Key Points
The FAB type (ie, M0-M7) does not provide prognostic information for cases of “AML, NOS” in the 2008 WHO classification.
Abstract
The World Health Organization (WHO) classifies acute myeloid leukemia (AML) via genetic, immunophenotypic, biological, and clinical features. Still, “AML, not otherwise specified (NOS)” is further subdivided based on morphologic criteria similar to those of the French-American-British (FAB) classification. We analyzed the relevance of this practice in patients with newly diagnosed “AML, NOS” with available FAB information undergoing curative-intent therapy in trials of 3 cooperative study groups (Dutch-Belgian Cooperative Trial Group for Hematology/Oncology [HOVON], UK Medical Research Council/National Cancer Research Institute [MRC/NCRI], and the US cooperative group Southwest Oncology Group [SWOG]) or at MD Anderson Cancer Center. Ignoring information on NPM1 and CEBPA, 5848 patients met criteria for “AML, NOS.” After multivariate adjustment, FAB M0 was independently associated with significantly lower likelihood of achieving complete remission and inferior relapse-free and overall survival as compared with FAB M1, M2, M4, M5, and M6, with inconclusive data regarding M7. However, restricting attention to known NPM1neg patients, FAB M0 was no longer associated with worse outcomes; restricting attention to patients known to be NPM1neg/CEPBAneg (ie, honoring the provisional entities of “AML with mutated NPM1” and “AML with mutated CEBPA”) did not affect this result. In conclusion, in the 2008 WHO classification scheme, FAB subclassification does not provide prognostic information for “AML, NOS” cases if data on NPM1 and CEBPA mutations are available.
Introduction
Acute myeloid leukemia (AML) comprises a heterogeneous group of disorders.1,2 Initial recognition of this heterogeneity was based largely on morphology. Thus, several decades ago, the French-American-British (FAB) Cooperative Group developed a classification system based on conventional, readily available morphologic and cytochemical characteristics.3-6 The group’s proposal aimed to provide objectivity in the diagnosis of AML that would facilitate comparisons between series of cases.3,4 However, as the importance of cytogenetics became apparent, it was unclear whether data on FAB subtypes (M0-M7) added prognostic information beyond that available from cytogenetics [eg, t(15;17) for M3 or inv(16) for M4Eo]. Despite this uncertainty, AML cases have generally been categorized according to the FAB classification for many years.
More recently, recognition of the diversity of cytogenetic and molecular abnormalities and better understanding of the disease biology of AML prompted the World Health Organization (WHO) to develop a new classification that integrated genetic, immunophenotypic, biological, and clinical features to define specific disease entities.7,8 The WHO classification scheme essentially replaced the FAB system with the exception that the latter remains embedded in the WHO’s “AML, not otherwise specified (NOS)” category, which encompasses cases that do not fulfill WHO criteria for other categories such as “AML with recurrent genetic abnormalities,” “AML with myelodysplasia-related changes,” and so forth.8,9 Whether this recapitulation of FAB is prognostically useful is unclear. Assuming that part of a classification system’s value derives from its clinical relevance, we used data from adults with newly diagnosed AML treated in trials conducted by the Dutch-Belgian Cooperative Trial Group for Hematology/Oncology (HOVON), the UK Medical Research Council/National Cancer Research Institute (MRC/NCRI), the US cooperative group Southwest Oncology Group (SWOG), and MD Anderson Cancer Center (MDA) to assess the prognostic significance of FAB in the WHO’s “AML, NOS” category.
Patients and methods
Study population
Our analyses included data on predominantly adult patients with newly diagnosed AML based on WHO 2008 classification criteria9 that were treated with curative intent in any of 6 HOVON trials from 1987 to 2008, 9 SWOG trials from 1987 to 2009, or 6 MRC/NCRI trials from 1988 to 2010, or that received treatment in various protocols at MDA from 2000 to 2011. Institutional review boards of participating institutions approved all protocols, and patients were treated according to the Declaration of Helsinki. All patients had data available on FAB type and routine cytogenetics (G-banding) from the time of diagnosis. Information on FAB type was used as provided by the cooperative study groups and MDA. For patient categorization, 2 authors (R.B.W. and E.H.E.) reviewed the cytogenetic information of all patients, and discordant interpretations were resolved with the help of a cytogeneticist (Dr Min Fang, Seattle Cancer Care Alliance, Seattle, WA). We excluded patients classified as “AML with recurrent genetic abnormalities” based on information from routine karyotyping. We excluded patients classified as “AML with myelodysplasia-related changes” based on history of antecedent hematologic disorder or presence of cytogenetic abnormalities sufficient for the diagnosis of this disease subgroup.9 We excluded patients with therapy-related myeloid neoplasms based on a history of cancer with use of chemo- and/or radiotherapy. After these exclusions, our data set encompassed 5848 patients (Table 1). For subset analyses, we further excluded patients with NPM1 or CEBPA mutations or whose mutation status for these genes was unknown.
Characteristics of study population
| . | HOVON . | MDA . | MRC/NCRI . | SWOG . | All . |
|---|---|---|---|---|---|
| Parameter . | n = 578 . | n = 1003 . | n = 3666 . | n = 601 . | n = 5848 . |
| Age in y, median (range) | 48 (16-77) | 58 (12-88) | 54 (16-91) | 53 (16-84) | 54 (12-91) |
| Patients aged ≥60 y, n (%) | 44 (8) | 466 (46) | 1364 (37) | 192 (32) | 2066 (35) |
| Male gender, n (%) | 300 (52) | 534 (53) | 1879 (51) | 319 (53) | 3032 (52) |
| Karyotype,* n (%) | |||||
| Normal | 451 (78) | 456 (45) | 2749 (75) | 432 (72) | 4088 (70) |
| Abnormal | 127 (22) | 547 (55) | 917 (25) | 169 (28) | 1760 (30) |
| NPM1 mutation,* n (%) | |||||
| Yes | 293 (51) | 97 (26) | 520 (50) | 77 (28) | 987 (44) |
| No | 278 (49) | 275 (74) | 525 (50) | 199 (72) | 1277 (56) |
| Unknown | 7 | 631 | 2621 | 325 | 3584 |
| CEBPA mutation,* n (%) | |||||
| Yes | 61 (11) | 10 (12) | 42 (5) | 41 (48) | 154 (10) |
| No | 500 (89) | 76 (88) | 771 (95) | 45 (52) | 1392 (90) |
| Unknown | 17 | 917 | 2853 | 515 | 4302 |
| WBC* [×103/µL], median (range) | 29 (0-510) | 7 (0-390) | 15 (0-559) | 17 (0-1300) | 13 (0-1300) |
| Platelets* [×103/µL], median (range) | 69 (5-778) | 49 (0-635) | 65 (0-903) | 58 (3-9300) | 62 (0-9300) |
| Bone marrow blasts,* median % (range) | 68 (0-98) | 58 (0-99) | 80 (0-100) | 70 (1-100) | 73 (0-100) |
| FAB subclassification,* n (%) | |||||
| M0 | 28 (5) | 76 (8) | 215 (6) | 34 (6) | 353 (6) |
| M1 | 151 (26) | 162 (16) | 977 (27) | 152 (25) | 1442 (25) |
| M2 | 158 (27) | 340 (34) | 999 (27) | 161 (27) | 1658 (28) |
| M4 | 74 (13) | 244 (24) | 766 (21) | 160 (27) | 1244 (21) |
| M5 | 148 (26) | 116 (12) | 523 (14) | 81 (13) | 868 (15) |
| M6 | 19 (3) | 55 (5) | 158 (4) | 9 (1) | 241 (4) |
| M7 | 0 (0) | 10 (1) | 28 (1) | 4 (1) | 42 (1) |
| PS,* n (%) | |||||
| 0 | 250 (43) | 172 (17) | 1841 (50) | 231 (38) | 2494 (43) |
| 1 | 279 (48) | 628 (63) | 1090 (30) | 296 (49) | 2293 (39) |
| ≥2 | 49 (8) | 203 (20) | 735 (20) | 74 (12) | 1061 (18) |
| CR, n (%) | 499 (86) | 649 (65) | 2929 (80) | 383 (64) | 4460 (76) |
| Early death,† n (%) | 19 (3) | 55 (5) | 300 (8) | 37 (6) | 411 (7) |
| . | HOVON . | MDA . | MRC/NCRI . | SWOG . | All . |
|---|---|---|---|---|---|
| Parameter . | n = 578 . | n = 1003 . | n = 3666 . | n = 601 . | n = 5848 . |
| Age in y, median (range) | 48 (16-77) | 58 (12-88) | 54 (16-91) | 53 (16-84) | 54 (12-91) |
| Patients aged ≥60 y, n (%) | 44 (8) | 466 (46) | 1364 (37) | 192 (32) | 2066 (35) |
| Male gender, n (%) | 300 (52) | 534 (53) | 1879 (51) | 319 (53) | 3032 (52) |
| Karyotype,* n (%) | |||||
| Normal | 451 (78) | 456 (45) | 2749 (75) | 432 (72) | 4088 (70) |
| Abnormal | 127 (22) | 547 (55) | 917 (25) | 169 (28) | 1760 (30) |
| NPM1 mutation,* n (%) | |||||
| Yes | 293 (51) | 97 (26) | 520 (50) | 77 (28) | 987 (44) |
| No | 278 (49) | 275 (74) | 525 (50) | 199 (72) | 1277 (56) |
| Unknown | 7 | 631 | 2621 | 325 | 3584 |
| CEBPA mutation,* n (%) | |||||
| Yes | 61 (11) | 10 (12) | 42 (5) | 41 (48) | 154 (10) |
| No | 500 (89) | 76 (88) | 771 (95) | 45 (52) | 1392 (90) |
| Unknown | 17 | 917 | 2853 | 515 | 4302 |
| WBC* [×103/µL], median (range) | 29 (0-510) | 7 (0-390) | 15 (0-559) | 17 (0-1300) | 13 (0-1300) |
| Platelets* [×103/µL], median (range) | 69 (5-778) | 49 (0-635) | 65 (0-903) | 58 (3-9300) | 62 (0-9300) |
| Bone marrow blasts,* median % (range) | 68 (0-98) | 58 (0-99) | 80 (0-100) | 70 (1-100) | 73 (0-100) |
| FAB subclassification,* n (%) | |||||
| M0 | 28 (5) | 76 (8) | 215 (6) | 34 (6) | 353 (6) |
| M1 | 151 (26) | 162 (16) | 977 (27) | 152 (25) | 1442 (25) |
| M2 | 158 (27) | 340 (34) | 999 (27) | 161 (27) | 1658 (28) |
| M4 | 74 (13) | 244 (24) | 766 (21) | 160 (27) | 1244 (21) |
| M5 | 148 (26) | 116 (12) | 523 (14) | 81 (13) | 868 (15) |
| M6 | 19 (3) | 55 (5) | 158 (4) | 9 (1) | 241 (4) |
| M7 | 0 (0) | 10 (1) | 28 (1) | 4 (1) | 42 (1) |
| PS,* n (%) | |||||
| 0 | 250 (43) | 172 (17) | 1841 (50) | 231 (38) | 2494 (43) |
| 1 | 279 (48) | 628 (63) | 1090 (30) | 296 (49) | 2293 (39) |
| ≥2 | 49 (8) | 203 (20) | 735 (20) | 74 (12) | 1061 (18) |
| CR, n (%) | 499 (86) | 649 (65) | 2929 (80) | 383 (64) | 4460 (76) |
| Early death,† n (%) | 19 (3) | 55 (5) | 300 (8) | 37 (6) | 411 (7) |
At diagnosis.
Death within 28 d after initiation of therapy or study registration (if exact date of initiation of therapy was unknown).
Definitions of outcomes
Based on earlier work, early death was defined as death within 28 days after initiating therapy10 or study registration, if the exact date of initiation of therapy was unknown. Designation of complete remission (CR) required the achievement of a morphologic leukemia-free state (bone marrow blasts, <5%; absence of extramedullary disease) and recovery of peripheral blood counts (absolute neutrophil count, >1000/µL; platelet count, >100 000/µL).2,11 Overall survival (OS) was defined as the time from initiation of therapy or study registration (if the date of initiation of therapy was unknown) to death, with censoring on the day they were last known to be alive. For patients who achieved CR, relapse-free survival (RFS) was defined as the time from achievement of remission until relapse or death from any cause, with patients not known to have relapsed or died being censored at last follow-up.
Statistical analysis
OS and RFS were estimated using the Kaplan-Meier method.12 χ2 tests and the Kruskal-Wallis test were used to assess differences between categorical variables and median values of numeric variables across categories, respectively. Cox models were used for regression modeling of OS and RFS, whereas logistic models were used for regression modeling of binary outcomes (early death and CR). The c-statistic was used to quantify the predictive ability of regression models for OS and RFS, whereas the area under the receiver operator characteristic curve (AUC) was used to quantify the predictive ability of regression models for early death and achievement of CR. All analyses were performed using R (http://www.r-project.org).
Results
Characterization of study population
After exclusion of patients with therapy-related neoplasms, AMLs with myelodysplasia-related changes, and leukemias with recurrent cytogenetic abnormalities but ignoring information on NPM1 and CEBPA, our cohort included 5848 predominantly adult patients with newly diagnosed AML that were treated in HOVON, MRC/NCRI, or SWOG protocols or received treatment at MDA and had information on FAB classification available; their baseline characteristics are summarized in Table 1. M0, M6, and particularly M7 were consistently rare, whereas there was more variability in the more common subtypes: M1 (range, 16% to 27%), M4 (13% to 27%), and M5 (12% to 26%). There were 2264 (38.7%) patients that had information on NPM1 mutational status available (44% of these had a mutation), whereas 1546 (26.4%) had data on CEPBA status (10% of which had a mutation); 683 (11.7%) patients had information on both genes. HOVON patients tended to be younger and MDA patients older than MRC/NCRI and SWOG patients. The proportion of patients with abnormal karyotypes [recalling that inv(16), t(8;21), t(15;17), and myelodysplastic syndrome (MDS)-associated karyotypes are excluded from “AML, NOS”] was higher among MDA patients, whereas performance status (PS) 0 was more frequent among MRC/NCRI and HOVON patients. HOVON and MRC/NCRI patients appeared much more likely to be NPM1 mutated. Although induction treatment generally contained cytarabine and daunorubicin or idarubicin, there were expected intergroup differences; for example, induction therapy at MDA more often included cytarabine at >1 g/m2 per dose, whereas at MRC/NCRI, cytarabine at 100 to 200 mg/m2 per dose was typically used for 10 rather than 7 days. Furthermore, HOVON postremission therapy often included amsacrine. Overall, 4460 (76%) of the patients achieved CR, whereas 411 (7%) died within 28 days after initiation of therapy or study registration.
Association of FAB category with outcome in entire study cohort
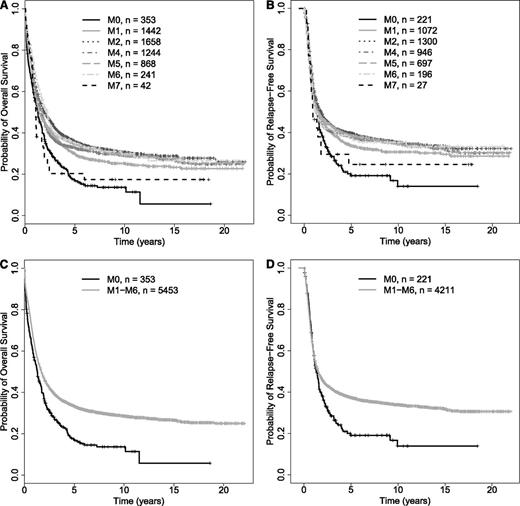
In initial analyses, we included our entire study cohort irrespective of NPM1 and CEBPA status (ie, mutated, not mutated, or mutational status unknown); in other words, we ignored the existence of the 2 WHO provisional entities “AML with mutated NPM1” and “AML with mutated CEBPA” among patients in the “AML, NOS” group. The patient- and disease-specific characteristics, as well as treatment outcomes by FAB category, are summarized in Table 2. Of note, patients with M0 were significantly older and more likely had abnormal karyotypes than other patients; they were also less likely to have NPM1 mutations. Patients with M0 and M7 had significantly lower rates of achieving CR than patients with M1, M2, M4, M5, or M6 AML; this did not reflect a greater early death rate. As shown in Figure 1A-B, patients with M0 had significantly worse OS and RFS than the other patients. Given the differences in distribution of other covariates possibly associated with outcome, we performed multivariate analyses using the following pretreatment covariates in the regression modeling: FAB category, age at diagnosis, white blood cell count (WBC), platelet count, bone marrow blast percentage, gender, PS (0 vs 1 vs ≥2), karyotype (normal vs abnormal), and treatment site (HOVON vs MDA vs MRC/NCRI vs SWOG). As detailed in Table 3, in which M0 serves as the reference considered to have an odds ratio (OR; for early death and CR) and a hazard ratio (HR; for RFS and OS) of 1.0, older age, a worse PS, and a higher WBC but not FAB category were independently associated with early death. On the other hand, after multivariate adjustment, FAB M0 was independently associated with lower likelihood of achieving CR than M1, M2, M4, M5, and M6, but not M7, although the small number of patients (n = 42) in the M7 category may limit the latter comparison. Similarly, after adjustment, FAB M0 was also independently associated with worse OS relative to M1, M2, M4, and M5, but not M6 and M7, and FAB M0 tended to be independently associated with worse RFS relative to M1, M2, M4, and M5. We then used the AUC to quantify the ability of FAB and other covariates to predict early death, achievement of CR, RFS, and OS. As summarized in supplemental Table 1 (see the Blood Web site), the predictive accuracy of these models was relatively limited as indicated by AUCs ranging from 0.58 to 0.77; removal of age had the largest effect on accuracy, whereas removal of FAB and cytogenetics had similar and lesser effects.
Patient/disease-specific characteristics and outcome by FAB category
| . | M0 . | M1 . | M2 . | M4 . | M5 . | M6 . | M7 . | . |
|---|---|---|---|---|---|---|---|---|
| Parameter . | n = 353 . | n = 1442 . | n = 1658 . | n = 1244 . | n = 868 . | n = 241 . | n = 42 . | P value* . |
| Age in y, median (range) | 60 (16-86) | 55 (14-86) | 54 (16-91) | 52 (16-88) | 52 (12-86) | 52 (16-87) | 50 (21-84) | < .001 |
| Patients aged ≥60 y, n (%) | 182 (52) | 568 (39) | 588 (35) | 391 (31) | 252 (29) | 73 (30) | 12 (29) | < .001 |
| Male gender, n (%) | 209 (59) | 762 (53) | 835 (50) | 621 (50) | 432 (50) | 153 (63) | 20 (48) | < .001 |
| Abnormal karyotype,† n (%) | 155 (44) | 442 (31) | 470 (28) | 349 (28) | 261 (30) | 69 (29) | 14 (33) | < .001 |
| NPM1 mutation,† n (%) | ||||||||
| Yes | 13 (11) | 234 (41) | 231 (40) | 245 (50) | 237 (60) | 20 (24) | 7 (50) | < .001 |
| No | 110 (89) | 336 (59) | 353 (60) | 249 (50) | 159 (40) | 63 (76) | 7 (50) | |
| Unknown | 230 | 872 | 1074 | 750 | 472 | 158 | 28 | |
| CEBPA mutation,† n (%) | ||||||||
| Yes | 5 (7) | 50 (13) | 54 (13) | 26 (8) | 14 (5) | 4 (7) | 1 (11) | < .001 |
| No | 71 (93) | 325 (87) | 361 (87) | 294 (92) | 282 (95) | 51 (93) | 8 (89) | |
| Unknown | 277 | 1067 | 1243 | 924 | 572 | 186 | 33 | |
| WBC† [×103/µL], median (range) | 5 (0-559) | 12 (0-435) | 9 (0-1100) | 26 (0-1300) | 39 (0-1104) | 2 (0-169) | 4 (1-35) | < .001 |
| Platelets† [×103/µL], median (range) | 66 (4-358) | 59 (2-7900) | 59 (0-1052) | 64 (0-5300) | 73 (1-9300) | 48 (0-335) | 61 (7-481) | < .001 |
| Bone marrow blasts,† median % (range) | 80 (0-100) | 86 (0-100) | 60 (0-100) | 70 (0-100) | 82 (0-100) | 24 (2-95) | 56 (0-99) | < .001 |
| PS,† n (%) | ||||||||
| 0 | 153 (43) | 640 (44) | 762 (46) | 475 (38) | 325 (37) | 125 (52) | 14 (33) | < .001 |
| 1 | 149 (42) | 572 (40) | 641 (39) | 473 (38) | 354 (41) | 85 (35) | 19 (45) | |
| ≥2 | 51 (14) | 230 (16) | 225 (15) | 296 (24) | 189 (22) | 31 (13) | 9 (21) | |
| CR, n (%) | 221 (63) | 1072 (74) | 1301 (78) | 946 (76) | 697 (80) | 196 (81) | 27 (64) | < .001 |
| Early death,‡ n (%) | 29 (8) | 119 (8) | 84 (5) | 92 (7) | 80 (9) | 6 (2) | 1 (2) | < .001 |
| . | M0 . | M1 . | M2 . | M4 . | M5 . | M6 . | M7 . | . |
|---|---|---|---|---|---|---|---|---|
| Parameter . | n = 353 . | n = 1442 . | n = 1658 . | n = 1244 . | n = 868 . | n = 241 . | n = 42 . | P value* . |
| Age in y, median (range) | 60 (16-86) | 55 (14-86) | 54 (16-91) | 52 (16-88) | 52 (12-86) | 52 (16-87) | 50 (21-84) | < .001 |
| Patients aged ≥60 y, n (%) | 182 (52) | 568 (39) | 588 (35) | 391 (31) | 252 (29) | 73 (30) | 12 (29) | < .001 |
| Male gender, n (%) | 209 (59) | 762 (53) | 835 (50) | 621 (50) | 432 (50) | 153 (63) | 20 (48) | < .001 |
| Abnormal karyotype,† n (%) | 155 (44) | 442 (31) | 470 (28) | 349 (28) | 261 (30) | 69 (29) | 14 (33) | < .001 |
| NPM1 mutation,† n (%) | ||||||||
| Yes | 13 (11) | 234 (41) | 231 (40) | 245 (50) | 237 (60) | 20 (24) | 7 (50) | < .001 |
| No | 110 (89) | 336 (59) | 353 (60) | 249 (50) | 159 (40) | 63 (76) | 7 (50) | |
| Unknown | 230 | 872 | 1074 | 750 | 472 | 158 | 28 | |
| CEBPA mutation,† n (%) | ||||||||
| Yes | 5 (7) | 50 (13) | 54 (13) | 26 (8) | 14 (5) | 4 (7) | 1 (11) | < .001 |
| No | 71 (93) | 325 (87) | 361 (87) | 294 (92) | 282 (95) | 51 (93) | 8 (89) | |
| Unknown | 277 | 1067 | 1243 | 924 | 572 | 186 | 33 | |
| WBC† [×103/µL], median (range) | 5 (0-559) | 12 (0-435) | 9 (0-1100) | 26 (0-1300) | 39 (0-1104) | 2 (0-169) | 4 (1-35) | < .001 |
| Platelets† [×103/µL], median (range) | 66 (4-358) | 59 (2-7900) | 59 (0-1052) | 64 (0-5300) | 73 (1-9300) | 48 (0-335) | 61 (7-481) | < .001 |
| Bone marrow blasts,† median % (range) | 80 (0-100) | 86 (0-100) | 60 (0-100) | 70 (0-100) | 82 (0-100) | 24 (2-95) | 56 (0-99) | < .001 |
| PS,† n (%) | ||||||||
| 0 | 153 (43) | 640 (44) | 762 (46) | 475 (38) | 325 (37) | 125 (52) | 14 (33) | < .001 |
| 1 | 149 (42) | 572 (40) | 641 (39) | 473 (38) | 354 (41) | 85 (35) | 19 (45) | |
| ≥2 | 51 (14) | 230 (16) | 225 (15) | 296 (24) | 189 (22) | 31 (13) | 9 (21) | |
| CR, n (%) | 221 (63) | 1072 (74) | 1301 (78) | 946 (76) | 697 (80) | 196 (81) | 27 (64) | < .001 |
| Early death,‡ n (%) | 29 (8) | 119 (8) | 84 (5) | 92 (7) | 80 (9) | 6 (2) | 1 (2) | < .001 |
P value of a test of the null hypothesis that all the groups are the same (eg, distribution of age is the same across all cohorts).
At diagnosis.
Death within 28 d after initiation of therapy or study registration (if exact date of initiation of therapy was unknown).
Survival estimates in newly diagnosed patients with “AML, NOS.” Kaplan-Meier estimates of OS (A) of 5848 and RFS (B) of 4458 patients with “AML, NOS” based on WHO 2008 classification by individual FAB category. (C-D) Survival in patients with FAB M0 and M1-M6. Kaplan-Meier estimates of OS (C) and RFS (D) of patients with newly diagnosed “AML, NOS” based on WHO 2008 classification and subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis.
Survival estimates in newly diagnosed patients with “AML, NOS.” Kaplan-Meier estimates of OS (A) of 5848 and RFS (B) of 4458 patients with “AML, NOS” based on WHO 2008 classification by individual FAB category. (C-D) Survival in patients with FAB M0 and M1-M6. Kaplan-Meier estimates of OS (C) and RFS (D) of patients with newly diagnosed “AML, NOS” based on WHO 2008 classification and subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis.
Multivariate Cox regression models for early death, achievement of CR, and RFS/OS among all patients with “AML, NOS” regardless of NPM1 and CEBPA status
| Parameter . | Early death*,† . | CR* . | RFS* . | OS* . |
|---|---|---|---|---|
| M1 | 1.06 (0.68-1.65), P = .79 | 1.47 (1.13-1.93), P = .0047 | 0.85 (0.71-1.00), P = .057 | 0.82 (0.72-0.94), P = .0033 |
| M2 | 0.83 (0.52-1.31), P = .42 | 1.73 (1.32-2.26), P < .001 | 0.83 (0.70-0.98), P = .029 | 0.78 (0.68-0.89), P < .001 |
| M4 | 1.05 (0.66-1.66), P = .84 | 1.64 (1.24-2.17), P < .001 | 0.91 (0.76-1.08), P = .28 | 0.86 (0.75-0.98), P = .027 |
| M5 | 1.28 (0.80-2.05), P = .30 | 2.02 (1.50-2.73), P < .001 | 0.88 (0.73-1.05), P = .16 | 0.83 (0.72-0.96), P = .011 |
| M6 | 0.50 (0.19-1.27), P = .14 | 1.70 (1.10-2.63), P = .017 | 1.00 (0.78-1.28), P = .99 | 0.89 (0.73-1.10), P = .28 |
| M7 | 0.41 (0.05-3.18), P = .39 | 0.71 (0.34-1.47), P = .35 | 1.22 (0.76-1.97), P = .42 | 1.34 (0.94-1.92), P = .11 |
| Age | 1.06 (1.05-1.07), P < .001 | 0.95 (0.95-0.96), P < .001 | 1.02 (1.02-1.02), P < .001 | 1.03 (1.03-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P < .001 | 1.00 (1.00-1.00), P < .001 | 1.00 (1.00-1.00), P < .001 | 1.00 (1.00-1.00), P < .001 |
| Platelets | 1.00 (1.00-1.00), P = .64 | 1.00 (1.00-1.00), P = .28 | 1.00 (1.00-1.00), P = .012 | 1.00 (1.00-1.00), P = .077 |
| Bone marrow blasts | 1.00 (1.00-1.01), P = .13 | 1.00 (1.00-1.00), P = .50 | 1.00 (1.00-1.00), P = .0074 | 1.00 (1.00-1.00), P < .001 |
| Male gender | 1.09 (0.88-1.34), P = .44 | 0.91 (0.80-1.04), P = .18 | 1.09 (1.02-1.18), P = .018 | 1.05 (0.98-1.11), P = .16 |
| PS = 1 | 1.14 (0.88-1.48), P = .32 | 0.75 (0.65-0.88), P < .001 | 0.98 (0.90-1.07), P = .62 | 1.05 (0.97-1.13), P = .22 |
| PS ≥ 2 | 2.95 (2.25-3.87), P < .001 | 0.49 (0.41-0.60), P < .001 | 1.15 (1.03-1.28), P = .013 | 1.44 (1.32-1.57), P < .001 |
| Abnormal karyotype | 1.13 (0.89-1.43), P = .32 | 0.67 (0.58-0.78), P < .001 | 1.15 (1.06-1.25), P = .0013 | 1.25 (1.16-1.34), P < .001 |
| Parameter . | Early death*,† . | CR* . | RFS* . | OS* . |
|---|---|---|---|---|
| M1 | 1.06 (0.68-1.65), P = .79 | 1.47 (1.13-1.93), P = .0047 | 0.85 (0.71-1.00), P = .057 | 0.82 (0.72-0.94), P = .0033 |
| M2 | 0.83 (0.52-1.31), P = .42 | 1.73 (1.32-2.26), P < .001 | 0.83 (0.70-0.98), P = .029 | 0.78 (0.68-0.89), P < .001 |
| M4 | 1.05 (0.66-1.66), P = .84 | 1.64 (1.24-2.17), P < .001 | 0.91 (0.76-1.08), P = .28 | 0.86 (0.75-0.98), P = .027 |
| M5 | 1.28 (0.80-2.05), P = .30 | 2.02 (1.50-2.73), P < .001 | 0.88 (0.73-1.05), P = .16 | 0.83 (0.72-0.96), P = .011 |
| M6 | 0.50 (0.19-1.27), P = .14 | 1.70 (1.10-2.63), P = .017 | 1.00 (0.78-1.28), P = .99 | 0.89 (0.73-1.10), P = .28 |
| M7 | 0.41 (0.05-3.18), P = .39 | 0.71 (0.34-1.47), P = .35 | 1.22 (0.76-1.97), P = .42 | 1.34 (0.94-1.92), P = .11 |
| Age | 1.06 (1.05-1.07), P < .001 | 0.95 (0.95-0.96), P < .001 | 1.02 (1.02-1.02), P < .001 | 1.03 (1.03-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P < .001 | 1.00 (1.00-1.00), P < .001 | 1.00 (1.00-1.00), P < .001 | 1.00 (1.00-1.00), P < .001 |
| Platelets | 1.00 (1.00-1.00), P = .64 | 1.00 (1.00-1.00), P = .28 | 1.00 (1.00-1.00), P = .012 | 1.00 (1.00-1.00), P = .077 |
| Bone marrow blasts | 1.00 (1.00-1.01), P = .13 | 1.00 (1.00-1.00), P = .50 | 1.00 (1.00-1.00), P = .0074 | 1.00 (1.00-1.00), P < .001 |
| Male gender | 1.09 (0.88-1.34), P = .44 | 0.91 (0.80-1.04), P = .18 | 1.09 (1.02-1.18), P = .018 | 1.05 (0.98-1.11), P = .16 |
| PS = 1 | 1.14 (0.88-1.48), P = .32 | 0.75 (0.65-0.88), P < .001 | 0.98 (0.90-1.07), P = .62 | 1.05 (0.97-1.13), P = .22 |
| PS ≥ 2 | 2.95 (2.25-3.87), P < .001 | 0.49 (0.41-0.60), P < .001 | 1.15 (1.03-1.28), P = .013 | 1.44 (1.32-1.57), P < .001 |
| Abnormal karyotype | 1.13 (0.89-1.43), P = .32 | 0.67 (0.58-0.78), P < .001 | 1.15 (1.06-1.25), P = .0013 | 1.25 (1.16-1.34), P < .001 |
Data are presented as OR or HR (95% CI), as appropriate.
Death within 28 d after initiation of therapy or study registration (if exact date of initiation of therapy was unknown).
These data suggested that FAB M0 is associated with worse outcomes, both short term (achievement of CR) and long term (survival) than the other FAB categories except possibly M7. We thus combined FAB M1, M2, M4, M5, and M6 into 1 category (“M1-M6”); given the uncertainty and small sample size with M7, we excluded such cases from this analysis. As shown in Figure 1 C-D, patients with M0 had inferior OS and RFS than those with FAB M1-M6. In multivariate regression models, adjusting for the same covariates as discussed previously, FAB M0 was associated with a similar risk of early death (OR = 1.01 [0.67-1.53], P = .96) but lower likelihood of achieving CR (OR = 1.66 [1.30-2.12], P < .001) and worse RFS (HR = 0.86 [0.73-1.01], P = .068) and OS (HR = 0.82 [0.72-0.92], P = .0013) than FAB M1-M6 (supplemental Table 2).
Association of FAB type with outcome in “AML, NOS” after restriction to patients with NPM1– and NPM1–/CEBPA– leukemias
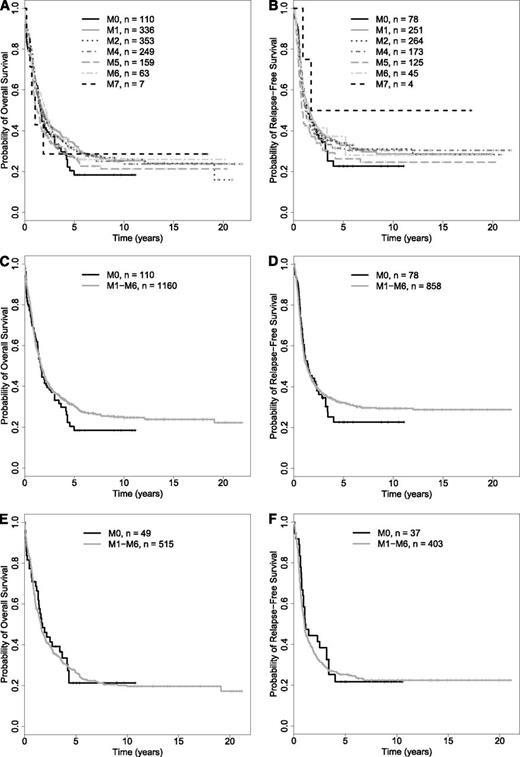
As noted previously, the 2008 WHO classification recognizes the 2 provisional entities “AML with mutated NPM1” and “AML with mutated CEBPA” within the subset of “AML with recurrent genetic abnormalities”; of note, patients with CEBPA mutations are placed in the “AML with mutated CEBPA” entity regardless of the specifics of the mutational status (ie, single or double CEBPA mutation).8,9 We therefore wondered about the association between FAB type and outcome among “AML, NOS” cases after restriction of our data set to patients that were NPM1– (in a first analysis) and NPM1–/CEBPA– (in a second analysis). We were particularly interested in these analyses as we found favorable effects of NPM1 and/or CEBPA mutations in our patient cohort. Specifically, multivariate models, in which we only excluded patients with unknown NPM1 or CEBPA status, indicated that being NPM1+ was associated with a higher likelihood of achieving CR and a better RFS and OS than being NPM1–, whereas being CEBPA+ (single or double mutation) was associated with better RFS and OS than being CEBPA– (data not shown). This favorable effect of NPM1 and/or CEBPA mutations together with the relative infrequency of the former in patients with FAB M0 suggested that the effects of FAB M0 and NPM1 mutations were confounded and provide an empirical rationale for exclusion of patients with NPM1 (and CEBPA) mutations in the WHO classification. To investigate this further, we therefore performed analyses in which we stepwise excluded patients based on information on the mutational status of NPM1 and CEBPA to reproduce the current WHO classification. As testing for these mutations has only recently become routine, many patients lacked molecular analyses of these genes and thus had to be a priori excluded from these analyses (Table 1). In a first step, we assessed the effect of NPM1 mutations and disregarded CEBPA data. Among 5848 patients, 2264 had available NPM1 data (39%), and 987 of those 2264 patients (44%) with known NPM1 mutation status were found to be NPM1+. In contrast, 1277 of the 2294 patients with available NPM1 data (56%) tested negative for the NPM1 mutation. As shown in Table 2, the prevalence of NPM1+ leukemias was lowest (11%) among cases classified as FAB M0. Supplemental Table 3 and Figure 2A-B summarize the disease characteristics and treatment outcome after exclusion of patients in whom an NPM1 mutation was present or whose NPM1 status was unknown (ie, restriction to NPM1– patients). Unlike the situation where all patients were included regardless of NPM1 status, CR rates were relatively similar across all FAB categories in this NPM1– patient subset, as were estimates of OS and RFS. Consistently, as shown in Figure 2C-D, OS and RFS of NPM1– patients with M0 AML were not significantly different from that of NPM1– patients with M1-M6 AMLs. In our cohort, this convergence was due to both worsening of outcome in the M1-M6 group and improvement in outcome in the M0 group (supplemental Figure 1). Consistent with this observation, in multivariate regression models, FAB M0 was associated with a similar risk of early death (OR = 1.03 [0.35-3.03], P = .95), likelihood of achieving CR (OR = 1.19 [0.75-1.91], P = .46), RFS (HR= 0.98 [0.74-1.31], P = .89), and OS (HR = 0.91 [0.72-1.16], P = .45) as FAB M1-M6 (Table 4). Similar to the models built with all “AML, NOS” patients, the accuracies of models predicting early death, achievement of CR, RFS, and OS were relatively limited, as indicated by AUCs ranging from 0.58 to 0.78, and neither removal of FAB nor karyotypic information substantially altered the AUCs of models containing these covariates (supplemental Table 4).
Survival estimates of NPM1– and NPM1–/CEBPA– patients with newly diagnosed “AML, NOS.” (A-D) NPM1– patients. OS (A) and RFS (B) of NPM1– patients by individual FAB category. OS (C) and RFS (D) of NPM1– patients subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis. (E-F) NPM1–/CEBPA– patients. OS (E) and RFS (F) of NPM1–/CEBPA– patients subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis.
Survival estimates of NPM1– and NPM1–/CEBPA– patients with newly diagnosed “AML, NOS.” (A-D) NPM1– patients. OS (A) and RFS (B) of NPM1– patients by individual FAB category. OS (C) and RFS (D) of NPM1– patients subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis. (E-F) NPM1–/CEBPA– patients. OS (E) and RFS (F) of NPM1–/CEBPA– patients subclassified as either FAB M0 or M1-M6; patients with FAB M7 were excluded from this analysis.
Multivariate Cox regression models for early death, achievement of CR, and disease-free OS for FAB M0 vs FAB M1-M6 among 1277 NPM1– patients and 564 NPM1–/CEBPA– patients with “AML, NOS”
| Parameter . | Early death*,† . | CR* . | RFS* . | OS* . |
|---|---|---|---|---|
| NPM1– | ||||
| M1-M6 | 1.03 (0.35-3.03), P = .95 | 1.19 (0.75-1.91), P = .46 | 0.98 (0.74-1.31), P = .89 | 0.91 (0.72-1.16), P = .45 |
| Age | 1.04 (1.02-1.06), P < .001 | 0.96 (0.96-0.97), P < .001 | 1.02 (1.01-1.02), P < .001 | 1.03 (1.02-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P = .11 | 0.99 (0.99-1.00), P < .001 | 1.00 (1.00-1.00), P = .096 | 1.00 (1.00-1.00), P < .001 |
| Platelets | 1.00 (0.99-1.00), P = .13 | 1.00 (1.00-1.00), P = .53 | 1.00 (1.00-1.00), P = .72 | 1.00 (1.00-1.00), P = .75 |
| Bone marrow blasts | 1.00 (0.99-1.02), P = .062 | 1.00 (1.00-1.01), P = .49 | 1.00 (1.00-1.00), P = .46 | 1.00 (1.00-1.01), P = .14 |
| Male gender | 0.81 (0.46-1.44), P = .48 | 0.78 (0.60-1.03), P = .078 | 1.14 (0.97-1.34), P = .11 | 1.06 (0.93-1.22), P = .37 |
| PS = 1 | 1.74 (0.77-3.95), P = .19 | 0.77 (0.56-1.06), P = .11 | 0.97 (0.81-1.17), P = .78 | 1.09 (0.93-1.27), P = .29 |
| PS ≥ 2 | 6.02 (2.64-13.72), P < .001 | 0.54 (0.36-0.81), P = .0034 | 1.26 (0.98-1.62), P = .069 | 1.50 (1.23-1.84), P < .001 |
| Abnormal karyotype | 1.59 (0.88-2.88), P = .13 | 0.85 (0.64-1.12), P = .24 | 1.20 (1.01-1.42), P = .039 | 1.29 (1.12-1.49), P < .001 |
| NPM1–/CEBPA– | ||||
| M1-M6 | 0.59 (0.12-2.97), P = .52 | 1.16 (0.56-2.42), P = .69 | 1.11 (0.74-1.66), P = .62 | 1.00 (0.70-1.43), P = .99 |
| Age | 1.06 (1.02-1.09), P < .001 | 0.98 (0.96-0.99), P = .0012 | 1.01 (1.00-1.01), P = .17 | 1.02 (1.01-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P = .35 | 0.99 (0.99-1.00), P < .001 | 1.00 (1.00-1.00), P = .25 | 1.00 (1.00-1.00), P = .0055 |
| Platelets | 1.00 (0.99-1.00), P = .66 | 1.00 (1.00-1.00), P = .098 | 1.00 (1.00-1.00), P = .21 | 1.00 (1.00-1.00), P = .066 |
| Bone marrow blasts | 1.01 (0.98-1.03), P = .51 | 1.00 (0.99-1.01), P = .92 | 1.00 (0.99-1.00), P = .48 | 1.00 (1.00-1.01), P = .81 |
| Male gender | 0.78 (0.31-1.95), P = .60 | 0.66 (0.43-1.03), P = .067 | 1.19 (0.95-1.50), P = .13 | 1.14 (0.93-1.40), P = .20 |
| PS = 1 | 1.27 (0.35-4.68), P = .71 | 0.73 (0.44-1.19), P = .21 | 0.85 (0.66-1.10), P = .22 | 0.98 (0.78-1.23), P = .87 |
| PS ≥ 2 | 6.11 (1.75-21.28), P = .0045 | 0.58 (0.31-1.07), P = .083 | 1.07 (0.77-1.48), P = .68 | 1.25 (0.94-1.65), P = .13 |
| Abnormal karyotype | 2.34 (0.91-6.04), P = .077 | 0.89 (0.57-1.38), P = .59 | 1.07 (0.85-1.35), P = .58 | 1.21 (1.12-1.49), P = .068 |
| Parameter . | Early death*,† . | CR* . | RFS* . | OS* . |
|---|---|---|---|---|
| NPM1– | ||||
| M1-M6 | 1.03 (0.35-3.03), P = .95 | 1.19 (0.75-1.91), P = .46 | 0.98 (0.74-1.31), P = .89 | 0.91 (0.72-1.16), P = .45 |
| Age | 1.04 (1.02-1.06), P < .001 | 0.96 (0.96-0.97), P < .001 | 1.02 (1.01-1.02), P < .001 | 1.03 (1.02-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P = .11 | 0.99 (0.99-1.00), P < .001 | 1.00 (1.00-1.00), P = .096 | 1.00 (1.00-1.00), P < .001 |
| Platelets | 1.00 (0.99-1.00), P = .13 | 1.00 (1.00-1.00), P = .53 | 1.00 (1.00-1.00), P = .72 | 1.00 (1.00-1.00), P = .75 |
| Bone marrow blasts | 1.00 (0.99-1.02), P = .062 | 1.00 (1.00-1.01), P = .49 | 1.00 (1.00-1.00), P = .46 | 1.00 (1.00-1.01), P = .14 |
| Male gender | 0.81 (0.46-1.44), P = .48 | 0.78 (0.60-1.03), P = .078 | 1.14 (0.97-1.34), P = .11 | 1.06 (0.93-1.22), P = .37 |
| PS = 1 | 1.74 (0.77-3.95), P = .19 | 0.77 (0.56-1.06), P = .11 | 0.97 (0.81-1.17), P = .78 | 1.09 (0.93-1.27), P = .29 |
| PS ≥ 2 | 6.02 (2.64-13.72), P < .001 | 0.54 (0.36-0.81), P = .0034 | 1.26 (0.98-1.62), P = .069 | 1.50 (1.23-1.84), P < .001 |
| Abnormal karyotype | 1.59 (0.88-2.88), P = .13 | 0.85 (0.64-1.12), P = .24 | 1.20 (1.01-1.42), P = .039 | 1.29 (1.12-1.49), P < .001 |
| NPM1–/CEBPA– | ||||
| M1-M6 | 0.59 (0.12-2.97), P = .52 | 1.16 (0.56-2.42), P = .69 | 1.11 (0.74-1.66), P = .62 | 1.00 (0.70-1.43), P = .99 |
| Age | 1.06 (1.02-1.09), P < .001 | 0.98 (0.96-0.99), P = .0012 | 1.01 (1.00-1.01), P = .17 | 1.02 (1.01-1.03), P < .001 |
| WBC | 1.00 (1.00-1.01), P = .35 | 0.99 (0.99-1.00), P < .001 | 1.00 (1.00-1.00), P = .25 | 1.00 (1.00-1.00), P = .0055 |
| Platelets | 1.00 (0.99-1.00), P = .66 | 1.00 (1.00-1.00), P = .098 | 1.00 (1.00-1.00), P = .21 | 1.00 (1.00-1.00), P = .066 |
| Bone marrow blasts | 1.01 (0.98-1.03), P = .51 | 1.00 (0.99-1.01), P = .92 | 1.00 (0.99-1.00), P = .48 | 1.00 (1.00-1.01), P = .81 |
| Male gender | 0.78 (0.31-1.95), P = .60 | 0.66 (0.43-1.03), P = .067 | 1.19 (0.95-1.50), P = .13 | 1.14 (0.93-1.40), P = .20 |
| PS = 1 | 1.27 (0.35-4.68), P = .71 | 0.73 (0.44-1.19), P = .21 | 0.85 (0.66-1.10), P = .22 | 0.98 (0.78-1.23), P = .87 |
| PS ≥ 2 | 6.11 (1.75-21.28), P = .0045 | 0.58 (0.31-1.07), P = .083 | 1.07 (0.77-1.48), P = .68 | 1.25 (0.94-1.65), P = .13 |
| Abnormal karyotype | 2.34 (0.91-6.04), P = .077 | 0.89 (0.57-1.38), P = .59 | 1.07 (0.85-1.35), P = .58 | 1.21 (1.12-1.49), P = .068 |
Data are presented as OR or HR (95% CI), as appropriate.
Death within 28 d after initiation of therapy or study registration (if exact date of initiation of therapy was unknown).
We then performed exploratory analysis on the subset of 564 NPM1– patients who had CEBPA information available and were CEBPA–. Within the limitations of a relatively small data set, the results were essentially identical to those obtained in the NPM1– patient subset. Specifically, OS and RFS of NPM1–/CEBPA– patients with M0 AML were not different from those with M1-M6 AMLs (Figure 2E-F). Likewise, in multivariate regression models, FAB M0 was associated with a similar likelihood of achieving CR (OR = 1.16 [0.56-2.42], P = .69), RFS (HR = 1.11 [0.74-1.66], P = .62), and OS (HR = 1.00 [0.70-1.43], P = .99) as FAB M1-M6 in NPM1–/CEBPA– patients with “AML, NOS” (Table 4).
Finally, given the differences among HOVON, MDA, MRC/NCRI, and SWOG with respect to the covariates noted in the first paragraph of the “Results” section and Table 1, we examined whether our general conclusions were similarly applicable in each of these groups. To accomplish this, we constructed terms describing the multivariate interaction between FAB M1 at HOVON, FAB M1 at MDA, FAB M1 at MRC/NCRI, M1 in SWOG, and so on for FAB M2-M6. There were insufficient patients to do the same for FAB M7, and too few events to do this for early death and RFS. Within these constraints, and acknowledging the low power even for the end points of CR and OS, there was no evidence to suggest that the effect of FAB differed at different sites (P = .19 for CR; P = .12 for OS). Of note, only 21 of 578 (3.6%) HOVON patients had missing NPM1 and/or CEBPA status information, and analysis of the HOVON data set confirmed the prognostic irrelevance of FAB M0 once analyses were restricted to NPM1– and CEBPA– patients (data not shown).
Discussion
The classification of AML has evolved over time, departing from schemes such as FAB that relied principally on morphology and instead including cytogenetic and molecular data as the possible pathogenic and obvious prognostic role of these become clearer. The current WHO classification recognizes such defined disease entities as AMLs “with recurrent genetic abnormalities,” a category that will likely further expand in the future to encompass leukemias currently included in the “AML, NOS” category, much as has happened with “AML with mutated NPM1” and “AML with mutated CEBPA.” Therefore, the composition of leukemias remaining in the “AML, NOS” category will change, calling for periodic reassessment of prognosis in such patients.
The findings from the present study directly support this notion. Specifically, our retrospective data derived from a large number of patients indicate that morphologic features among cases of “AML, NOS” provided independent prognostic information before the recognition of AMLs with mutated NPM1 and CEBPA as separate entities. Thus, although “AML, NOS” patients classified as FAB M0 had significantly worse outcomes than patients classified as FAB M1, M2, M4, M5, or M6, removal of NPM1+ cases, as in the current WHO scheme, as well as cases with unknown NPM1 status, eliminated the effect of morphology. This observation is likely explained by the higher prevalence of NPM1+ cases among non-M0 AMLs in our data set and the fact that NPM1+ leukemias generally have a more favorable outcome.13 Consequently, outcomes of cases with FAB M0 and non-M0 are expected to converge after removal of NPM1+ patients. Indeed, although there were only 13 NPM1+ patients vs 230 patients with unknown NPM1 status among the 243 FAB M0 patients that were removed for analysis in which NPM1 was accounted for, analysis of HOVON data (only 1% of FAB M0 patients with missing NPM1 data) confirmed that removal of NPM1+ patients eliminated the unfavorable effect of FAB M0. The same was true for CEBPA+ patients, where again HOVON had relatively few cases whose CEBPA status was unknown.
Our ability to predict important outcomes in AML, such as early death, achievement of CR, or long-term outcome remains limited, as indicated by our multicomponent models (see Table 4). This finding is consistent with our previous observations.10,14 Interestingly, although data on FAB subtype provided prognostic information across our cohort of “AML, NOS” patients, at least as long as data on NPM1 were ignored, the accuracy of models predicting achievement of CR or survival did not substantially change when FAB information was removed from such models. This suggests that for individual patients FAB information is not that helpful in predicting outcomes and is replaced by information provided by other covariates. Similarly, the accuracy of our models was not substantially different after removal of cytogenetic information from these models, possibly reflective of the fact that the spectrum of cytogenetic abnormalities within the “AML, NOS” subgroup is more limited than in AML at large, as cases at either end of the prognostic spectrum are not included in this category.
Several limitations need to be acknowledged. First, although a substantial number of patients had molecular information on NPM1 available, only a relatively small subset of patients had molecular data on both NPM1 and CEBPA available, limiting the conclusions that can be drawn regarding the importance of CEBPA. However, consistent with previous observations, CEBPA+ leukemias were relatively rare in our study population. Thus, removal of CEBPA+ patients, reflecting their current classification under AMLs with recurrent genetic abnormalities, is unlikely to significantly change the prognostic role of FAB information in the remaining patients. Second, despite the use of a large study cohort, the rarity of “AML, NOS” FAB M7 cases precluded any firm conclusions regarding the significance of this FAB subcategory, but our data suggested that patients with AML FAB M7 may have outcomes more similar to patients with FAB M0 AML than those with non-M0 AMLs. Third, our data sets did not contain information on cellular dysplasia. We were therefore only able to exclude cases of “AML with myelodysplasia-related changes” based on history of antecedent hematologic disorder or presence of cytogenetic abnormalities sufficient for the diagnosis of this disease subgroup but not based on the presence of dysplastic features. And fourth, our data sets were incomplete regarding the use of hematopoietic cell transplantation (HCT), and we could therefore not assess what potential role HCT played in the relationship between FAB type and outcome among patients with “AML, NOS.” However, because adverse disease features were more common in those diagnosed as FAB M0, we can speculate that patients with FAB M0 AMLs more frequently underwent HCT, a possibility that could have led us to underestimate the difference in outcome between patients with FAB M0 and those with other FAB types.
In summary, our data suggest that morphologic subclassification of “AML, NOS” cases according to FAB criteria does not provide prognostic information if molecular data on NPM1 and CEBPA mutations are available. In turn, this finding leads us to question the utility of continued use of the FAB type in the relatively small subset of AML patients with “AML, NOS” at least for prognostic purposes, although FAB may remain useful for pathological discrimination of these cases awaiting further cytogenetic/molecular characterization.
Presented in part at the 54th annual meeting of the American Society of Hematology, Atlanta, GA, December 8-11, 2012.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Min Fang (Seattle Cancer Care Alliance) for help in the cytogenetic classification of our study population.
This work was supported by grants from the National Cancer Institute/National Institutes of Health (NCI/NIH) (P30-CA015704-35S6, R.B.W.; and R01-CA090998-09, M.O.). SWOG trials were supported in part by the following Public Health Service Cooperative Agreement grant numbers awarded by the NCI/NIH (U10-CA032102, U10-CA038926, and U10-CA105409).
Authorship
Contribution: R.B.W., M.O., and E.H.E. contributed to the conception and design of the study; A.K.B., B.L., H.M.K, G.J.O., F.R., and F.R.A. contributed to the provision of study material, patient recruitment, and acquisition of data; M.O., R.K.H, K.G.M.v.M., A.E., and S.R.P. participated in the collection and assembly of data; R.B.W., M.O., A.K.B., B.L., H.M.K., R.K.H., F.R.A., and E.H.E. participated in the data analysis and interpretation; R.B.W., M.O., and E.H.E. participated in drafting of the manuscript; and all authors revised the manuscript critically and gave final approval to submit for publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Roland B. Walter, Clinical Research Division, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, D2-190, Seattle, WA 98109-1024; e-mail: rwalter@fhcrc.org.