Abstract
We have used whole exome sequencing to compare a group of presentation t(4;14) with t(11;14) cases of myeloma to define the mutational landscape. Each case was characterized by a median of 24.5 exonic nonsynonymous single-nucleotide variations, and there was a consistently higher number of mutations in the t(4;14) group, but this number did not reach statistical significance. We show that the transition and transversion rates in the 2 subgroups are similar, suggesting that there was no specific mechanism leading to mutation differentiating the 2 groups. Only 3% of mutations were seen in both groups, and recurrently mutated genes include NRAS, KRAS, BRAF, and DIS3 as well as DNAH5, a member of the axonemal dynein family. The pattern of mutation in each group was distinct, with the t(4;14) group being characterized by deregulation of chromatin organization, actin filament, and microfilament movement. Recurrent RAS pathway mutations identified subclonal heterogeneity at a mutational level in both groups, with mutations being present as either dominant or minor subclones. The presence of subclonal diversity was confirmed at a single-cell level using other tumor-acquired mutations. These results are consistent with a distinct molecular pathogenesis underlying each subgroup and have important impacts on targeted treatment strategies. The Medical Research Council Myeloma IX trial is registered under ISRCTN68454111.
Introduction
The t(4;14) is seen in 11% of presenting myeloma and defines a unique subtype associated with a poor clinical outcome.1 In contrast to other molecular subtypes, it is seen at lower frequencies in MGUS, suggesting that the genes deregulated by the t(4;14) rapidly push plasma cells to a more aggressive phenotype.2,3 Although FGFR3 was initially suggested to be the critical driver gene in t(4;14) myeloma, it is lost by deletion of the der(14) chromosome in 25% to 30% of cases, focusing attention on the other deregulated gene MMSET.4,5 MMSET is a member of a family of genes that have histone methyltransferase activity. Other members of the family are known oncogenes relevant to the pathogenesis of acute leukemia, providing strong support for the central importance of this gene in myeloma.6 Pathologically, MMSET has been shown to disrupt the distribution of dimethylation marks on histone H3 throughout the genome and is relevant to disease progression.7-9 A specific DNA methylation pattern associated with t(4;14) samples also is thought to be pathologically relevant, although the relationship of these patterns to changes in histone methylation marks is uncertain.10
The t(4;14) alone is insufficient to lead directly to myeloma, being seen in monoclonal gammopathy of undetermined significance (MGUS) and smoldering myeloma.2,3 Therefore, it is likely that there are collaborating mutations at other sites throughout the genome and that the spectrum of these mutations will be unique to this subtype of disease. In this respect, the changes in histone and DNA methylation seen in this subset may affect both the spectrum and number of mutations. It also has been suggested that MMSET functions in DNA repair.8 Consequently, based on these arguments we hypothesized that there may be a unique mutational spectrum associated with t(4;14) myeloma.
Until recently, the optimum way of identifying collaborating mutations has been to study regions of copy number abnormality (CNA) within which there are relevant mutational events.5,11,12 Comprehensive mapping analyses show that CNA are common, the most frequent being deletions of 1p, 6q, 8p, 12p, 13q, 14q, 16q, and 17p.5,13 Mutational analysis of genes within regions of homozygous deletion has led to the discovery of key genes involved in the pathogenesis of myeloma, including FAM46C on 1p, BIRC2/BIRC3 on 11q, RB1 on 13q, TRAF3 on 14q, CYLD on 16q, and TP53 on 17p.5,14-19 Despite the success of this approach, we have not, as yet, identified a specific pattern associated with the t(4;14) cases. More recently, whole genome and whole exome–based sequencing (WES) approaches have been developed that provide a means of global mutational screening that have given us the ability to define mutations outside of regions of CNA. In addition to identifying the range of mutations present within a tumor, this technology can provide semiquantitative analysis of the size of the clonal population carrying the abnormality.20 This technical advantage allows for the characterization of the tumor substructure, whereas previous tools such as gene mapping arrays have only analyzed the predominant clone.
The first results generated from the application of next generation sequencing approaches were carried out on a mix of presentation and relapse samples, and they confirmed previous hypotheses suggesting that there are critical pathways that are deregulated leading to the malignant transformation of plasma cells.14 This study did not focus on the t(4;14) subgroup for which we understand in more detail the basis of its molecular etiology, and as such the changes associated with this group remain unknown. We have integrated genome-wide single-nucleotide polymorphism (SNP)–based mapping array data with WES derived from a series of presenting cases of t(4;14) myeloma and compared the mutations present with a group of cases defined by the t(11;14). The t(11;14) is a good comparator group because it also has a clear pathogenesis that is defined by the deregulation of cyclin D1. The aim of this work was to inform whether the collaborating mutational events in these different etiological settings are the same or different as well as how these events are acquired leading to tumor evolution and treatment resistance.
Methods
Exome sequencing
WES was performed on 22 presentation myeloma samples entered in the Medical Research Council (MRC) Myeloma IX trial consisting of 10 t(4;14) samples and 12 t(11;14) samples. All patients consented to storage and use of tissue in research studies, and the study was approved by the National Research Ethics Service. The MRC Myeloma IX trial is registered under ISRCTN68454111. CD138-positive bone marrow plasma cells were selected to a purity of more than 95%, as determined by cytospin, using magnetic-assisted cell sorting (Miltenyi Biotec). Tumor DNA and RNA were extracted using the AllPrep kit (QIAGEN), and the DNA concentration was assessed using Pico-green (Invitrogen). Nontumor DNA was extracted from a white cell pellet from peripheral blood samples using the FlexiGene kit (QIAGEN).
Libraries were prepared from tumor and nontumor DNA from the same patient and were sequenced to identify acquired single-nucleotide variations (SNVs) and indels in the tumor sample. We used 50 ng of genomic DNA to capture the exome using the SureSelect Human All Exon 50Mb target enrichment set (Agilent Technologies). We have validated this approach previously and shown it to have parity with approaches using larger starting amounts of DNA.21 In brief, 50 ng of DNA was fragmented on a Covaris E-series sonicator to a mean length of 200 to 400 bp, as assessed on a High Sensitivity DNA Bio-analyzer chip (Agilent Technologies). Fragmented DNA underwent end repair and A-tailing before ligation of Illumina adaptor sequences (Illumina). The prep underwent subsequent PCR amplification for 8 cycles, and then 250 ng of amplified product was hybridized overnight to the SureSelect Human All Exon baits (Agilent Technologies). After hybridization and washing, the captured DNA underwent a further 8 cycles of PCR. Purified libraries were sequenced on a GAIIx sequencer (Illumina), giving 76-bp paired end reads. Samples were sequenced to a median depth after deduplication of 61 ×, with 99% more than 1 × and 85% more than 20 × exomic coverage. After base calling and quality control metrics, the raw fastq reads were aligned to the reference human genome.
Exome analysis
The Genome Analysis Toolkit was used to call SNVs.22 These variant calls were recalibrated, and soft filters were applied to remove potential false-positives using dbSNP 132, HapMap 3.3, and the 1000 Genomes Project as truth sets. Variants that occurred in both the normal and tumor samples were filtered out, and the tumor-specific variants were annotated using ANNOVAR.23 Tumor-acquired indels and deleted exons also were called. Full details of data analysis are in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In addition to the identification of commonly affected genes, functional annotation enrichment analysis was used to identify commonly affected pathways.
Mutation validation
Confirmation of WES variants was performed on whole genome–amplified DNA. We amplified 50 ng of genomic DNA in a linear manner using the Repli-g kit (QIAGEN) to generate ∼ 40 μg of amplified DNA. Samples included the 22 samples that had undergone WES as well as an additional 127 samples from the MRC Myeloma IX study. Individual PCR primer sequences and conditions can be found in supplemental Methods. Mutations in NRAS, KRAS, and BRAF were confirmed on an individual sample basis.
Copy number and gene expression analysis
GeneChip Mapping 500K Array sets (Affymetrix) were performed as described previously.5,15,24 The SNP genotypes and inferred copy numbers were obtained using GTYPE and dChip. Data have been deposited into Gene Expression Omnibus under accession GSE21349.5 U133 Plus 2.0 gene expression arrays (Affymetrix) had been performed previously using total RNA from these samples. In total, 260 samples were available to determine the proportion of samples expressing that gene. Genes were placed in bins according to the proportion of samples expressing them, and then the number of variants was counted for each bin. Genes defined as having spiked expression in myeloma were defined according to the criteria outlined in a prior publication.25
Single-cell sorting and genotyping
Single cells from a patient were sorted on an FACSAria cell sorter (BD Biosciences) using a panel of antibodies known to discriminate abnormal from normal plasma cells26 : anti–CD45-PB (Dako UK), anti–CD38-FITC (Caltag MedSystems), anti–CD138-PO (Caltag MedSystems), anti–CD56-PE (Caltag MedSystems), anti–CD19-PE–Cy7 (Beckman Coulter), anti–IGK-APC (Dako UK), and anti–λ-APC–Cy7 (BD Biosciences). Abnormal plasma cells were identified as CD38+, CD138+, CD45−, CD56−, CD19−, and IGK+. We sorted 270 single cells on 3 96-well plates (90 cells/plate). In parallel, lymphocytes from a healthy donor patient also were sorted to serve as wild-type control for further genotyping analysis 5 cells/plate). Finally, 1 well/plate was left empty to serve as no-template control for the analysis.
This novel approach for single-cell multiplex real-time quantitative (Q)–PCR analysis was followed (N.E.P., L. Ermini, E. Papaemmanuil, G. Vijayaraghavan, I. Titley, A. Ford, L. Kearney, M. Greaves, Multiplexing genetic analysis of single cancer cells; manuscript in preparation). In brief, single cells were sorted directly into lysis buffer. Specific (DNA) target amplification (STA) was performed before Q-PCR. This multiplex STA reaction involves the simultaneous amplification of target regions of interest using custom-designed TaqMan assays. Genotyping assays specific for the mutations of interest were custom-designed according to manufacturer's guidelines. The STA product was diluted before Q-PCR interrogation for mutations using the 96.96 dynamic microfluidic array and the BioMark HD (Fluidigm) as recommended by the manufacturer. Genotyping analysis was performed by Genotyping SNP Version 3.0.2 software (Fluidigm). Hierarchical clustering was performed using Pearson correlation and average linkage on the Rock platform.27
Results
Background rates of mutation in the 2 molecular subgroups
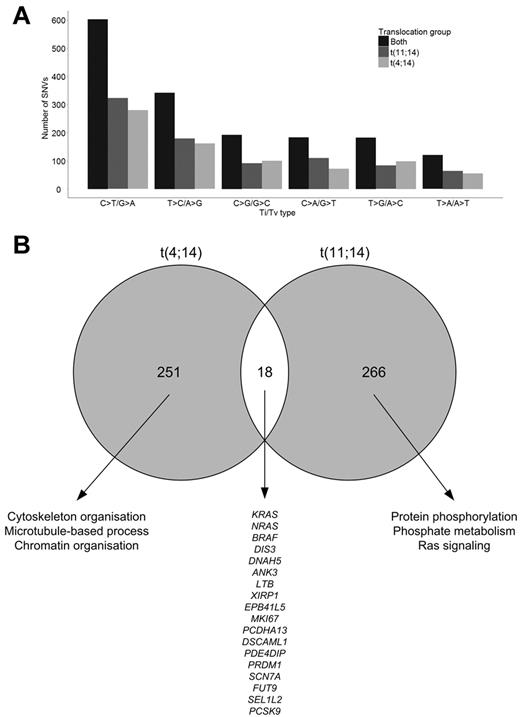
After accounting for the variants in the nontumor DNA, there were, on average, 74 tumor-specific SNVs per sample (range, 54-99), including all captured sequences comprising exonic and surrounding intronic regions. There was a median of 36 exonic SNVs per sample (range, 27-64), and of these 24.5 were nonsynonymous (NS; range, 15-41). Although there was not a marked difference in the frequency of SNVs between the t(11;14) and t(4;14) groups, the number of SNVs was consistently higher in the t(4;14) samples (Table 1; supplemental Figure 3). The composition of somatic variations across both groups shows a predominance of C·G→T·A transitions (37%) and T·A→C·G transitions (20%; Figure 1A). A high proportion of C·G→T·A transitions has been noted previously14 and is because of spontaneous deamination of methyl-C residues to uracil that is subsequently replaced by thymine. Although we have shown previously that t(4;14) samples have more CpG methylation than other cytogenetic groups, in this analysis we found no significant difference in the rates of transition mutations between t(4;14) and t(11;14) samples, suggesting that this is not responsible for the pattern of mutations seen. There were in total 39 acquired indels over the 22 samples (mean = 1.77; range, 0-4). Of these indels, all were exonic and 28 (71.8%) introduced a frameshift and 11 (28.2%) were in-frame with the coding sequence. All 39 indels affected unique genes (supplemental Table 4).
Comparison of mutation rates between the t(11;14) and t(4;14) subgroups
| . | t(4;14) (n = 10) . | t(11;14) (n = 12) . | Total (n = 22) . |
|---|---|---|---|
| SNVs | |||
| Median acquired exonic SNVs (range) | 38 (28-63) | 33.5 (26-50) | 36 (26-63) |
| Median acquired NS exonic SNVs | 27 | 23.5 | 24.5 |
| Acquired NS:S | 2.51 | 2.59 | 2.55 |
| Indels | |||
| Median acquired exonic indels (range) | 1.5 (0-4) | 2 (0-4) | 2 (0-4) |
| Median acquired frameshift indels | 1 | 1.5 | 1 |
| Median acquired nonframeshift indels | 0 | 0 | 0 |
| Exonic homozygous deletions | |||
| Average acquired exonic deletions (range) | 3 (1-19) | 3.5 (0-181) | 3 (0-181) |
| . | t(4;14) (n = 10) . | t(11;14) (n = 12) . | Total (n = 22) . |
|---|---|---|---|
| SNVs | |||
| Median acquired exonic SNVs (range) | 38 (28-63) | 33.5 (26-50) | 36 (26-63) |
| Median acquired NS exonic SNVs | 27 | 23.5 | 24.5 |
| Acquired NS:S | 2.51 | 2.59 | 2.55 |
| Indels | |||
| Median acquired exonic indels (range) | 1.5 (0-4) | 2 (0-4) | 2 (0-4) |
| Median acquired frameshift indels | 1 | 1.5 | 1 |
| Median acquired nonframeshift indels | 0 | 0 | 0 |
| Exonic homozygous deletions | |||
| Average acquired exonic deletions (range) | 3 (1-19) | 3.5 (0-181) | 3 (0-181) |
Composition of somatic variations. (A) Transition and transversion rates in sample groups. (B) Intersection of t(4;14) and t(11;14) genes with NS SNVs and indels.
Composition of somatic variations. (A) Transition and transversion rates in sample groups. (B) Intersection of t(4;14) and t(11;14) genes with NS SNVs and indels.
Because intragenic deletions have been reported in myeloma for key genes such as KDM6A (UTX), we investigated the prevalence of these deletions. This analysis identified 276 regions of deletion in the tumor samples, excluding physiologic immunoglobulin gene deletions. There was a median of 4 exonic deletions per sample, but 1 sample had an abnormally high number of deletions (n = 181) because of a large homozygous deletion on chromosome 17 at the keratin cluster; this deletion also was seen by mapping array.
There were significantly more mutations within genes that were not commonly expressed (P = .022, χ2 test), and there was an increasing mutation rate as the proportion of samples that did not express the gene increased (supplemental Figure 4). However, for the subset of genes expressed in more than 75% of myeloma samples, an elevated mutation rate was seen. Genes in this subset include known oncogenes (KRAS, NRAS, and MYC) and tumor suppressor genes (CYLD and FAM46C). We also examined genes with spiked expression in myeloma, and we found that PTPRG, FGFR3, ICAM1, LGMN, BMP2K, BCL6, MYC, and ROBO1 were mutated.
To gain mechanistic insight into the relationship of copy number abnormalities and mutations, we integrated the mutation data with genome mapping data. There were 102 distinct regions of copy number loss (totaling 3678 Mb) and 91 regions of copy number gain (4296 Mb) containing 33 and 48 NS-acquired SNVs, respectively. In regions of normal copy number, there were 517 mutations over 51 070 Mb of DNA. Per Mb of DNA, the mutation rate is 1/111.4 Mb in regions of loss, 1/110.3 Mb in regions of normal copy number, and 1/89.5 Mb in regions of gain. Taking into account the number of alleles available for analysis, the results are consistent with a mutation rate in regions of loss of heterozygosity of 1 SNV/111 Mb compared with 1/220 Mb in regions of normal copy number and 1/268 Mb in regions of gain, consistent with an increased mutation rate in regions of loss (Table 2). There were 33 NS-acquired mutations in regions of loss of heterozygosity (LOH) within the 22 samples. These were found in the common regions of LOH, namely, 8p, 13q, 14q, 16q, 17p, 22, and X in females. There was only 1 recurrently mutated gene within a region of LOH, and this gene was DIS3 on 13q. Other nonrecurrently mutated genes of interest in regions of LOH include MYC, CYLD, and FLT3 (Table 3).
Mutations in regions of copy number change
| . | Gain . | Loss . | Normal . |
|---|---|---|---|
| No. of NS mutations | 48 | 33 | 517 |
| No. of regions | 91 | 102 | NA |
| Total size of regions (Mb) | 4296 | 3678 | 57 010 |
| Mb/mutation | 89.5 | 111.4 | 110.3 |
| . | Gain . | Loss . | Normal . |
|---|---|---|---|
| No. of NS mutations | 48 | 33 | 517 |
| No. of regions | 91 | 102 | NA |
| Total size of regions (Mb) | 4296 | 3678 | 57 010 |
| Mb/mutation | 89.5 | 111.4 | 110.3 |
NA indicates not applicable.
List of mutated genes in regions of LOH
| Gene . | Sample . | Chr . | Base position . | Amino acid change . |
|---|---|---|---|---|
| ZNF474 | 91 603 | 5 | 121488236 | p.H184R |
| PHAX | 91 603 | 5 | 125960427 | p.N359S |
| FUT9 | 91 603 | 6 | 96651572 | p.E181X |
| SLC18A1 | 90 738 | 8 | 20038466 | p.T4P |
| MYC | 91 890 | 8 | 128752881 | p.L348V |
| FLT3 | 91 577 | 13 | 28592689 | p.V819A |
| COG3 | 91 827 | 13 | 46057379 | p.D244E |
| DIS3 | 90 468 | 13 | 73333955 | p.K922T |
| DIS3 | 90 405 | 13 | 73345947 | p.Y501D |
| DIS3 | 90 738 | 13 | 73347941 | p.T344P |
| UGGT2 | 90 357 | 13 | 96511903 | p.N1256T |
| STK24 | 90 482 | 13 | 99109461 | p.S407F |
| CHD8 | 90 245 | 14 | 21862547 | p.R1830C |
| MAX | 90 245 | 14 | 65560493 | p.R26H |
| TCL1A | 90 142 | 14 | 96180394 | p.C4S |
| CYLD | 90 273 | 16 | 50783949 | p.K114X |
| PKD1L2 | 90 273 | 16 | 81209247 | p.R849H |
| MYH4 | 90 482 | 17 | 10354723 | p.E1262G |
| KRTAP16-1 | 90 405 | 17 | 39464278 | p.T410P |
| ACTG1 | 90 983 | 17 | 79478992 | p.E100D |
| ZCCHC2 | 90 142 | 18 | 60242631 | p.R1106K |
| CRYBB2 | 91 577 | 22 | 25620907 | p.N26I |
| SEC14L3 | 91 603 | 22 | 30857373 | p.D335E |
| FAM47B | 90 738 | X | 34962196 | p.K416N |
| USP11 | 90 226 | X | 47092384 | p.V24A |
| MORC4 | 90 226 | X | 106200218 | p.E468X |
| IL13RA2 | 90 738 | X | 114251797 | p.Y12X |
| MAP7D3 | 90 738 | X | 135318488 | fs |
| Gene . | Sample . | Chr . | Base position . | Amino acid change . |
|---|---|---|---|---|
| ZNF474 | 91 603 | 5 | 121488236 | p.H184R |
| PHAX | 91 603 | 5 | 125960427 | p.N359S |
| FUT9 | 91 603 | 6 | 96651572 | p.E181X |
| SLC18A1 | 90 738 | 8 | 20038466 | p.T4P |
| MYC | 91 890 | 8 | 128752881 | p.L348V |
| FLT3 | 91 577 | 13 | 28592689 | p.V819A |
| COG3 | 91 827 | 13 | 46057379 | p.D244E |
| DIS3 | 90 468 | 13 | 73333955 | p.K922T |
| DIS3 | 90 405 | 13 | 73345947 | p.Y501D |
| DIS3 | 90 738 | 13 | 73347941 | p.T344P |
| UGGT2 | 90 357 | 13 | 96511903 | p.N1256T |
| STK24 | 90 482 | 13 | 99109461 | p.S407F |
| CHD8 | 90 245 | 14 | 21862547 | p.R1830C |
| MAX | 90 245 | 14 | 65560493 | p.R26H |
| TCL1A | 90 142 | 14 | 96180394 | p.C4S |
| CYLD | 90 273 | 16 | 50783949 | p.K114X |
| PKD1L2 | 90 273 | 16 | 81209247 | p.R849H |
| MYH4 | 90 482 | 17 | 10354723 | p.E1262G |
| KRTAP16-1 | 90 405 | 17 | 39464278 | p.T410P |
| ACTG1 | 90 983 | 17 | 79478992 | p.E100D |
| ZCCHC2 | 90 142 | 18 | 60242631 | p.R1106K |
| CRYBB2 | 91 577 | 22 | 25620907 | p.N26I |
| SEC14L3 | 91 603 | 22 | 30857373 | p.D335E |
| FAM47B | 90 738 | X | 34962196 | p.K416N |
| USP11 | 90 226 | X | 47092384 | p.V24A |
| MORC4 | 90 226 | X | 106200218 | p.E468X |
| IL13RA2 | 90 738 | X | 114251797 | p.Y12X |
| MAP7D3 | 90 738 | X | 135318488 | fs |
Chr indicates chromosome.
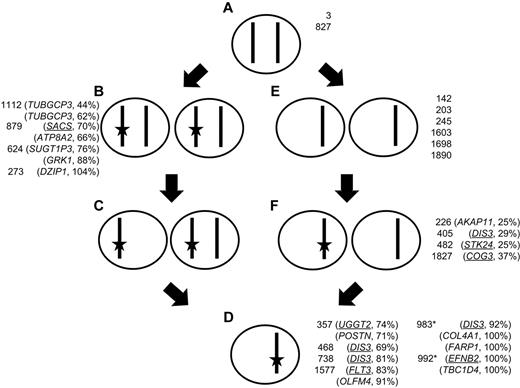
Almost all t(4;14) cases have del(13), and using acquired SNVs on chromosome 13 as a tool, we have investigated the dynamics of the development of del(13) and the mutation of genes carried on it (Figure 2; see supplemental Figures). Mutational frequency was determined by mutant sequence reads, and copy number was determined by a combination of SNP array copy number and LOH. We show that in the majority of instances del(13) occurs first and is followed by the acquisition of a mutation, but when mutations occur first these are more likely to be NS. Samples in which uniparental disomy (UPD) and acquired mutations occur together can be used to validate this finding. Three samples had UPD and NS mutations. The first is on 1q that has LOH and is also trisomic and has 2 mutations at frequencies of 14% (FCRL5) and 27% (IL6R). The second on chromosome 7 has 2 mutations present in 49% (BUD31) and 38% (CTTNBP2) of reads. The third is on chromosome 8 and has 43% mutated reads (CSMD3). These data suggest that deletion and subsequent duplication, to create UPD, occurred before mutation, which is the order determined by our analysis of chromosome 13.
Dynamic relationship of mutation to LOH on chromosome 13. A cell with 2 copies of a given chromosome (A) can gain a mutation (star) in 1 copy (B) and subsequently delete the unaltered copy, giving rise to a mixed population of cells (C) that then, through selection, results in a clonal population of cells with a mutation and deletion (D); or alternatively, the original cell (A) could delete 1 copy of the chromosome (E) and subsequently a subpopulation mutates the remaining allele (F) that is the selected for giving rise to a clonal population with both deletion and mutation (D). Samples are shown next to the cell genotype, as defined by sequencing and copy number data, along with the mutated gene on chromosome 13. Synonymous and NS (underlined) SNVs are shown. Asterisk indicates interstitial deletion of RB1 region detected with mutation in a region of normal copy number.
Dynamic relationship of mutation to LOH on chromosome 13. A cell with 2 copies of a given chromosome (A) can gain a mutation (star) in 1 copy (B) and subsequently delete the unaltered copy, giving rise to a mixed population of cells (C) that then, through selection, results in a clonal population of cells with a mutation and deletion (D); or alternatively, the original cell (A) could delete 1 copy of the chromosome (E) and subsequently a subpopulation mutates the remaining allele (F) that is the selected for giving rise to a clonal population with both deletion and mutation (D). Samples are shown next to the cell genotype, as defined by sequencing and copy number data, along with the mutated gene on chromosome 13. Synonymous and NS (underlined) SNVs are shown. Asterisk indicates interstitial deletion of RB1 region detected with mutation in a region of normal copy number.
DIS3 is located on chromosome 13 and is recurrently mutated in myeloma (10% reported previously and 18% in this set). We have identified 4 mutations in this gene, 1 mutation of which has been described previously, R750K, suggesting a gain in function of this gene. Interestingly, to date, mutation of DIS3 has only been seen in cases with either a t(4;14) or t(11;14). In samples with deletion of the whole chromosome mutation of DIS3 was seen as both a subclonal minority (29%) and as a majority population (69% and 81%; Table 4). In contrast, in the sample with an isolated interstitial deletion of RB1, the DIS3 mutation was present at 92%. It seems that DIS3 mutation always occurs in parallel with deletion of the RB1 region (13q14), raising the possibility that these are collaborating events. We also noted that of the 4 samples with DIS3 mutations in a previous dataset14 2 samples had del(13q) and the remaining 2 samples a t(4;14), which is highly associated with del(13q). Thus, it is possible that mutation and selection of DIS3, as a driver mutation, is dependent on deletion of 13q14.
Frequency of clonal subpopulations carrying a mutation
| Sample . | Translocation group . | Gene . | SNV . | Copy no. . | Frequency of SNV, % . | Estimated % of tumor population* . | Major or minor clone component . |
|---|---|---|---|---|---|---|---|
| 142 | t(4;14) | KRAS | c.A182G | 2 | 36 | 72 | Major |
| 738 | t(4;14) | KRAS | c.G436C | 2 | 36 | 72 | Major |
| 879 | t(11;14) | KRAS | c.A199C | 2 | 35 | 70 | Major |
| 879 | t(11;14) | KRAS | c.A183C | 2 | 34 | 68 | Major |
| 245 | t(4;14) | KRAS | c.G38A | 2 | 33 | 66 | Major |
| 3 | t(11;14) | KRAS | c.A182G | 2 | 10 | 20 | Minor |
| 3 | t(11;14) | KRAS | c.G34C | 2 | 24 | 48 | Minor |
| 827 | t(11;14) | KRAS | c.A182G | 2 | 14 | 28 | Minor |
| 1112 | t(11;14) | KRAS | c.G38A | 2 | 14 | 28 | Minor |
| 983 | t(11;14) | NRAS | c.C181A | 2 | 47 | 94 | Major |
| 203 | t(11;14) | NRAS | c.C181A | 2 | 48 | 96 | Major |
| 482 | t(11;14) | NRAS | c.A182G | 2 | 16 | 32 | Minor |
| 1890 | t(4;14) | NRAS | c.A182G | 2 | 17 | 34 | Minor |
| 468 | t(11;14) | BRAF | c.T1799A | 2 | 46 | 92 | Major |
| 405 | t(4;14) | BRAF | c.T1799A | 3 | 12 | 36 | Minor |
| 983 | t(11;14) | DIS3 | c.G2249A | 2 | 46 | 92 | Major |
| 738 | t(4;14) | DIS3 | c.A1030C | 1 | 81 | 81 | Major |
| 468 | t(11;14) | DIS3 | c.A2765C | 1 | 69 | 69 | Major |
| 405 | t(4;14) | DIS3 | c.T1501G | 1 | 29 | 29 | Minor |
| Sample . | Translocation group . | Gene . | SNV . | Copy no. . | Frequency of SNV, % . | Estimated % of tumor population* . | Major or minor clone component . |
|---|---|---|---|---|---|---|---|
| 142 | t(4;14) | KRAS | c.A182G | 2 | 36 | 72 | Major |
| 738 | t(4;14) | KRAS | c.G436C | 2 | 36 | 72 | Major |
| 879 | t(11;14) | KRAS | c.A199C | 2 | 35 | 70 | Major |
| 879 | t(11;14) | KRAS | c.A183C | 2 | 34 | 68 | Major |
| 245 | t(4;14) | KRAS | c.G38A | 2 | 33 | 66 | Major |
| 3 | t(11;14) | KRAS | c.A182G | 2 | 10 | 20 | Minor |
| 3 | t(11;14) | KRAS | c.G34C | 2 | 24 | 48 | Minor |
| 827 | t(11;14) | KRAS | c.A182G | 2 | 14 | 28 | Minor |
| 1112 | t(11;14) | KRAS | c.G38A | 2 | 14 | 28 | Minor |
| 983 | t(11;14) | NRAS | c.C181A | 2 | 47 | 94 | Major |
| 203 | t(11;14) | NRAS | c.C181A | 2 | 48 | 96 | Major |
| 482 | t(11;14) | NRAS | c.A182G | 2 | 16 | 32 | Minor |
| 1890 | t(4;14) | NRAS | c.A182G | 2 | 17 | 34 | Minor |
| 468 | t(11;14) | BRAF | c.T1799A | 2 | 46 | 92 | Major |
| 405 | t(4;14) | BRAF | c.T1799A | 3 | 12 | 36 | Minor |
| 983 | t(11;14) | DIS3 | c.G2249A | 2 | 46 | 92 | Major |
| 738 | t(4;14) | DIS3 | c.A1030C | 1 | 81 | 81 | Major |
| 468 | t(11;14) | DIS3 | c.A2765C | 1 | 69 | 69 | Major |
| 405 | t(4;14) | DIS3 | c.T1501G | 1 | 29 | 29 | Minor |
Calculated using copy number and SNV frequency.
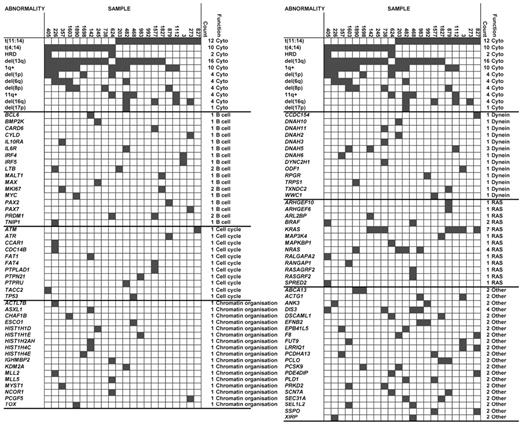
Recurrently mutated genes and families
We identified 28 genes with recurrent mutations, NS SNVs, and indels (Figure 3; supplemental Table 1). The most frequently mutated genes were KRAS (n = 7), DIS3 (n = 4), NRAS (n = 4), and DNAH5 (n = 3). In total, 5 genes were recurrently mutated in the t(4;14) samples (KRAS, ABCA13, DIS3, DNAH5, and PRKD2) and 11 genes in the t(11;14) samples (KRAS, NRAS, ACTG1, DIS3, EFNB2, F8, LRR1Q1, PCLO, PLD1, SEC31A, and SSPO). The genes mutated in the 2 cytogenetic groups were distinct, and of the 284 genes mutated in t(4;14) and 305 genes in t(11;14), samples only 18 (3.0%) genes were found mutated in both cytogenetic groups (Figure 1B). The majority of mutations occurred exclusively in either subgroup, and we looked at the pathways deregulated in each of the subgroups to determine whether they defined different pathobiologic routes to myeloma. Gene ontology analysis of genes disrupted in the t(4;14) group suggests an enrichment for genes involved in cytoskeleton organization, microtubule movement, and actin filament–based processes as well as chromatin organization, Table 5. The t(11;14) group showed an enrichment for protein phosphorylation, phosphate metabolism, and Ras signaling.
Association of abnormalities and mutations. Cytogenetic data repeated at the top of each column.
Association of abnormalities and mutations. Cytogenetic data repeated at the top of each column.
Pathway analysis for genes mutated in t(4;14) and t(11;14)
| Term . | Count . | % . | P . |
|---|---|---|---|
| t(4;14) | |||
| GO:0007010 cytoskeleton organization | 17 | 8.6 | 1.92 × 10−5 |
| GO:0030036 actin cytoskeleton organization | 11 | 5. 6 | 1.70 × 10−4 |
| GO:0030029 actin filament-based process | 11 | 5.6 | 2.85 × 10−4 |
| GO:0006325 chromatin organization | 13 | 6.6 | 8.10 × 10−4 |
| GO:0007017 microtubule-based process | 10 | 5.1 | .001 |
| GO:0051276 chromosome organization | 13 | 6.6 | .006 |
| GO:0016568 chromatin modification | 9 | 4.6 | .009 |
| t(11;14) | |||
| GO:0006468 protein amino acid phosphorylation | 17 | 9.3 | .001 |
| GO:0016044 membrane organization | 12 | 6.6 | .002 |
| GO:0006796 phosphate metabolic process | 21 | 11.5 | .003 |
| GO:0006793 phosphorus metabolic process | 21 | 11.5 | .003 |
| GO:0007265 Ras protein signal transduction | 6 | 3.3 | .005 |
| Term . | Count . | % . | P . |
|---|---|---|---|
| t(4;14) | |||
| GO:0007010 cytoskeleton organization | 17 | 8.6 | 1.92 × 10−5 |
| GO:0030036 actin cytoskeleton organization | 11 | 5. 6 | 1.70 × 10−4 |
| GO:0030029 actin filament-based process | 11 | 5.6 | 2.85 × 10−4 |
| GO:0006325 chromatin organization | 13 | 6.6 | 8.10 × 10−4 |
| GO:0007017 microtubule-based process | 10 | 5.1 | .001 |
| GO:0051276 chromosome organization | 13 | 6.6 | .006 |
| GO:0016568 chromatin modification | 9 | 4.6 | .009 |
| t(11;14) | |||
| GO:0006468 protein amino acid phosphorylation | 17 | 9.3 | .001 |
| GO:0016044 membrane organization | 12 | 6.6 | .002 |
| GO:0006796 phosphate metabolic process | 21 | 11.5 | .003 |
| GO:0006793 phosphorus metabolic process | 21 | 11.5 | .003 |
| GO:0007265 Ras protein signal transduction | 6 | 3.3 | .005 |
GO indicates gene ontology.
Mutated genes driving the different pattern in the t(4;14) group included chromatin modulating and cellular motility genes. Interstitial loss of exons of the histone demethylase KDM6A (UTX) have been described in myeloma, and we identified a mutated case in this analysis. MMSET is a histone methyltransferase gene, but in an extended analysis we did not identify an inverse relationship between MMSET and interstitial deletions of KDM6A. Other genes involved in DNA or histone modifications also were mutated and include ACTL7B, ASXL1, CHAF1B, ESCO1, KDM2A, KDM4B, KDM6A, MLL, MLL2, MLL5, MYST1, PCGF5, SETD1B, SETD6, and TOX. DNAH5, which encodes an axonemal dynein heavy chain, was mutated in 3 of the 22 cases and also was mutated in 2 cases in a previous report, accounting for 8% of myeloma samples sequenced to date. Several other genes encoding dynein-associated proteins also were mutated, including DNAH2, DNAH3, DNAH10, DNAH11, DYNC2H1, TXNDC2, CCDC154, WWC1, TRPS1, RPRG, and ODF1.
In the t(11;14) group, Ras signaling seemed prevalent, including NRAS, KRAS, and BRAF mutations that were validated by orthogonal technologies in an extended dataset, confirming the prevalence at 22%, 18%, and 4%, respectively (supplemental Figure 5). We show that the RAS pathway mutations (KRAS, NRAS, and BRAF) are mutually exclusive (Figure 3) and that deregulation of the pathway at any point is sufficient to lead to a clonal growth advantage. Interestingly, the mutations were not associated with any specific prognosis (supplemental Figure 5). In addition to these RAS–MAPK pathway variants, other RAS-related genes were found to have acquired mutations or deletions, including ARHGEF10, ARHGEF6, ARL2BP, MAP3K4, MAPKBP1, RALGAPA2, RANGAP1, RASA4, RASGRF2, and SPRED2, both in our dataset and the Broad dataset. These findings reflect the importance of deregulation of this pathway in presenting myeloma linking to the phenotypic function deregulated by these events in myeloma.
Both molecular subgroups showed deregulation of plasma cell biology and cell cycle–related genes. Genes involved in B-cell biology that were identified as being mutated include PRDM1, PAX2, PAX7, IL6R, IRF4, IL10RA, BCL6, MAX, MYC, CYLD, MALT1, BMP2K, CARD6, TNIP1, IRF5, MKI67, and LTB. Of these genes, only PRDM1, MKI67, and LTB carried recurrent mutations. Several cell cycle genes have mutations, including ATM, ATR, CCAR1, CDC14B, PTPLAD1, PTPN21, PTPRU, TP53, TACC2, FAT1, and FAT4.
Intraclonal heterogeneity at the level of mutation
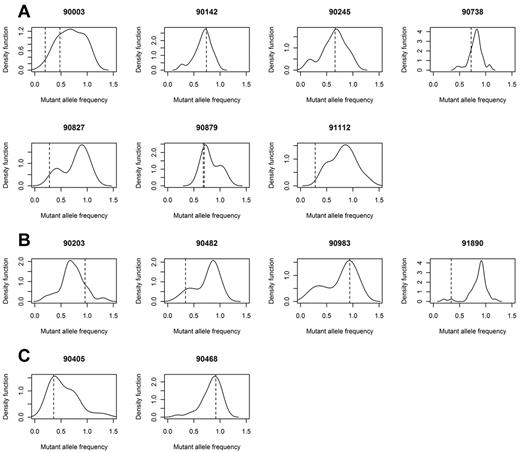
Deregulation of the RAS–MAPK pathway is clearly a central feature of both groups. Importantly, despite their driver status, clones with a RAS pathway mutation were not always present in the dominant clone. The subclonal populations can be visualized by calculating the percentage of mutant reads for all acquired mutations within sequence of depth more than 30 × in a sample and adjusting for copy number and cell purity to generate a frequency of mutated cells for each mutation. These data can be used to generate kernel density plots, with peaks showing the dominant clone and any subclones that may be present. In doing this, we see that in all samples with a RAS pathway mutation, there is at least 1 subclonal population, and frequently more than 1 subclonal population (Figure 4).
Gaussian kernel density plots indicating the frequency of cells carrying all acquired mutations. Frequency is calculated by adjusting mutant allele burden by copy number of the loci mutated. Samples with mutated KRAS (A), NRAS (B), or BRAF (C) indicates that driver mutations may be in major (> 0.5) or minor (< 0.5) clone populations. Frequency of the mutated RAS pathway gene is indicated by a dotted line.
Gaussian kernel density plots indicating the frequency of cells carrying all acquired mutations. Frequency is calculated by adjusting mutant allele burden by copy number of the loci mutated. Samples with mutated KRAS (A), NRAS (B), or BRAF (C) indicates that driver mutations may be in major (> 0.5) or minor (< 0.5) clone populations. Frequency of the mutated RAS pathway gene is indicated by a dotted line.
In NRAS, in the 4 samples with mutations, mutated reads were seen at frequencies varying from 16% to 48%, equating to 32% to 96% of the tumor population (Table 4). Examples of the 2 extremes are provided by samples 983 and 1890. Sample 983 has a mutation in NRAS (c.C181A) that is present in 47% of the sequencing reads, equating to 94% of the clonal cells. In contrast, sample 1890 has a mutation (c.A182G) in 17% of the sequencing reads, consistent with 34% of the clone. Similarly in the case of KRAS, we found mutated reads in 7 samples at a frequency between 10% and 36% of all reads or 20% and 72% of the tumor cell population (Table 4; Figure 4). Such intraclonal heterogeneity also was seen in KRAS where 2 samples had 2 acquired mutations each. In the first (sample 3), both SNVs were present in minor clones (10% and 24% of reads), yet both were activating KRAS mutations (c.G34C p.Q61R and c.A182G p.G12R). Thus, it is likely that these 2 mutations were present in different cell populations and had evolved independently. Conversely, in the second example (sample 879 that harbors both c.A199C p.M67L and c.A183C p.Q61H), the 2 mutations were present at similar frequencies, 70%, and are present in the same cells (both mutations are present in the same sequencing reads). However, M67L is not a known activating mutation; therefore, it is likely that the M67L mutation preceded or occurred concurrently with Q61H mutation and its clonal expansion is because of the activating mutation. BRAF was mutated in 2 samples and was also present as a major and minor clone (92% and 36% of cells, 46% and 12% of reads). The sample with only 12% of mutated reads was trisomic for chromosome 7, the location of BRAF, consistent with the mutation occurring after the chromosome became trisomic. Previous data on BRAF in myeloma reported 2.4% of samples with a V600E mutation and an additional 1.8% with a K601N.14 In our dataset, we identified 4% with the V600E mutation and no samples with the K601N mutation.
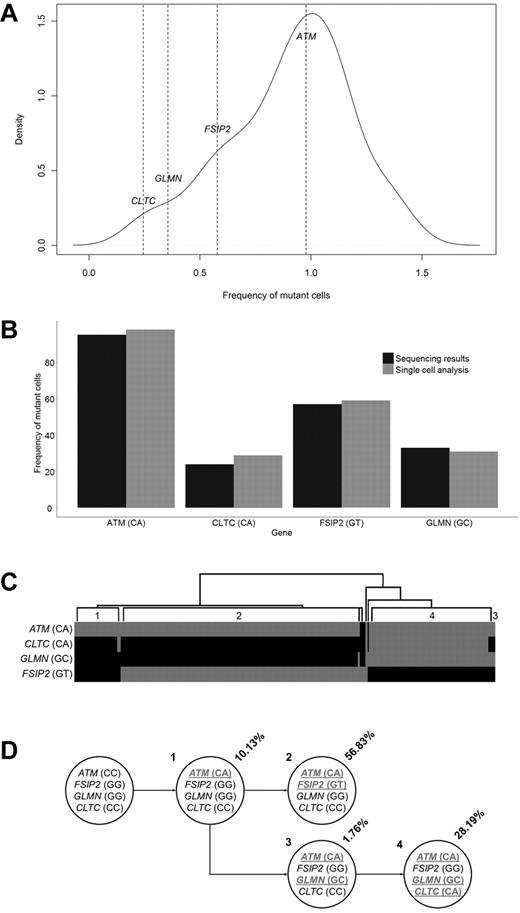
To provide further information on how the subclonal architecture at a mutational level develops, we studied a case of t(11;14) for which we had exome sequencing data and single cells stored. Exome sequencing identified 38 acquired NS mutations. On integration with copy number data from a SNP 6.0 array (Affymetrix), the frequency of each SNV in the cell population was calculated, and a mutation kernel plot was produced (Figure 5A). This identified 3 potential populations that could be summarized through analysis of 4 mutations, ATM (estimated at 95% of population), FSIP2 (57%), CLTC (24%), and GLMN (33%). These 4 mutations were picked for confirmation of clonal frequency by single-cell analysis. Genotyping for all 4 variants was performed on 270 single cells and showed the presence of the ATM mutation in 97% of single cells, FSIP2 in 59%, CLTC in 30%, and GLMN in 27% (Figure 5B). This confirmed that sequencing allelic reads can be used to accurately estimate the frequency of the mutation in the cell population. In addition to this, the single-cell analysis also indicated that all the cells contained the ATM mutation, and there were 3 subpopulations, 1 subpopulation of which only contained the ATM mutations; another subpopulation with an ATM and FSIP2 mutation; and a third subpopuation with ATM, CLTC, and GLMN mutations (Figure 5C-D).
Clonal heterogeneity determined by single cell analysis. (A) Mutant allele frequencies of 47 exonic synonymous and NS variants identified through exome sequencing. The frequencies of 4 variants are shown. (B) Proportion of cells identified with a mutated genotype for 4 genes as determined by exome sequencing allelic frequencies and single cell genotyping. (C) Hierarchical clustering of the genotypes generated from the analysis of 270 single cells. Grey indicates mutated; and black, reference. Individual populations are identified (1-4). (D) Determined cell lineages showing the proportion of tumor population and genotypes for the 4 genes in the identified populations (mutated genotypes underlined).
Clonal heterogeneity determined by single cell analysis. (A) Mutant allele frequencies of 47 exonic synonymous and NS variants identified through exome sequencing. The frequencies of 4 variants are shown. (B) Proportion of cells identified with a mutated genotype for 4 genes as determined by exome sequencing allelic frequencies and single cell genotyping. (C) Hierarchical clustering of the genotypes generated from the analysis of 270 single cells. Grey indicates mutated; and black, reference. Individual populations are identified (1-4). (D) Determined cell lineages showing the proportion of tumor population and genotypes for the 4 genes in the identified populations (mutated genotypes underlined).
Discussion
All myeloma passes through a MGUS phase; yet, there are well-described differences in the pathogenesis and clinical outcome of the t(4;14) and t(11;14) subgroups of myeloma.28,29 It has been estimated that the transition from MGUS to myeloma takes a median of 25 years30 during which time mutations may accumulate.10 The frequency at which the t(4;14) translocation occurs in MGUS patients is markedly lower than that seen in myeloma patients (4% vs 11%), whereas the t(11;14) occurs in relatively equal proportions (10%-14%).2,3 This difference in frequency reflects the unstable phenotype induced by the t(4;14) that decreases the time to develop myeloma and could affect mutational load at presentation. We show that t(4;14) samples at presentation have an increased mutational load compared with t(11;14) samples (median, 27 vs 23.5 NS mutations, respectively), and with increased sample sizes this trend may become significant. We can conclude that the number and spectrum of mutations necessary to be a presenting case of myeloma is similar in the 2 molecular subgroups. Because it is likely that cases with t(4;14) develop myeloma over a shorter period than those with t(11;14), we conclude that the genome of the t(4;14) cases is more unstable.
In addition, transition and transversion rates between the 2 groups were equivalent. Previously, we have shown that the t(4;14) subgroup has increased DNA hypermethylation compared with other subgroups.10 It may be expected therefore that there would be an increase in transversions in the t(4;14) group, because of the deamination of 5-methylcytosine to uracil that is replaced by thymine. However, this was not the case, but a previous study had identified an increased mutation rate at CpG islands,14 and it may be that the exome capture, which has a bias against first exons and CpG-rich sequences, has not detected this.
We incorporated our data with data from the other myeloma genome sequencing dataset that together identified 228 recurrently mutated genes in 60 samples. The most frequently mutated genes were KRAS, NRAS, DIS3, FAM46C, LRP1B, DNAH5, and WHSC1. There were some differences in the genes identified between the datasets, but this be accounted for by the techniques used (whole genome vs whole exome; WHSC1 and LRP1B) or the cytogenetic groups studied (random sampling versus translocations; DIS3) or treated and nontreated patients (FAM46C). To address the passenger or driver status of the genes identified, we used the COSMIC database to identify recurrently mutated genes in other cancer cell types. Of the 571 genes found mutated in our own dataset, 63 genes have not been identified as mutated in any cancer type. Twenty-one genes were identified as being mutated in hematologic cancers, and of these 6 genes were recurrently mutated in our dataset (NRAS, KRAS, BRAF, PRDM1, SEC31A, and PDE4DIP; supplemental Table 3).
One of the novel findings of the first myeloma genome publication was the identification of DIS3 mutations in 10% of myeloma samples. We have confirmed the presence of DIS3 mutations in myeloma, and we found that the proportion of affected samples was higher (18%). This difference is because of the samples analyzed, because DIS3 mutations have been found exclusively in t(4;14) or t(11;14) samples, of which our dataset is composed. In addition, we found that all samples with DIS3 mutations also have deletion of 13q14, the region surrounding RB1, and so deletion of 13q14 and mutation of DIS3 may be collaborating lesions. The gene of interest at 13q14 has been debated for many years and may be RB1 or 1 of the microRNAs implicated in chronic lymphocytic leukemia (miR-15a/16).31,32 It remains unclear which region(s) of chromosome 13 is important, but the discovery of yet another locus will confound the situation further and only through analyses of large datasets, comprising copy number and mutational data, will the true nature of these abnormalities be discovered. DIS3 mutations also have been found in relapse acute myeloid leukemia, known to have deletion or uniparental disomy of chromosome 13.33,34
When we compare the intersection of mutations in the 2 groups, we see that only 3% are shared. The majority of mutations in each group are distinct, suggesting that the pathologic contribution of the genes mutated in each group is different. We show that genes expressed at more than 75% have a greater rate of mutation and could explain some of the differences between the subgroups. Gene ontology analysis of genes disrupted in the t(4;14) group suggests an enrichment for genes involved in cytoskeleton organization, microtubule movement, and actin filament–based processes as well as chromatin organization (Table 5). The t(11;14) group showed an enrichment for protein phosphorylation and Ras signaling. This observation is consistent with the idea that collaborating mutations are likely to interact with the initiating events in each molecular subtypes of myeloma to hijack the normal physiologic role of plasma cells, contributing to their malignant transformation. RAS pathway mutations have been shown to occur in myeloma,35 and here we show convincingly that RAS-MAPK pathway mutations tend to be mutually exclusive, consistent with the lack of a clonal survival advantage for cells acquiring additional mutations within a pathway already deregulated.
We demonstrate that driver mutations are present as minor subclones in half of the samples. One sample was identified that contained 2 activating KRAS mutations present in different subclonal populations, comprising 20% and 48% of cells. This suggests that subclones are continually at risk of developing driver mutations that can confer a growth and survival advantage, leading to clonal dominance over time. Single-cell analysis confirmed the presence of subclonal substructure at a mutational level. In 1 example, we showed that all cells contained an ATM mutation, but 2 subpopulations existed with mutually exclusive mutations in FSIP2 and GLMN/CLTC. The mutant cell frequency plot for this sample shows 3 distinct peaks (at 0.3, 0.5, and 1.0), corresponding to the 3 main clones in this sample (mutated at ATM, ATM + FSIP2, or ATM + GLMN/CLTC). This type of subclonal structure has been described for other hematologic malignancies such as acute myeloid leukemia, acute lymphoblastic leukemia, breast cancer and prostate cancer using FISH, single cell sequencing or ultra-deep sequencing.34,36-39 The realization that tumor biology consists of multiple independent subclones has important ramifications for tumor progression and personalized treatment strategies.
We provide evidence that there is a distinct mutational landscape in the t(4;14) and t(11;14) groups of myeloma. After initial immortalization by the acquisition of a translocation, each group follows a distinct pathway to becoming a presenting case of myeloma. Copy number abnormalities may occur relatively early on in the pathogenesis of myeloma, and regions of LOH undergo mutation more frequently. Mutations in regions of LOH tend to be NS, pathologically relevant variants that contribute to clonal progression. The rate of mutation seems to be higher in the t(4;14) group, which may lead to their more rapid progression to myeloma. The net mutational load, however, that is required to develop presenting myeloma is consistent in the 2 groups. We demonstrate intraclonal heterogeneity and the continued acquisition of mutations within subclones that leads to clonal outgrowth and disease progression. The clonal architecture described here, with multiple subclones vying for dominance, is reminiscent of the Darwinian natural selection process with a branching, nonlinear accumulation of mutations in malignant plasma cells.
There is an Inside Blood commentary on this article in this issue.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ian Titley for expert assistance in flow cytometry sorting of single cells and Ricardo Morilla for assistance in setting up the myeloma flow panel.
This study was supported by grants from Myeloma UK and the National Institutes of Health Biomedical Research Center at the Royal Marsden Hospital as well as Children with Cancer UK.
Authorship
Contribution: B.A.W. designed and performed research, analyzed data, and wrote the paper; C.P.W. performed computational research, analyzed data, and wrote the paper; L.M. performed research and analyzed data; S.H. and K.F. performed research; N.E.P. developed single-cell protocols; D.C.J. and D.G. designed research; I.K. designed and performed research; C.J.L. and A.A. provided resources; F.E.D. designed research; and G.J.M. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gareth J. Morgan, Haemato-Oncology Research Unit, Division of Molecular Pathology, The Institute of Cancer Research, 15 Cotswold Road, Sutton, United Kingdom, SM2 5NG; e-mail: gareth.morgan@icr.ac.uk.
References
Author notes
B.A.W. and C.P.W. contributed equally to this work.