In this issue of Blood, Dastugue and colleagues report their findings examining the impact of hyperdiploidy on the outcome of children with B-cell acute lymphoblastic leukemia (ALL). Since the 1980s, it has been appreciated that 25% to 30% of B-cell ALL cases have a “high” hyperdiploid karyotype (generally defined as >50-67 chromosomes) and that this subset has an unusually good prognosis (reviewed by Paulsson and Johansson1 ). This feature has been used routinely to stratify patients into treatment groups. However, in spite of major progress understanding other biological subtypes of ALL, the underlying transforming pathways that define this major subgroup and those that account for the good response to therapy remain largely unknown. Moreover, controversy exists on whether the driver in both cases is the gain of specific chromosomes or whether it is due to the overall gain in chromosome number. Dastugue et al report that the best indicator of overall prognosis is ploidy assessed by karyotype and that prognosis is improved at higher modal chromosome numbers.2
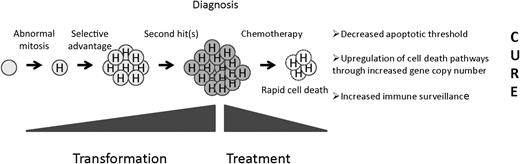
A precursor B cell undergoes an abnormal mitosis that results in hyperdiploidy. A selective advantage is conferred, resulting in a preleukemic clone, and second “hits” result in the fully malignant phenotype. Hyperdiploid cells display unusual sensitivity to chemotherapy through as-yet-undefined mechanisms.
A precursor B cell undergoes an abnormal mitosis that results in hyperdiploidy. A selective advantage is conferred, resulting in a preleukemic clone, and second “hits” result in the fully malignant phenotype. Hyperdiploid cells display unusual sensitivity to chemotherapy through as-yet-undefined mechanisms.
Aneuploidy, an abnormal number of chromosomes, is a frequent observation in human cancer (reviewed by Gordon et al3 ). Evolution has led to protective mechanisms to ensure faithful replication and inheritance of genetic information so that cells with abnormal chromosome content are removed. Compensatory mechanisms must exist in cancer cells to compensate for the stress of aneuploidy. Cancer cells often display defects in pathways that regulate genome stability that result in aneuploidy. Thus, hyperdiploidy may be a consequence rather than a driver of malignancy. However, available data indicate the genomes of hyperdiploid ALL cells (and all childhood ALL cells) are relatively stable (note chromosome instability refers to the rate of karyotypic change). The great majority of cases of hyperdiploid ALL appear to be the result of the simultaneous gain of extra chromosomes in a single abnormal cell division.4
Evidence seems to favor a beneficial impact of hyperdiploidy in leukemogenesis (see Figure). Like most other forms of ALL, hyperdiploid clones appear to have a prenatal origin followed a long latency until the development of the truly transformed clone though acquisition of “second hits.” What accounts for this selective advantage? The repertoire of altered expression of key growth and survival pathways could provide cells with a selective advantage. Clues to the identification of such pathways come from the pattern of chromosome gains in hyperdiploid ALL. It is important to note that chromosomes 21, X, 14, 6, 18, 4, 17, and 10 tend to be gained in blasts associated with lower modal karyotypes and retained as modal chromosome number increases.5 Such selection could be due to the acquisition of novel beneficial properties associated with this subset of chromosomes or selection against other chromosomes because of negative attributes. Acquisition of chromosome 21 is seen in 95% of hyperdiploid cases, and children with constitutional trisomy 21 do display an increase in hematologic malignancies (but a decrease in solid tumors, curiously).
Although hyperdiploidy may be involved with leukemogenesis, it is clearly disadvantageous to the tumor after application of chemotherapy. This is in sharp contrast to the unfavorable impact of aneuploidy in many solid tumors, likely because of the presence of chromosomal instability. Numerous reports, including the paper by Dastugue et al in this issue, show 5-year event-free and survival rates of 84% to 90% and 93% to 95%, respectively, using contemporary treatments.6,7 Hyperdiploidy is associated with other features associated with a good prognosis but it carries independent prognostic significance in multivariate analyses. The unique chemosensitivity of these cells has been demonstrated in ex vivo assays and by the high rate of end-induction remission. The study by Dastugue et al also showed that hyperdiploidy was associated with low minimal residual disease burden at end induction. Some disagreement exists about the prognostic relevance of individual chromosomes. Combined analysis of Pediatric Oncology Group and Children’s Oncology Group studies showed the powerful effect of trisomies of chromosomes 4, 10, and 17 (triple trisomy) that led to routine incorporation into risk stratification (trisomies of 4 and 10 are in current use), while investigators from the Medical Research Council could not confirm this finding but showed that among high hyperdiploid cases those with trisomy 18 had a lower risk of relapse.7,8 Much has been made of these discrepancies, but the incidence of trisomies 4, 10, 17, and 18 is proportional to increases in chromosome number. Thus it might be predicted that modal chromosome number should emerge as the most important variable and that is exactly the conclusion of the study by Dastugue et al.
The mechanism of enhanced chemosensitivity is unknown. Cells might be primed for cell death because of a decreased apoptotic threshold due to the stress associated with aneuploidy. Alternatively, increased gene and protein expression associated with increased copy number of individual genes involved in drug transport, metabolism, or common to many different agents (eg, executors of cell death) might enhance the effect of chemotherapy. In this case, “passenger lesions” associated with transformation could function as “drivers” of chemosensitivity. For example, elevated levels of the reduced folate carrier (hRFC, SLC19A1) located on chromosome 21 has been associated with a decreased risk of relapse. As expected, levels are higher in hyperdiploid cases, although this difference did not reach statistical significance compared with National Cancer Institute standard-risk patients without hyperdiploidy in one study.9 Nonetheless, subtle differences in therapy among treatment groups may account for the relative importance of certain trisomies in published studies. Another provocative mechanism of enhanced cell death was illustrated by a recent study showing that immunosurveillance mechanisms exist to control ploidy.10
In summary, analysis of modal chromosome number remains a powerful predictor of outcome in childhood B-cell ALL and should be used routinely to stratify patients. The importance of individual trisomies may relate to subtle differences in treatment. DNA index may be used in place of karyotype in resource-poor areas, and newer technology may be integrated in the future. Finally, we await a more detailed understanding of the biology of hyperdiploid ALL in the quest to develop targeted therapy.
Conflict-of-interest disclosure: The author declares no competing financial interests.