 (94.4) | 38 (77.6) | ||
| ≥ 10 | 12 (17.9) | 75 (37.3) | 1 (5.6) | 11 (22.4) | ||
| WBC count (%) | .561 | .872 | ||||
| < 50 000/μL | 55 (82.1) | 171 (85.1) | 15 (83.3) | 40 (81.6) | ||
| ≥ 50 000/μL | 12 (17.9) | 30 (14.9) | 3 (16.7) | 9 (18.4) | ||
| Pred response (%) | .279 | .796 | ||||
| Good | 64 (95.5) | 183 (91.0) | 17 (94.4) | 47 (95.9) | ||
| Poor | 3 (4.5) | 17 (8.5) | 1 (5.6) | 2 (4.1) | ||
| Not available | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | ||
| Treatment arm (%) | .343 | .671 | ||||
| Standard risk | 19 (28.4) | 39 (19.4) | 6 (33.3) | 13 (26.6) | ||
| Intermediate risk | 39 (58.2) | 129 (64.2) | 11 (61.1) | 28 (57.1) | ||
| High risk | 7 (10.4) | 29 (14.4) | 1 (5.6) | 6 (12.2) | ||
| Death before risk stratification | 2 (3.0) | 3 (1.5) | 0 (0.0) | 2 (4.1) | ||
| Other | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | ||
| DS (%) | < .001 | .009 | ||||
| Yes | 15 (22.4) | 5 (2.5) | 8 (44.4) | 7 (14.3) | ||
| No | 52 (77.6) | 196 (97.5) | 10 (55.6) | 42 (85.7) | ||
| Relapse (%) | .006 | .137 | ||||
| Yes | 16 (23.9) | 21 (10.4) | 2 (11.1) | 14 (28.6) | ||
| No | 51 (76.1) | 180 (89.6) | 16 (88.9) | 35 (71.4) | ||
| Incidence of relapse non-DS cases (CIR)† (%) | 13/52 (35 ± 8) | 21/196 (13 ± 3) | .008 | 2/10 (38 ± 21) | 11/42 (35 ± 9) | .825 |
| Standard risk | 2/13 (24 ± 15) | 2/37 (7 ± 5) | .216 | 0/3 | 2/10 (24 ± 15) | .739 |
| Intermediate risk | 9/31 (38 ± 10) | 14/127 (13 ± 4) | .015 | 2/6 (40 ± 22) | 7/25 (38 ± 12) | .855 |
| High risk | 2/7 (50 ± 25) | 5/28 (22 ± 9) | .540 | 0/1 (0) | 2/6 (50 ± 25) | n.a. |
| Median follow-up, y (range) | 5.1 (1.1-10.9) | 5.2 (1.1-10.6) | n.a. | 5.4 (1.5-9.8) | 5.1 (1.1-10.9) | n.a. |
| . | P2RY8-CRLF2 . | P2RY8-CRLF2 + . | ||||
|---|---|---|---|---|---|---|
| Positive . | Negative . | P . | Major sub/clone* . | Minor subclone* . | P . | |
| No. of patients (%) | 67 (25.0) | 201 (75.0) | 18 (6.7) | 49 (18.3) | ||
| Sex (%) | .724 | .974 | ||||
| Male | 37 (55.2) | 106 (52.7) | 10 (55.6) | 27 (55.1) | ||
| Female | 30 (44.8) | 95 (47.3) | 8 (44.4) | 22 (44.9) | ||
| Age, y (%) | .003 | .11 | ||||
| 1-10 | 55 (82.1) | 126 (62.7) | 17 (94.4) | 38 (77.6) | ||
| ≥ 10 | 12 (17.9) | 75 (37.3) | 1 (5.6) | 11 (22.4) | ||
| WBC count (%) | .561 | .872 | ||||
| < 50 000/μL | 55 (82.1) | 171 (85.1) | 15 (83.3) | 40 (81.6) | ||
| ≥ 50 000/μL | 12 (17.9) | 30 (14.9) | 3 (16.7) | 9 (18.4) | ||
| Pred response (%) | .279 | .796 | ||||
| Good | 64 (95.5) | 183 (91.0) | 17 (94.4) | 47 (95.9) | ||
| Poor | 3 (4.5) | 17 (8.5) | 1 (5.6) | 2 (4.1) | ||
| Not available | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | ||
| Treatment arm (%) | .343 | .671 | ||||
| Standard risk | 19 (28.4) | 39 (19.4) | 6 (33.3) | 13 (26.6) | ||
| Intermediate risk | 39 (58.2) | 129 (64.2) | 11 (61.1) | 28 (57.1) | ||
| High risk | 7 (10.4) | 29 (14.4) | 1 (5.6) | 6 (12.2) | ||
| Death before risk stratification | 2 (3.0) | 3 (1.5) | 0 (0.0) | 2 (4.1) | ||
| Other | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0.0) | ||
| DS (%) | < .001 | .009 | ||||
| Yes | 15 (22.4) | 5 (2.5) | 8 (44.4) | 7 (14.3) | ||
| No | 52 (77.6) | 196 (97.5) | 10 (55.6) | 42 (85.7) | ||
| Relapse (%) | .006 | .137 | ||||
| Yes | 16 (23.9) | 21 (10.4) | 2 (11.1) | 14 (28.6) | ||
| No | 51 (76.1) | 180 (89.6) | 16 (88.9) | 35 (71.4) | ||
| Incidence of relapse non-DS cases (CIR)† (%) | 13/52 (35 ± 8) | 21/196 (13 ± 3) | .008 | 2/10 (38 ± 21) | 11/42 (35 ± 9) | .825 |
| Standard risk | 2/13 (24 ± 15) | 2/37 (7 ± 5) | .216 | 0/3 | 2/10 (24 ± 15) | .739 |
| Intermediate risk | 9/31 (38 ± 10) | 14/127 (13 ± 4) | .015 | 2/6 (40 ± 22) | 7/25 (38 ± 12) | .855 |
| High risk | 2/7 (50 ± 25) | 5/28 (22 ± 9) | .540 | 0/1 (0) | 2/6 (50 ± 25) | n.a. |
| Median follow-up, y (range) | 5.1 (1.1-10.9) | 5.2 (1.1-10.6) | n.a. | 5.4 (1.5-9.8) | 5.1 (1.1-10.9) | n.a. |
n.a. indicates not analyzed.
Clonal size definition: major, ≥ 20%; minor, < 20%.
Five-year cumulative incidence of relapse is given in parentheses. Gray test was used to estimate strata homogeneity.
Applying a breakpoint region-specific genomic PCR, we primarily identified 61/268 (22.8%) cases with a P2RY8-CRLF2 fusion (Figure 1A). In line with previous reports, DNA sequence analysis revealed unique breakpoints with the insertion of template independent nucleotides.2,10,22
Identification of P2RY8-CRLF2 in childhood BCP-ALL cases. (A) PCR products of the genomic P2RY8-CRLF2 fusion of representative cases. Patient identification is indicated at the top of the gel: ad, nontemplate control; pB, peripheral blood MNCs; and SM, size marker. Left part of the gel (samples 360-903) shows PCR products from cases that were later defined as harboring the P2RY8-CRLF2 fusion in a major clone. Right part of the gel (samples 460-906) shows PCR products from ALL cases with P2RY8-CRLF2 in a minor subclone. (B) PCR for the fusion transcripts of corresponding cases. Note that sample 365 had no genomic PCR product but a distinct band for the transcript. (C) PCR products of the 6 cases with the newly discovered breakpoints upstream of CRLF2. A vertical line was inserted to indicate that different gels, run in parallel, from identical experiments were pasted together.
Identification of P2RY8-CRLF2 in childhood BCP-ALL cases. (A) PCR products of the genomic P2RY8-CRLF2 fusion of representative cases. Patient identification is indicated at the top of the gel: ad, nontemplate control; pB, peripheral blood MNCs; and SM, size marker. Left part of the gel (samples 360-903) shows PCR products from cases that were later defined as harboring the P2RY8-CRLF2 fusion in a major clone. Right part of the gel (samples 460-906) shows PCR products from ALL cases with P2RY8-CRLF2 in a minor subclone. (B) PCR for the fusion transcripts of corresponding cases. Note that sample 365 had no genomic PCR product but a distinct band for the transcript. (C) PCR products of the 6 cases with the newly discovered breakpoints upstream of CRLF2. A vertical line was inserted to indicate that different gels, run in parallel, from identical experiments were pasted together.
Screening for the corresponding P2RY8-CRLF2 transcripts in 234/268 (87%) cases with available RNA confirmed the presence of the genomic fusion in all instances, but this screening also revealed a distinct RT-PCR product in another 6 (including 3 with DS), apparently fusion breakpoint-negative, cases (Figure 1B). With the help of long-distance multiplex PCR, we subsequently discovered 2 new breakpoints 2.1 kb and 0.1 kb upstream of CRLF2 exon 1 (Figure 1C and supplemental Figure 1, sequences submitted to dbVar, study accession nstd72). In line with the previously described breakpoints,2,22 they had a sequence pattern that indicates that they were also V(D)J recombination mediated. Comprising approximately 10% of the entire fusion-positive cohort, these new breakpoints increased the frequency of P2RY8-CRLF2 cases to 67/268 (25%), translating into an overall frequency of 12.1% in BCP-ALL cases, a figure that is approximately twice as high as the 5% to 7% reported by others.1,3,5,11 Fifteen of those P2RY8-CRLF2–positive cases had a DS, relating to 60% of the entire ALL DS group, a rate that is comparable to the 50% reported previously.2,10
In contrast to other studies,1,5,8 we found that P2RY8-CRLF2–positive cases were considerably younger than fusion-negative cases (Table 1), a finding that can be explained by the fact that we had excluded ETV6-RUNX1–positive and HD cases with a relatively younger median age, from our fusion-negative control population.
Not only did we detect more P2RY8-CRLF2–positive cases than groups that used probably less sensitive RT-PCR– or FISH-based screening techniques,1,3,11 we also noted that, irrespective of identical DNA and RNA/cDNA inputs, the amount of derived PCR products varied substantially. This observation led us to investigate whether these discrepancies might result from dissimilar proportions of P2RY8-CRLF2–positive cell clones in the various samples (Figure 1A-B).
Quantification of P2RY8-CRLF2 genomic fusions and CRLF2 transcripts
We determined the respective sizes of the P2RY8-CRLF2–positive clones by quantifying the genomic breakpoint region with a qRT-PCR whose sensitivity reached a detection limit of 1 × 10-5 (supplemental Figure 2). To enable the comparison of the ensuing results with other less sensitive methods, such as FISH (general detection limit of 5%-10%; a respective comparison is shown in supplemental Table 2) and multiplex ligation–dependent probe amplification (detection limit of 20%), we split the samples into 2 groups using a 20% cut-off. Cases with equal or > 20% of genomic fusion-positive cells are referred to as major clones, whereas those with < 20% (including only 1 case with ∼ 10% cells), which may be below the detection limit using the aforementioned methods, are referred to as minor subclones. The major clone-positive group comprised 18 (27%) and the minor subclone-positive group 49 (73%) of all fusion-positive cases (Table 1).
Next, we precisely quantified CRLF2 transcripts under standardized conditions to enable their comparison with the quantity of fusion-positive cells. As shown in Figure 2A, these 2 parameters are highly correlated (correlation coefficient [CC], 0.43; P = .0018).
Quantification of P2RY8-CRLF2 genomic breakpoints and CRLF2 transcripts. (A) Relation between abundance of genomic fusion and CRLF2 expression in 56 P2RY8-CRLF2–positive ALL cases. Genomic DNA was used for the fusion (triangles; empty symbols, cases with P2RY8-CRLF2 in a minor subclone [< 20% of leukemia population]; filled symbols, cases with P2RY8-CRLF2 in a major clone [≥ 20% of leukemia population]); and mRNA for CRLF2 transcript quantification (squares). Black symbols indicate cases in remission; and red, relapse cases. (Left) Y-axis: normalized CRLF2 levels plotted relative to CRLF2 expression in peripheral blood MNC. (Right) Amounts of genomic fusion in percentage of the leukemia population. X-axis: ALL cases ordered from the lowest to highest CRLF2 expression. A dashed horizontal line has been inserted at the 50-fold CRLF2 overexpression defined by the lowest expression in cases with the fusion in a major population of the leukemia clone and no expression in all other genetic ALL subtypes. The black vertical line separates cases with low (left) and high (right) CRLF2 expression. (B) Quantification of CRLF2 expression by real-time PCR: comparison of 2 methods. (Left) Y-axis: ΔCt values of CRLF2 expression using the Applied Biosystems kit and the criteria for high expression of ΔCt ≤ 8.0. (Right) Fold changes of CRLF2 expression, as in panel A. X-axis: ALL cases ordered from the lowest to highest percentage of CRLF2 expression as in panel A. Two horizontal lines were inserted to show the respective limits for overexpression by each method; dashed, the 50-fold overexpression as in panel A; dotted, CRLF2 overexpression at ΔCt ≤ 8.0. (C) Relation between abundance of genomic fusion and CRLF2 expression by the inventoried assays for CRLF2 and EEF2. The dotted horizontal line indicates the CLRF2 expression at ΔCt 8.0 as in panel B. ALL cases ordered from the lowest to highest CRLF2 expression as in panel B. The black vertical line separates cases with low (left) and high (right) CRLF2 expression.
Quantification of P2RY8-CRLF2 genomic breakpoints and CRLF2 transcripts. (A) Relation between abundance of genomic fusion and CRLF2 expression in 56 P2RY8-CRLF2–positive ALL cases. Genomic DNA was used for the fusion (triangles; empty symbols, cases with P2RY8-CRLF2 in a minor subclone [< 20% of leukemia population]; filled symbols, cases with P2RY8-CRLF2 in a major clone [≥ 20% of leukemia population]); and mRNA for CRLF2 transcript quantification (squares). Black symbols indicate cases in remission; and red, relapse cases. (Left) Y-axis: normalized CRLF2 levels plotted relative to CRLF2 expression in peripheral blood MNC. (Right) Amounts of genomic fusion in percentage of the leukemia population. X-axis: ALL cases ordered from the lowest to highest CRLF2 expression. A dashed horizontal line has been inserted at the 50-fold CRLF2 overexpression defined by the lowest expression in cases with the fusion in a major population of the leukemia clone and no expression in all other genetic ALL subtypes. The black vertical line separates cases with low (left) and high (right) CRLF2 expression. (B) Quantification of CRLF2 expression by real-time PCR: comparison of 2 methods. (Left) Y-axis: ΔCt values of CRLF2 expression using the Applied Biosystems kit and the criteria for high expression of ΔCt ≤ 8.0. (Right) Fold changes of CRLF2 expression, as in panel A. X-axis: ALL cases ordered from the lowest to highest percentage of CRLF2 expression as in panel A. Two horizontal lines were inserted to show the respective limits for overexpression by each method; dashed, the 50-fold overexpression as in panel A; dotted, CRLF2 overexpression at ΔCt ≤ 8.0. (C) Relation between abundance of genomic fusion and CRLF2 expression by the inventoried assays for CRLF2 and EEF2. The dotted horizontal line indicates the CLRF2 expression at ΔCt 8.0 as in panel B. ALL cases ordered from the lowest to highest CRLF2 expression as in panel B. The black vertical line separates cases with low (left) and high (right) CRLF2 expression.
Moreover, evaluation of the CRLF2 expression levels in various subgroups revealed that a 50-fold overexpression clearly demarcates major clone-positive P2RY8-CRLF2 cases from all P2RY8-CRLF2–negative cases. Only 20% to 30% of those with a minor subclone also have expression levels above this threshold, indicating that CRLF2 in these leukemic clones might either be extremely up-regulated or, alternatively, that CRLF2 expression in such instances is not entirely governed by the P2RY8-CRLF2–positive clone alone. The expression levels of the remaining minor subclone-positive cases largely overlap with those of HD cases. The lowest expression levels, in contrast, are seen in ETV6-RUNX1–positive cases, whereas those in a genetically undefined control group were marginally higher (supplemental Figure 3).
We also validated the results with a second system for measuring and calculating the relative amounts of CRLF2 transcripts, and we found a nearly perfect match not only between the 2 methods (CC, −0.90; P = < .0001; Figure 2B) but also with the second method and the corresponding amounts of genomic fusion products (CC, −0.33; P = .0274; Figure 2C).
Predictive value of P2RY8-CRLF2 in BCP-ALL cases
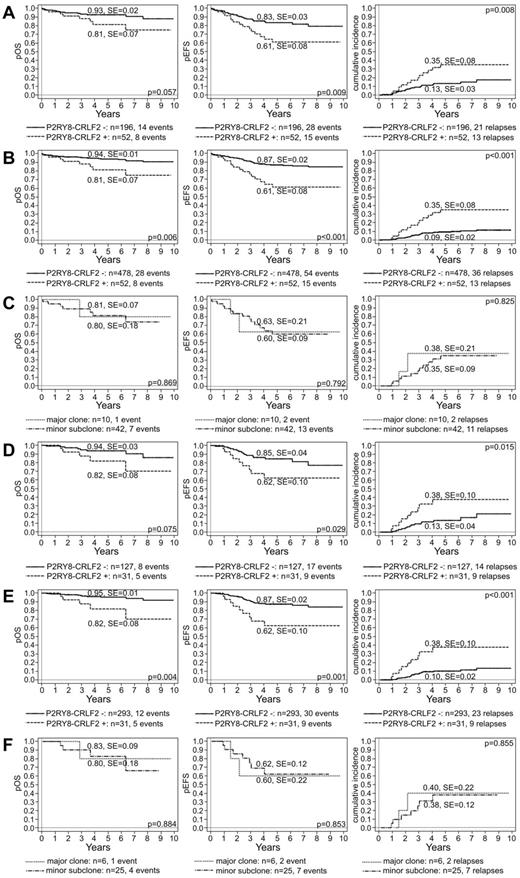
Because of the small number of DS cases in our study and the reported lack of a relapse predictive power of P2RY8-CRLF2 in this particular entity, we excluded them from further analysis.2,10 We first evaluated whether P2RY8-CRLF2–positive cases have indeed an increased relapse risk and how this may impair the OS in the selected cohort of our 248 cases. We found that P2RY8-CRLF2–positive non-DS ALL cases had a worse 5-year probability for overall survival (pOS), event-free survival (pEFS), and CIR than fusion-negative cases (pOS, 81% ± 7% vs 93% ± 2%, P = .057; pEFS, 61% ± 8% vs 83% ± 3%, P = .009; and CIR, 35% ± 8% vs 13 ± 3%, P = .008; Figure 3A). The worse prognosis also remained valid when the total study cohort was analyzed (pOS, 81% ± 7% vs 94% ± 1%, P = .006; pEFS, 61% ± 8% vs 87% ± 2%, P < .001; and CIR, 35% ± 8% vs 9 ± 2%, P < .001; Figure 3B). Next, we checked to which extent the size of the fusion-positive clone might modify the relapse risk, and we found that ALL cases with a fusion-positive minor subclone did as poorly as those with a major involvement (pOS, 80% ± 18% and 81% ± 7%, P = .869; pEFS = 60% ± 9% and 63% ± 21%, P = .792; and CIR, 38% ± 21% and 35% ± 9%, P = .825, for major and minor fusion-positive subclones, respectively; Figure 3C). We were not able to associate any particular cytogenetic abnormality with this increased relapse risk.
Clinical outcome of children with BCP-ALL enrolled in the BFM ALL 2000 protocol in Austria according to P2RY8-CRLF2. Kaplan-Meier estimates of 5-year pOS (left), pEFS (middle), and CIR (cumulative incidence; right) according to the presence of P2RY8-CRLF2 and its proportion in the leukemia of 248 screened non-DS cases (A,C) and of all 530 non-DS cases (B) enrolled in the treatment study. Analogous analyses of ALL cases treated according to the IR treatment arm (D-F).
Clinical outcome of children with BCP-ALL enrolled in the BFM ALL 2000 protocol in Austria according to P2RY8-CRLF2. Kaplan-Meier estimates of 5-year pOS (left), pEFS (middle), and CIR (cumulative incidence; right) according to the presence of P2RY8-CRLF2 and its proportion in the leukemia of 248 screened non-DS cases (A,C) and of all 530 non-DS cases (B) enrolled in the treatment study. Analogous analyses of ALL cases treated according to the IR treatment arm (D-F).
Because the predictive value of P2RY8-CRLF2 is particularly pronounced in the non-SR/non-very-HR groups in the COG and the IR group in the AIEOP-BFM protocols,1,2,6-9 we also analyzed the role of minimal residual disease risk group assignment as well as of applied treatment. We found that fusion-positive patients in the IR group had a significantly higher relapse risk than fusion-negative patients (CIR, 38% ± 10% vs 13% ± 4%; P = .015; pOS, 82% ± 8% vs 94% ± 3%, P = .075; pEFS, 62% ± 10% vs 85% ± 4%, P = .029; Figure 3D). Again, the results remained valid when the fusion-positive cases were compared with the entire IR cohort of 324 cases (pOS, 82% ± 8% vs 95% ± 1%, P = .004; pEFS, 62% ± 10% vs 87% ± 2%, P = .001; and CIR, 38% ± 10% vs 10 ± 2%, P < .001; Figure 3E). In addition, within the IR group, the size of the fusion harboring clone was completely irrelevant for the respective relapse risk and overall survival (pOS, 80% ± 18% vs 83% ± 9%, P = .884; pEFS, 60% ± 22% vs 62% ± 12%, P = .853; and CIR, 40% ± 22% vs 38 ± 12%; P = .855; for major vs minor clone fusion-positive ALL cases; Figure 3F). Comparison of fusion-positive and fusion-negative ALL cases in the SR group revealed a similar trend but did not reach the necessary statistical significance (pOS, 92% ± 8% vs 100% ± 0%, P = .092; pEFS, 68% ± 16% vs 93% ± 5%, P = .046; and CIR, 24% ± 15% vs 7% ± 5%, P = .216; supplemental Figure 3A). HR cases were not analyzed because the number was too small (Table 1).
Assessment of the P2RY8-CRLF2–harboring clone stability in ALL relapse cases
Presuming that P2RY8-CRLF2–positive cell clones are directly involved in relapse development, they should have a certain proliferative or selective advantage, or both and it would be expected that they prevail in the respective relapses. We therefore studied 22 relapse samples that derived from 8 ALL cases with a fusion-positive major clone (36.4%) and 14 with a fusion-positive minor subclone at diagnosis. The clinical and biologic data of these cases are summarized in Table 2.
Clinical characteristic of 22 P2RY8-CRLF2–positive relapse cases
| Pt ID . | Sex . | Age at dx, y . | MRD . | Rem, mo . | Rel site . | Source . | Clonal stability of P2RY8-CRLF2 . |
|---|---|---|---|---|---|---|---|
| P2RY8-CRLF2 major clone | |||||||
| 715 | M | 8.90 | IR | 17 | BM | AT | Conserved |
| 737 | M | 9.50 | IR | 25 | BM + CNS | AT | Reduced to minor subclone |
| 1* | F | 2.00 | IR | 33 | BM | DE | Lost |
| 4 | F | 1.40 | IR | 42 | BM | DE | Lost |
| 2* | M | 8.70 | IR | 63 | BM | DE | Conserved |
| 3 | M | 4.60 | LR | 65 | BM | DE | Lost |
| 5* | F | 4.10 | IR | 67 | BM | DE | Conserved |
| 6 | F | 2.70 | LR | 98 | BM | DE | Conserved |
| P2RY8-CRLF2 minor subclone | |||||||
| 775 | M | 13.20 | IR | 11 | BM | AT | Lost |
| 391 | F | 12.70 | IR | 12 | BM | AT | Conserved |
| 814 | F | 4.60 | LR | 18 | BM | AT | Lost |
| 721 | M | 2.50 | IR | 20 | testis | AT | Conserved |
| 778 | M | 4.20 | IR | 20 | BM | AT | Lost |
| 810 | M | 4.90 | IR | 28 | BM + CNS | AT | Lost |
| 579 | M | 1.30 | IR | 35 | BM | AT | Lost |
| 460 | F | 5.70 | IR | 36 | BM | AT | Lost |
| 571 | M | 9.20 | IR | 44 | BM | AT | Lost |
| 456 | M | 12.00 | IR | 48 | BM | AT | Conserved |
| 545 | M | 4.70 | LR | 55 | BM | AT | Lost |
| 564* | M | 8.25 | LR | 55 | BM | AT | Different minor subclone |
| 417* | F | 9.75 | 60 | BM | AT | Conserved | |
| 243* | F | 10.40 | IR | 71 | BM | AT | Conserved |
| Pt ID . | Sex . | Age at dx, y . | MRD . | Rem, mo . | Rel site . | Source . | Clonal stability of P2RY8-CRLF2 . |
|---|---|---|---|---|---|---|---|
| P2RY8-CRLF2 major clone | |||||||
| 715 | M | 8.90 | IR | 17 | BM | AT | Conserved |
| 737 | M | 9.50 | IR | 25 | BM + CNS | AT | Reduced to minor subclone |
| 1* | F | 2.00 | IR | 33 | BM | DE | Lost |
| 4 | F | 1.40 | IR | 42 | BM | DE | Lost |
| 2* | M | 8.70 | IR | 63 | BM | DE | Conserved |
| 3 | M | 4.60 | LR | 65 | BM | DE | Lost |
| 5* | F | 4.10 | IR | 67 | BM | DE | Conserved |
| 6 | F | 2.70 | LR | 98 | BM | DE | Conserved |
| P2RY8-CRLF2 minor subclone | |||||||
| 775 | M | 13.20 | IR | 11 | BM | AT | Lost |
| 391 | F | 12.70 | IR | 12 | BM | AT | Conserved |
| 814 | F | 4.60 | LR | 18 | BM | AT | Lost |
| 721 | M | 2.50 | IR | 20 | testis | AT | Conserved |
| 778 | M | 4.20 | IR | 20 | BM | AT | Lost |
| 810 | M | 4.90 | IR | 28 | BM + CNS | AT | Lost |
| 579 | M | 1.30 | IR | 35 | BM | AT | Lost |
| 460 | F | 5.70 | IR | 36 | BM | AT | Lost |
| 571 | M | 9.20 | IR | 44 | BM | AT | Lost |
| 456 | M | 12.00 | IR | 48 | BM | AT | Conserved |
| 545 | M | 4.70 | LR | 55 | BM | AT | Lost |
| 564* | M | 8.25 | LR | 55 | BM | AT | Different minor subclone |
| 417* | F | 9.75 | 60 | BM | AT | Conserved | |
| 243* | F | 10.40 | IR | 71 | BM | AT | Conserved |
Pt indicates patient; dx, initial diagnosis; MRD, minimal residual disease risk classification; Rem, remission; Rel, relapse; BM, bone marrow; and CNS, central nervous system.
Down syndrome.
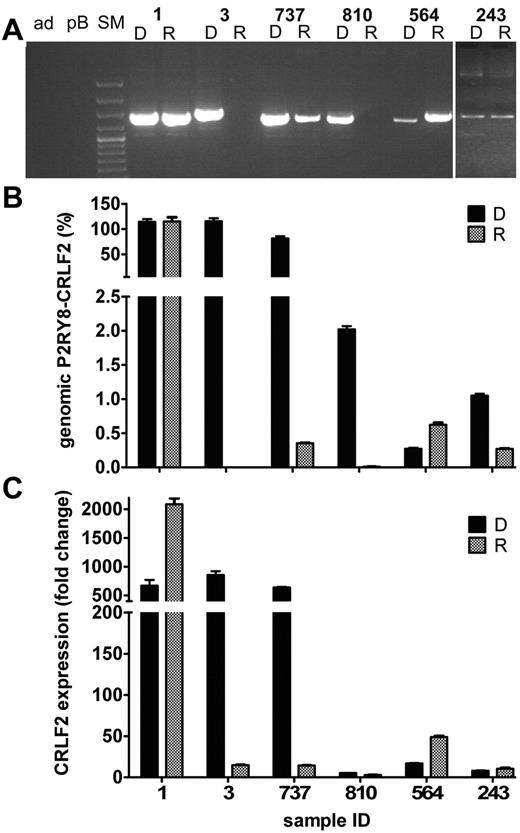
The behavior of P2RY8-CRLF2–positive clones was highly divergent and not at all predictable (Table 2 and supplemental Table 3). Only 4 of 8 cases with a fusion-positive major clone retained the same clone in a similar proportion at relapse. In 1 further case, the fusion-positive clone was still present at relapse but much smaller. In the remaining 3 cases a P2RY8-CRLF2 fusion was neither detected by genomic PCR nor by RT-PCR in the relapse samples anymore. However, also none of the small P2RY8-CRLF2–positive clones that were present at diagnosis in 14 ALL cases evolved into a dominant clone at relapse. Instead, in 7 of them the fusion-positive subclone had disappeared and in another case it was replaced by a novel fusion-positive clone that had an entirely different breakpoint sequence. The PAR1 deletion at relapse was smaller (breakpoint 451 bp distal to P2RY8 exon 1) than that at diagnosis (breakpoint 316 bp distal to P2RY8 exon 1), precluding that it had derived from the initial fusion. In the remaining 6 cases, the fusion-positive clone reappeared in relapse, albeit in an amount that was proportional to that at diagnosis. Representative examples of the various patterns of the respective quantities of fusion-positive clones and their corresponding CRLF2 transcript levels are depicted in Figure 4. The correlation between the CRLF2 expression levels and the amount of genomic fusion in the relapse samples was similar to the correlation established at diagnosis.
Patterns of genomic P2RY8-CRLF2 and CRLF2 transcripts of fusion-positive ALL relapse cases. The 6 different patterns of clonal relation between initial diagnosis (D) and relapse (R) leukemia are depicted. (A) Genomic PCR products for P2RY8-CRLF2 in representative cases. Sample identification is indicated at the top of the figure: ad, nontemplate control; pB, peripheral blood MNCs; and SM, size marker. Cases 1, 3, and 737 harbor the fusion in a major clone at initial diagnosis, and it was either conserved (1), lost (3), or reduced to a minor subclone (737) at relapse. In cases 810, 564, and 243 with a fusion-positive minor subclone at initial diagnosis, the fusion was also either lost (810), preserved (243), or replaced by a different fusion (564) at relapse. Quantification of the genomic P2RY8-CRLF2 fusion (indicated in percentage of leukemia cells at the y-axis; B) and the corresponding CRLF2 transcript levels (fold changes of normalized expression of the samples over the expression in peripheral blood MNCs; C). Sample identification is given at the bottom of the graph.
Patterns of genomic P2RY8-CRLF2 and CRLF2 transcripts of fusion-positive ALL relapse cases. The 6 different patterns of clonal relation between initial diagnosis (D) and relapse (R) leukemia are depicted. (A) Genomic PCR products for P2RY8-CRLF2 in representative cases. Sample identification is indicated at the top of the figure: ad, nontemplate control; pB, peripheral blood MNCs; and SM, size marker. Cases 1, 3, and 737 harbor the fusion in a major clone at initial diagnosis, and it was either conserved (1), lost (3), or reduced to a minor subclone (737) at relapse. In cases 810, 564, and 243 with a fusion-positive minor subclone at initial diagnosis, the fusion was also either lost (810), preserved (243), or replaced by a different fusion (564) at relapse. Quantification of the genomic P2RY8-CRLF2 fusion (indicated in percentage of leukemia cells at the y-axis; B) and the corresponding CRLF2 transcript levels (fold changes of normalized expression of the samples over the expression in peripheral blood MNCs; C). Sample identification is given at the bottom of the graph.
Backtracking the P2RY8-CRLF2 fusion to birth
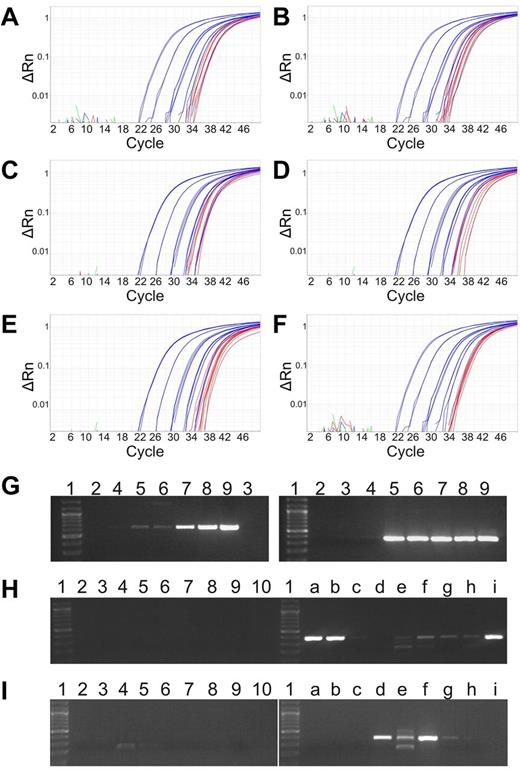
Given the apparent secondary nature of the P2RY8-CRLF2 fusion and the relatively young age of the affected children at diagnosis (Table 1), we were particularly interested in checking whether this lesion might be already present at birth. We therefore used breakpoint-specific genomic PCR to amplify DNA that was extracted from the archived dried neonatal blood spots of 7 cases with a fusion-positive major clone, and we were able to identify the respective lesion in all of them. The amplification blots of qRT-PCR and gels of these cases are depicted in Figure 5A through I and in the corresponding standard curves in supplemental Figure 5A through F. In case 887, the 2 clonotypic TRDV2-TRAJ29 and IGHV2-IGHJ4 that were identified at initial diagnosis were also present at birth (supplemental Figure 6). Moreover, in case 873 that had 1 of the novel fusion breakpoints and therefore needed different PCR conditions, the comparison of native and whole genome–amplified Guthrie card DNA yielded identical results (Figure 5H-I).
Detection of the P2RY8-CRLF2 in neonatal blood spots. Amplification plots of the qRT-PCR of genomic P2RY8-CRLF2 using whole genome–amplified genomic Guthrie card DNA of 6 ALL cases: 400 (A), 737 (B), 802 (C), 833 (D), 841 (E), and 887 (F). Standard curves showing a quantitative range of 1 × 10-5 (blue) and PCR products of Guthrie card DNA from 4 aliquots in triplicates (red). PCR products for P2RY8-CRLF2 with a newly identified genomic breakpoint of case 873. (G) Amplification of the genomic breakpoint. Dilution series of genomic leukemia DNA. (Left) First round. (Right) Second round. The fusion can be detected at the single cell level at 1 × 10-5 (lane 5). Lane 1, size marker; lane 2, H2O; lane 3, DNA mix from peripheral blood MNCs of healthy donors; and lanes 4 to 9, 10-log dilutions from 1 × 10-6 to 1 × 10-1. Amplification of P2RY8-CRLF2 in native (H) and whole genome–amplified Guthrie card (I) DNA. Lane 1, size marker; lane 2, H2O; lane 3, control DNA mix; and lanes 4 to 10, Guthrie cards DNA from a newborn who did not develop ALL later. a to i, PCR products from aliquots of the patients' Guthrie card DNA.
Detection of the P2RY8-CRLF2 in neonatal blood spots. Amplification plots of the qRT-PCR of genomic P2RY8-CRLF2 using whole genome–amplified genomic Guthrie card DNA of 6 ALL cases: 400 (A), 737 (B), 802 (C), 833 (D), 841 (E), and 887 (F). Standard curves showing a quantitative range of 1 × 10-5 (blue) and PCR products of Guthrie card DNA from 4 aliquots in triplicates (red). PCR products for P2RY8-CRLF2 with a newly identified genomic breakpoint of case 873. (G) Amplification of the genomic breakpoint. Dilution series of genomic leukemia DNA. (Left) First round. (Right) Second round. The fusion can be detected at the single cell level at 1 × 10-5 (lane 5). Lane 1, size marker; lane 2, H2O; lane 3, DNA mix from peripheral blood MNCs of healthy donors; and lanes 4 to 9, 10-log dilutions from 1 × 10-6 to 1 × 10-1. Amplification of P2RY8-CRLF2 in native (H) and whole genome–amplified Guthrie card (I) DNA. Lane 1, size marker; lane 2, H2O; lane 3, control DNA mix; and lanes 4 to 10, Guthrie cards DNA from a newborn who did not develop ALL later. a to i, PCR products from aliquots of the patients' Guthrie card DNA.
Discussion
Several study groups have analyzed the clinical and biologic relevance of P2RY8-CRLF2 positivity, CRLF2 overexpression, or both, albeit with only partly concordant results.1-8 The comparison and interpretation of these data remain difficult not only because treatment regimens, patient numbers and observation periods varied but also because heterogeneous target populations, variable screening techniques, and ascertainment algorithms were used for these analyses. Moreover, all conclusions are based on the mutual but unproven understanding that the P2RY8-CRLF2 fusion is a primary lesion that should mark the predominant leukemic clone both at diagnosis and, based on circumstantial evidence, also relapse. We therefore set out to explore these open issues and assumptions in an unbiased systematic and comprehensive manner.
Our analyses with PCR methods that were optimized for the high-sensitivity quantification of P2RY8-CRLF2 fusions at the DNA and RNA level, as well as the corresponding CRLF2 expression levels in a representative cohort of childhood ALL patients at diagnosis and relapse and also in DNA from neonatal blood spots, revealed a clear-cut relationship between these parameters. Our most important discovery was the unexpected heterogeneity of P2RY8-CRLF2–positive clone sizes and their astonishing inert behavior. This finding has important biologic and clinical implications, especially if size—these clones are usually very small—is being taken into account. It not only necessitates the reinterpretation of CRLF2 expression data and requires a critical reconsideration of the envisioned role of the respective clones in disease and relapse development as well as of P2RY8-CRLF2 as a clinical risk parameter but also demands a careful reassessment of the appropriateness of potential therapeutic interventions that might be developed to target P2RY8-CRLF2–associated signal transduction pathways.23
To increase the sensitivity of both DNA- and RNA-based P2RY8-CRLF2 detection methods, we first modified published primer sets2 and adapted the PCR conditions accordingly. The concomitant evaluation of the respective fusion products on the DNA and RNA level eventually led to the identification of 2 new breakpoints, which increased the number of cases that are now detectable on the genomic level by 10%. Moreover, the complete concordance between these 2 complementary approaches reassured us that we had indeed detected all cases. As evidenced by our discovery rate, which was approximately twice as high as the rate achieved in previous studies, the combined application of DNA/RNA screening definitely surpasses any other currently applied ascertainment procedures.1-5,8,10,11
The precise quantification of P2RY8-CRLF2–positive cell clones at the genomic level at diagnosis uncovered 2 distinct groups with large and small clone sizes, respectively. Their distribution at diagnosis as well as their involvement in disease recurrence was highly erratic. Out of the original 8 major clones 4 persisted, 3 disappeared, and 1 reverted to a minor clone. In contrast, none of the minor subclones evolved into a major clone, 8 were lost, 5 persisted, and 1 was replaced with a clone that had a different breakpoint (Table 2). Of the only 2 matched diagnosis/relapse pairs that had been analyzed previously, both had apparently also lost the P2RY8-CRLF2–positive clone at relapse.8 In line with various other leukemia clone-specific markers, we also confirmed that at least in cases with a major P2RY8-CRLF2–positive clone the fusion is already present at birth.16-18,24-27 Apart from that, however, P2RY8-CRLF2–positive clones do not seem to have any particular proliferative or selective advantage and therefore also not the necessary fitness to evolve into a disease-relevant relapse clone. This view is supported by experiments showing in various model systems that neither fusion nor CRLF2 overexpression alone is sufficient to transform cells without the concomitant expression of certain activating mutations, such as those affecting the JAK2, CRLF2, or IL7R genes.2-4,10,11,28
We also found a high concordance rate between P2RY8-CRLF2–positive clone sizes and the corresponding CRLF2 expression levels. The fact that this was achieved with 2 different transcript quantification techniques—one technique relating the CRLF2 normalized expression level to the expression levels of peripheral blood MNCs from healthy donors and the other relating the expression to the transcript level of an internal housekeeping control gene—proved that both approaches are valid and that the results are also interchangeable. All cases with a major clone as well as 20% to 30% of those with a minor clone expressed CRLF2 at least 50-fold higher than peripheral blood MNCs, a level that was not reached by any of the P2RY8-CRLF2-negative controls and therefore served as our cut-off for overexpression. Although our control group was relatively small (n = 54), we did not identify any CRLF2-overexpressing cases. This observation is in sharp contrast to several other studies that found that up to half of the CRLF2-overexpressing cases lack CRLF2 rearrangements.1,4-6,8,29 The common attribute of such CRLF2-overexpressing cases without CRLF2 rearrangement is a “BCR-ABL–like” gene expression profile whose special characteristic is an activated JAK/STAT pathway.4,7 Apart from this currently difficult to understand discrepancy, however, the established tight association between clone size and expression level can probably explain several other incongruities that are commonly encountered in studies that merely tried to correlate P2RY8-CRLF2 positivity with expression levels without the respective quantification.
Finally, our most perplexing result was that the overall disease recurrence risk of our P2RY8-CRLF2–positive cases was within the range that had been reported previously in other AIEOP/BFM studies1,8 ; notwithstanding the fact that we identified nearly twice as many cases, most of which had negligible clone sizes at diagnosis and no participation in disease recurrence.
We are now confronted with 3 partly conflicting and thus difficult to harmonize views of how P2RY8-CRLF2 fusions might shape the various levels of the respective disease process, namely a genomic, a clonal and a clinical level.
CRLF2 rearrangements are the first known step in a transformation process that further requires CRLF2 overexpression and at least certain cooperating mutations in specific genes to attain the biochemical and functional changes that, typically, also comprise activated JAK/STAT and PI3K/mTOR signaling pathways.2,10,23,28 The cases used in these functional studies were selected from expression profiles. It can therefore be safely assumed that they had either a P2RY8-CRLF2–positive major clone or an especially high expressing IGH@-CRLF2 fusion, the latter of which is also found in the cell lines used.3,23
In contrast to what might intuitively be expected from such transformed cells, our quantitative analyses of P2RY8-CRLF2–positive cell clones do not support the notion that these functional changes really equip these cells with the necessary fitness to successfully compete with their P2RY8-CRLF2–negative leukemic counterparts in vivo. Thus, although the P2RY8-CRLF2 fusions may drive the cellular transformation process, it seems highly improbable that they also drive the entire leukemic process. Instead, we suggest that the P2RY8-CRLF2 fusion is virtually always a secondary lesion in form of a passenger mutation, a notion that is strongly corroborated by the fact that these lesions always require concomitant mutations to even become functionally relevant. To explain the fact that, contrary to what would be expected and deduced from our laboratory results, P2RY8-CRLF2 fusions are nevertheless a clear clinical risk factor, 1 or several higher ranking yet unidentified primary changes have to be invoked that should be closely associated with or even trigger the manifestation of CRLF2 rearrangements. Considering the particularly strong tendency of ALL patients with a constitutional trisomy 21 to acquire CRLF2 rearrangements, such a concept is perhaps not as far-fetched as it might seem at first sight.2,10 It is worth noting that such an acquired initiating change probably already exists in the form of an intrachromosomal amplification of chromosome 21 (iAMP21). With a concurrence rate of 25%, it is at present the only known primary abnormality that has a nearly analogous affinity with P2RY8-CRLF2 fusions as a constitutional trisomy 21.5 A remote alternative explanation for an independent role of minor P2RY8-CRLF2–positive clones would be that they alone could perhaps render their fusion-negative leukemic counterparts resistant through a paracrine route in a similar manner as has been demonstrated for minor mutant BCR-ABL–positive clones in comparable situations.30 However, the absence of P2RY8-CRLF2–positive clones in most relapses hardly supports such a possibility.
We conclude that our comprehensive detailed quantitative analyses of P2RY8-CRLF2–positive leukemias revealed some novel and intriguing insight into the performance of the involved cell clones that were not readily deducible from the predicted function of fusion-carrying cells. Conversely, the respective distribution pattern and inert behavior of such clones still stands in sharp contrast with the clinical experience of P2RY8-CRLF2 as HR parameter. Thus, our findings deliver some valid answers and good explanations for some hitherto controversial issues that arose from the heterogeneous data sets, and they also open up new venues for further research with some important implications for the organization of experimental setups as well as the diagnostic workup and clinical management of patients. Taking into account that P2RY8-CRLF2 is most likely a pure secondary change that is usually present only in small clones and is commonly lost in relapses, it seems unlikely that therapies aiming to eliminate such clones can indeed prevent disease recurrences, at least in the currently proposed manner.23 Finally, we believe that precise quantitative information about clone sizes and expression levels will be indispensable in the future to reasonably and accurately interpret and compare experimentally, diagnostically and clinically obtained CRLF2 data.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all physicians, nurses, parents, and patients in Austria who made this work possible; Astrid Mecklenbräuker and Marion Zeginigg for providing clonotypic Ig/TCR rearrangements; and Evgenia Glogova for statistical analysis.
This work was supported in part by the ÖNB Jubiläumsfonds 14129 (M.M.), Clinical Investigator–driven grant 2010-03 (A.A., G.M., O.A.H., R.P.-G.), and the Help and Hope Foundation in collaboration with KIK Textilien und Non-Food GmbH Austria. This is a project of the international BFM study group.
Authorship
Contribution: M.M. designed and performed experiments, interpreted data, and drafted the manuscript; A.A. provided patient material and clinical data and performed respective analyses; S.F., C.N., R.G., S.B., and S.K. performed experiments; L.J.R. performed FISH; C.E., K.S., G.C., and M.S. contributed patient samples and clinical and biological data; D.K. provided Guthrie cards; U.P. performed statistical analyses; G.M. was responsible for the clinical study, provided patient samples and clinical data, and contributed to the design of the project; O.A.H. interpreted data, contributed to study design, and wrote the manuscript; R.G.-P. conceived and supervised the study, interpreted data, and wrote the manuscript; and all authors read and agreed to the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Renate Panzer-Grümayer, Leukemia Biology Group, Children's Cancer Research Institute, Zimmermannplatz 10, Room 110, 1090 Vienna, Austria; e-mail: renate.panzer@ccri.at.


![Figure 2. Quantification of P2RY8-CRLF2 genomic breakpoints and CRLF2 transcripts. (A) Relation between abundance of genomic fusion and CRLF2 expression in 56 P2RY8-CRLF2–positive ALL cases. Genomic DNA was used for the fusion (triangles; empty symbols, cases with P2RY8-CRLF2 in a minor subclone [< 20% of leukemia population]; filled symbols, cases with P2RY8-CRLF2 in a major clone [≥ 20% of leukemia population]); and mRNA for CRLF2 transcript quantification (squares). Black symbols indicate cases in remission; and red, relapse cases. (Left) Y-axis: normalized CRLF2 levels plotted relative to CRLF2 expression in peripheral blood MNC. (Right) Amounts of genomic fusion in percentage of the leukemia population. X-axis: ALL cases ordered from the lowest to highest CRLF2 expression. A dashed horizontal line has been inserted at the 50-fold CRLF2 overexpression defined by the lowest expression in cases with the fusion in a major population of the leukemia clone and no expression in all other genetic ALL subtypes. The black vertical line separates cases with low (left) and high (right) CRLF2 expression. (B) Quantification of CRLF2 expression by real-time PCR: comparison of 2 methods. (Left) Y-axis: ΔCt values of CRLF2 expression using the Applied Biosystems kit and the criteria for high expression of ΔCt ≤ 8.0. (Right) Fold changes of CRLF2 expression, as in panel A. X-axis: ALL cases ordered from the lowest to highest percentage of CRLF2 expression as in panel A. Two horizontal lines were inserted to show the respective limits for overexpression by each method; dashed, the 50-fold overexpression as in panel A; dotted, CRLF2 overexpression at ΔCt ≤ 8.0. (C) Relation between abundance of genomic fusion and CRLF2 expression by the inventoried assays for CRLF2 and EEF2. The dotted horizontal line indicates the CLRF2 expression at ΔCt 8.0 as in panel B. ALL cases ordered from the lowest to highest CRLF2 expression as in panel B. The black vertical line separates cases with low (left) and high (right) CRLF2 expression.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/26/10.1182_blood-2012-07-443218/4/m_zh89991200260002.jpeg?Expires=1767708246&Signature=119FouTkbpY0Moi7he~OjCj81BYsqSF2PG8VZSg4F4vSEbiyXWJ68a~~cQNAWUkZYMwPyWqv2h0PJFNZGLRN4kFxRo6arTnOnc~S87k5xVWMJ3HAHaICJnQCTJ8~LLAmCnyts5ZJCU~N4bPkBLLCsd6JmJR79Aj1O7ActjQDwCIxwSUuXFfCHKuJmAGFK0ulOh1czBabcJahUQSKW3YbzxFOATQOvyIdPBkuhnVZ4JM2HezzKvxlsRin3S23x-NFsNgLX0R-WAI96Tvxxu6~nUE2W3UzDQrmwyqoQ~FVbHoDJ65rLpml8Km9MC5XBnUric0LwX0duRCWKc0K8zlwzw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal