In this issue of Blood, Keats et al,1 Egan et al,2 and Walker et al3 provide a genome-wide snapshot of the clonal landscape in multiple myeloma (MM) illustrating the complexity of the evolutionary process and the dynamics of clonal evolution over time.
The advancement in cancer genomics technologies and analyses has challenged a longstanding cancer biology dogma, introduced in 1976 by Peter Nowell, that tumors are derived from a single cell with stepwise acquisition and accumulation of somatic cell mutations, sequential selections leading to malignant total domination by the fittest clone.4 This model of tumor evolution implies that all clones are linearly related to each other and homogenous in their mutational landscape. Massive genome deep-sequencing studies are now contradicting this model and revealing a more complex clonal architecture of Darwinian-like somatic evolution where tumor progression proceeds in a branching rather than in a linear manner, leading to substantial clonal diversity and coexistence of wide genetic heterogeneity.5-11
The genomic complexity in MM was recently corroborated by massive parallel-sequencing studies displaying an average of 35 amino acid changing point mutations per sample and the lack of a universal driving mutation in the 38 genomes analyzed.12 Here, these authors describe the intraclonal architectural heterogeneity at diagnosis and at different stages of disease progression over time.1-3 Clinical and phenotypic elements have long suspected this clonal diversity based on observations of biclonal disease on protein electrophoresis, the switch in the monoclonal protein subtype (light chain conversion), the presence of numeric and structural chromosomal aberrations only in a subset of purified MM cells, and occasionally the discordant therapeutic response (between extramedullary and bone marrow disease) within the same patient. As such, in the study by Walker and colleagues, despite the driver status of the RAS/MAPK pathway deregulation in t(4;14) and t(11;14) MM patients, these mutations were not always present in the dominant clone but rather in 1 or many minor subclonal populations.3 This subclonal diversity was also confirmed at a single cell level supporting the distinct pathogenesis underling the variable MM molecular subgroups as well as the contemporary clonal heterogeneity within subclones of any individual molecular subgroup.
Similar to this snapshot of intraclonal diversity at diagnosis and examining copy number alterations (CNAs) over time, Keats et al describe 3 temporal tumor types where the relapsing disease is identical to the diagnostic clone and thus genetically stable or has evolved linearly from the diagnostic clone with the acquisition of only 1 new CNA, or most commonly, as seen in nearly half of the patients, genetically distinct relapsed clones have emerged.1 Of interest in cytogenetically defined high-risk disease [t(4;14), t(14;16), t(14;20), del(17p)], an increase in CNA changes were observed over time, suggesting that the poor prognosis in this group is at least partially related to their increased clonal heterogeneity and genomic instability.1,3 Whole genome sequencing along with comparative genomic hybridization array examined the clonal architecture of 1 patient with t(4;14) over time and identified an alternating dominance of clones or clonal tiding as disease progressed from intramedullary MM to plasma cell leukemia.1,2 This complex clonal dynamic was also partially correlated to the administered therapy with the major clonal drift (highest frequency of CNA and nonsynonymous single nucleotide variants) observed after the administration of alkylating agent–containing regimens. This heterogeneous clonal mixture at relapse and clonal tiding over time supports the Darwinian branching model of tumor evolution with several clonal progenitors or tumor-initiating cells present at diagnosis with therapeutic or ecosystem-dependent selection pressures driving the alternating dominance of these clones over time. The Vk*MYC transgenic mouse model confirmed this purposeless spontaneous and branching clonal evolution with the coexistence of either competing or collaborating clones.1
Deep sequencing of the plasma cell genomes at different time points over the course of the disease has uncovered a previously unsuspected model of genomic evolution in MM. In contrast to the traditional model of linear cumulative genomic aberrations over time, these authors have described the presence of “clonal tides” with Darwinian mutational branching and shifting dominance of variable clones over time. These findings represent a novel paradigm in myeloma evolutionary biology and will revolutionize the current modeling of MM tumorogenesis and progression and are likely to have profound therapeutic implications. As such, the presence of clonal heterogeneity at diagnosis argues in favor of adopting combination rather than single-agent sequential therapies with the goal to eradicate dominant as well as minor clones that often emerge at relapse. In particular, such an approach may be required in cytogenetically defined high-risk MM where a clonal diversity is found to be more prevalent compared with low-risk disease.
The concept of clonal dominance illustrated in the Vk*MYC model also stresses the primordial need to avoid suboptimal therapies that may eradicate sensitive clones causing a “competitive release” allowing resistance clones to predominate within the bone marrow niches.11,13 These findings may also cast some uncertainty regarding the efficacy of highly targeted therapeutic approaches where the target may be a nonfounder mutation and in view of the likely coexistence at the time of diagnosis or emergence at relapse of an evolutionary-drifted minor clone with a nonlinear genomic alteration and thus independence of the targeted mutation. In addition, the tremendous variegation in the MM tumor-initiating clones likely represents a major roadblock for curative therapy targeting a single mutant molecule in this disease.
Furthermore, the results of the sequential genomic sequencing and structural studies1,2 of the patient with t(4;14) over time highlight the selective pressure that may be exerted by a class of DNA damaging agents (alkylating drugs) leading to the expansion of resistance clones that either existed before the onset of treatment or formed as a result of new genomic alterations gained during therapy. In such a scenario, one would question the potential harmful effects of DNA-damaging agents in cytogenetically high-risk MM groups prone to clonal divergence. Lastly, the understanding of the clonal dynamics in MM in particular with respect to clonal tides also challenges a widely adopted tenet in cancer therapeutics where a class of drugs should not be reintroduced at relapse because of acquired resistance in the relapsing clones. However, such dogma did not take into account the nonlinear and branching evolutionary model from a common ancestral clone(s) resulting in the re-emergence of a genetically diverse subclone at a later relapse with possibly retained sensitivity to previously administered chemotherapeutics.
These studies on the myeloma genomic landscape represent a major leap forward in our understanding of MM biogenesis and clonal evolution that is more consistent with Darwin's iconic evolutionary tree (see figure). The data presented in these articles may finally explain the shortcomings of the therapeutic approaches we have adopted thus far in this disease while providing a valuable genomically guided insight to our relentless search for a cure.
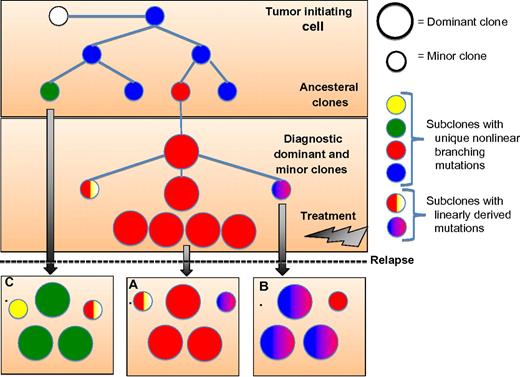
Schema illustrating the evolutional clonal architecture in multiple myeloma (MM) at diagnosis and relapse. Noted at diagnosis the clonal diversity with coexistence of dominant and minor subclones that have evolved from a common ancestral tumor-initiating cell or stem cell. The clonal disease at relapse may follow 1 of 3 evolutionary patterns with clones identical to the diagnostic sample and no newly acquired genomic alterations (A), or evolve from the diagnostic clone with linearly derived mutations (B), or, and as seen more commonly in cytogenetically high-risk disease, evolve from an ancestral minor clone(s) with newly acquired genomic mutations or structural rearrangements.
Schema illustrating the evolutional clonal architecture in multiple myeloma (MM) at diagnosis and relapse. Noted at diagnosis the clonal diversity with coexistence of dominant and minor subclones that have evolved from a common ancestral tumor-initiating cell or stem cell. The clonal disease at relapse may follow 1 of 3 evolutionary patterns with clones identical to the diagnostic sample and no newly acquired genomic alterations (A), or evolve from the diagnostic clone with linearly derived mutations (B), or, and as seen more commonly in cytogenetically high-risk disease, evolve from an ancestral minor clone(s) with newly acquired genomic mutations or structural rearrangements.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal