Key Points
Expression of RUNX1a, an isoform of RUNX1, enhances blood cell production from human pluripotent stem cells.
Abstract
Advancements in human pluripotent stem cell (hPSC) research have potential to revolutionize therapeutic transplantation. It has been demonstrated that transcription factors may play key roles in regulating maintenance, expansion, and differentiation of hPSCs. In addition to its regulatory functions in hematopoiesis and blood-related disorders, the transcription factor RUNX1 is also required for the formation of definitive blood stem cells. In this study, we demonstrated that expression of endogenous RUNX1a, an isoform of RUNX1, parallels with lineage commitment and hematopoietic emergence from hPSCs, including both human embryonic stem cells and inducible pluripotent stem cells. In a defined hematopoietic differentiation system, ectopic expression of RUNX1a facilitates emergence of hematopoietic progenitor cells (HPCs) and positively regulates expression of mesoderm and hematopoietic differentiation-related factors, including Brachyury, KDR, SCL, GATA2, and PU.1. HPCs derived from RUNX1a hPSCs show enhanced expansion ability, and the ex vivo–expanded cells are capable of differentiating into multiple lineages. Expression of RUNX1a in embryoid bodies (EBs) promotes definitive hematopoiesis that generates erythrocytes with β-globin production. Moreover, HPCs generated from RUNX1a EBs possess ≥9-week repopulation ability and show multilineage hematopoietic reconstitution in vivo. Together, our results suggest that RUNX1a facilitates the process of producing therapeutic HPCs from hPSCs.
Introduction
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) are pluripotent stem cells (PSCs), which can differentiate into nearly all types of cells and have self-renewal ability. Recent advancements in hESC and iPSC production have revolutionized their potential applications in regenerative medicine.1-5 Many patients with blood disorders can be treated or even cured with hematopoietic stem cell (HSC) transplantation.
In humans and mice, 3 RUNX genes have been identified, and all have been shown to play important roles in normal and cancer development.6-8 They, as the α subunit of the core binding factor (CBF) complex, share a high degree of homology in the runt homology domain, a homolog of the Drosophila runt protein, which is critical for both DNA binding and heterodimerization with CBFβ.9,10 Furthermore, their expression patterns are highly regulated temporally and spatially during embryogenesis. RUNX1 (also known as AML1, CBFA2, or PEBP2aB) is essential for the establishment of the definitive hematopoietic system during embryogenesis6,7,11 and is involved in the most frequent chromosomal translocation associated with human leukemia.10 It has been shown that mouse Runx1−/− ESCs are defective in hematopoietic differentiation in mice. This hematopoietic defect can be rescued by reexpression of Runx1 in a transcriptional activation domain–dependent manner.12,13 In the mouse embryo, Runx1 is expressed in all sites from which hematopoietic cells emerge. All the sources of definitive HSCs in the embryo express Runx1, which appears to regulate the specification of definitive HSCs in developing mouse embryos. It has been demonstrated that intraaortic hematopoietic clusters associated with the hemogenic endothelium, from which definitive HSCs emerge, are absent in Runx1−/− embryos.14 The observation that Runx1 is expressed in endothelial cells prior to the onset of definitive hematopoiesis supports the widely accepted hypothesis that blood cells develop from a “hemogenic endothelium.”15
The human RUNX1 gene has 12 exons, and its expression is controlled by 2 promoters that generate ≥12 different RUNX1 mRNA isoforms and 3 main protein isoforms.16-18 P1 and P2 promoters regulate expression of isoforms RUNX1c and RUNX1b, respectively. RUNX1b and RUNX1c both possess the DNA-binding region at the amino terminus and transcriptional regulatory domains at the carboxyl terminus. They are broadly considered similar in function, although RUNX1c has 27 extra amino acids at the amino terminus. Like RUNX1b, isoform RUNX1a is also regulated by the P2 promoter. However, RUNX1a retains the DNA-binding domain but lacks the transcriptional regulatory domains because of the alternative splicing. It has been reported that RUNX1 isoforms do not show any discernible functional difference in mouse hematopoietic stem/progenitor cells (HSPCs),18 and knockdown of RUNX1c does not affect normal hematopoietic development.19 However, enforced expression of RUNX1a in murine hematopoietic cells enhances engraftment after transplantation into mice, whereas RUNX1b/c does not,20 suggesting a critical role for RUNX1a in stem/progenitor cell expansion. These observations prompt the idea that RUNX1a may play a distinct role during hematopoiesis. In this report, we investigate the function of RUNX1a on hematopoietic cell commitment, expansion, and differentiation and aim to provide evidence demonstrating that expression of RUNX1a facilitates hematopoietic lineage commitment of hPSCs.
Methods
Refer to the supplemental Methods (on the Blood website) for additional details.
Long-term culture assays
M2-10B4 cells were treated with mitomycin C before plating on gelatin-coated 24-well plates at 1 × 105 to 2 × 105 cells per well.21 Cells were allowed to adhere overnight before adding umbilical cord blood– or hESC-derived hematopoietic progenitor cells (HPCs). For bulk culture, 5 × 103 CD34+ umbilical cord blood– or 1 × 104 hESC-derived cells were plated on top of mitotically inactivated M2-10B4 cells in 1 mL per well Myelocult H5100 media (StemCell Technologies) containing 1 μM hydrocortisone (Sigma).22 Cells were fed with fresh medium by half medium changes every 7 days. At the indicated time points, cells were harvested via collection of nonadherent cells and dissociation of the adherent cell layer with 0.05% trypsin. The nonadherent and adherent cells were combined and passed through a 70-μm filter (BD Biosciences) to remove clumps. After counting, cells were subjected to a hematopoietic colony forming cell (CFC) assay. Photos were taken by Nikon ECLIPSE TS100 microscope. For comparison of long-term culture-initiating cell (LTC-IC) frequency from CD34+CD45+ cells cultured on feeder layers, CD34+CD45+ hESC-derived cells were plated in limiting dilutions (semi-log dilution from 300 to 3 cells per well) in flat-bottom 96-well plates with a confluent monolayer of M2-10B4. Cells were cultured in the same media/incubation conditions as described above for bulk LTC-IC condition culture and were similarly provided with fresh media every 7 days. After 3 to 5 weeks of culture, cells were placed in hematopoietic CFC assay conditions by removing all but 20 μL of media from each well and replacing with 100 μL of Methocult 4436 SF (H4436; Stem Cell Technologies) per well. After 14 days in Methocult, wells were scored for the presence of hematopoietic colonies. Frequency of LTC-ICs from each feeder condition was calculated by Poisson distribution (L-Calc software; StemCell Technologies) based on the number of wells at each cell dose with ≥1 hematopoietic colony after 3 to 5 weeks of culture.
CFC assay
MACS beads (Mitenyi Biotec) sorted single CD34+ cells cultures in 0.1 mL Iscove modified Dulbecco medium with 2% fetal bovine serum were mixed with 1 mL MethoCult H4434 (Stemcell Technologies). Fluorescence-activated cell sorter (FACS)-sorted CD34+CD45+ cells cultures in 0.1 mL Stemspan were mixed with 1 mL MethoCult 4436 SF. The mixture was then transferred to 35-mm dishes and cultured for 14 days, followed by colony counting. Each type of colony was classified according to morphology. Each assay was performed in triplicate.
Intrafemoral bone marrow transplantation
H9-derived CD34+CD45+ cells were transplanted into irradiated (3.0 Gy) 8-week-old NOD-scid IL2rγnull (NSG) mice using the Intrafemoral bone marrow transplantation (IBMT) technique.23 For IBMT, the leg of the anesthetized mouse was flexed; a 27-gauge needle was inserted into the femur at the knee joint and then replaced with a 28-gauge insulin syringe containing 20 to 30 μL of cell suspension. Cell doses ranged from 1.5 × 105 to 3.0 × 105. All experiments using mice received approval from our local authority, the Animal Care and Veterinary Services of the University of California, San Diego. To prepare mouse bone marrow cells for flow cytometry analysis, red cells were lysed with 0.8% ammonium chloride solution. Human CD45+ cells were measured and analyzed from bone marrow. hCD45+ cells were further gated and analyzed for myeloid (hCD33+) and B lymphoid (hCD19+). MHC-I+ was gated for erythroid cells (CD36+) compositions (all conjugated antibodies from Becton Dickinson).23 In parallel, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analyses were performed to detect the human RUNX1a transcript in mouse bone marrow.
Results
Expression of endogenous RUNX1 is increased during spin embryoid body hematopoietic differentiation
We first examined whether expression of RUNX1 correlates with human embryoid body (hEB) development toward hematopoietic cells. To determine expression of endogenous RUNX1 isoforms throughout hESC differentiation and hEB development, we performed qRT-PCR analysis at various stages of EB development. Because RUNX1a and RUNX1b/c differ only in the C terminus, we used primer sets specific to the C terminus of either RUNX1a or RUNX1b/c in our qRT-PCR analysis to ensure target specificity. To this aim, H9 (hESC) and iCB5 (hiPSC) cells were analyzed. Both RUNX1 isoforms were barely detectable in undifferentiated ES cells and were strongly increased in parallel with EB development in both cells (Figure 1A-B). RUNX1a represents a fairly small portion of the total RUNX1 transcripts during EB development (supplemental Figure 1). Meanwhile, expression of 2 common pluripotency markers, OCT4 and NANOG, showed an opposite trend in H9 cells (Figure 1C), suggesting that steadily up-regulated expression of RUNX1 isoforms during EB development may play important roles during differentiation. This observation was further supported when we checked expression levels of several cell surface antigens related to definitive hematopoiesis, including CD31, CD34, CD43, and CD45 (Figure 1D). The increased expression of RUNX1 and these markers are parallel during EB development. These findings are in agreement with previous reports,11,15 which suggested that expression of RUNX1 correlates well with the establishment of definitive hematopoiesis and may function during further hematopoietic events.
Expression of endogenous RUNX1a and RUNX1b/c parallels lineage commitment and hematopoietic emergence from hPSCs. (A) RT-qPCR analysis of endogenous RUNX1a throughout EB development from hESC (H9) or hiPSC (iCB5). GAPDH is used as an internal control. Results from day 13 EBs are set as 1. Bar chart represents relative RUNX1a expression at various time points. Error bars represent standard deviations (SDs) of 3 independent experiments. (B) RT-qPCR analysis of endogenous RUNX1b/c throughout EB development from hESC (H9) or hiPSC (iCB5). GAPDH is used as an internal control. Results from day 13 EBs are set as 1. Bar chart represents relative RUNX1b/c expression at various time points. Error bars represent SDs of 3 independent experiments. (C) RT-qPCR analysis of endogenous OCT4 and NANOG throughout EB development from hESCs (H9). GAPDH is used as an internal control. Results from undifferentiated cells (D0) are set as 1. Bar chart represents relative OCT4 and NANOG expression at various time points. Error bars represent SDs of 3 independent experiments. (D) Developmental progression of hemato-endothelial and hematopoietic surface marker expressions on disaggregated H9 cells at various time points. y-axis represents percentage of cells with positive staining in FACS analysis. Each time point represents the mean of 3 to 5 independent experiments.
Expression of endogenous RUNX1a and RUNX1b/c parallels lineage commitment and hematopoietic emergence from hPSCs. (A) RT-qPCR analysis of endogenous RUNX1a throughout EB development from hESC (H9) or hiPSC (iCB5). GAPDH is used as an internal control. Results from day 13 EBs are set as 1. Bar chart represents relative RUNX1a expression at various time points. Error bars represent standard deviations (SDs) of 3 independent experiments. (B) RT-qPCR analysis of endogenous RUNX1b/c throughout EB development from hESC (H9) or hiPSC (iCB5). GAPDH is used as an internal control. Results from day 13 EBs are set as 1. Bar chart represents relative RUNX1b/c expression at various time points. Error bars represent SDs of 3 independent experiments. (C) RT-qPCR analysis of endogenous OCT4 and NANOG throughout EB development from hESCs (H9). GAPDH is used as an internal control. Results from undifferentiated cells (D0) are set as 1. Bar chart represents relative OCT4 and NANOG expression at various time points. Error bars represent SDs of 3 independent experiments. (D) Developmental progression of hemato-endothelial and hematopoietic surface marker expressions on disaggregated H9 cells at various time points. y-axis represents percentage of cells with positive staining in FACS analysis. Each time point represents the mean of 3 to 5 independent experiments.
Enforced expression of RUNX1a regulates hPSC pluripotency and differentiation
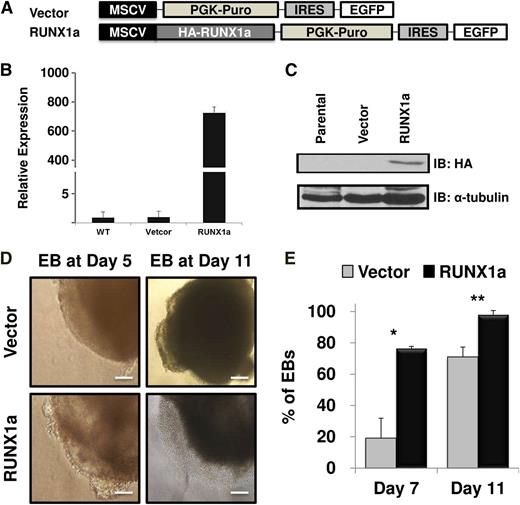
It has been reported that RUNX1a potentiates but RUNX1b abrogates mouse hematopoietic stem/progenitor cell engraftment and expansion.18,20 Based on the robust increase of RUNX1a expression during hPSC hematopoietic differentiation (Figure 1) and the different effects of RUNX1a and RUNX1b in mouse hematopoietic stem cells, we hypothesized that RUNX1a may enhance hematopoietic cell formation and expansion from hPSCs and that RUNX1a may be a useful tool for generation of more blood cells from hPSCs for therapeutic use. To test this hypothesis, we sought to enforce the expression of RUNX1a in both hESCs (H9) and hiPSCs (BC1 or iCB5). We first subcloned human RUNX1a cDNA into a lentiviral vector containing puromycin resistance and enhanced green fluorescent protein genes (Figure 2A). After lentiviral transduction and drug selection, infection was confirmed by checking green fluorescent protein expression in EBs by fluorescent microscopy (supplemental Figure 2). Furthermore, expression of RUNX1a was validated at both transcript (Figure 2B; supplemental Figure 3) and protein (Figure 2C; supplemental Figure 3B) levels with qRT-PCR and western blot, respectively. We performed spin EB hematopoietic differentiation after virus infection and puromycin resistance selection for 2 days. During EB differentiation, emergence of a sac structure predicts cell lineage commitment and germ layer development.24,25 As shown in Figure 2D, in RUNX1a-overexpressed H9 hESC-derived EBs, we consistently observed the appearance of typical sac structures at stages as early as EB day 5 and more obvious at EB day 11 in several independent experiments. Although formation of sac structure in control cells was also observed, it arose at a much later stage and with lower frequency (Figure 2E). Similarly, we also saw more EBs with typical and visible hemato-endothelial structure in RUNX1a-derived cells (supplemental Figure 4A). At day 5, this semiadherent structure appeared within or on the edge of RUNX1a EBs; at day 11, we started to see nonadherent cells appearing in culture (Figure 2D). Similar effects of RUNX1a on EB differentiation of hiPSCs (BC1 or iCB5) were also observed (supplemental Figure 3C). Interestingly, at day 13, >90% of these nonadherent cells were CD34+CD45+ (supplemental Figure 4B), which represented phenotypic HSCs.
Enforced expression of RUNX1a in hESC (H9) and sequential hematopoietic differentiation through spin EBs. (A) Schematic representation of lentiviral constructs used to express RUNX1a. EGFP, enhanced green fluorescent protein; HA, human influenza hemagglutinin; MSCV, murine stem cell virus; PGK, phosphoglycerate kinase; PURO, puromycin. (B) Expression of transduced RUNX1a in hESC (H9) cells detected by RT-qPCR after lentiviral infection and puromycin selection. GAPDH is used as an internal control. Results from parental cells (WT) that represent endogenous RUNX1a are set as 1. Bar chart represents RUNX1a expression in vector or RUNX1a transduced cells relative to parental cells. Error bars represent SDs of 3 independent experiments. (C) Expression of transduced RUNX1a in hESC (H9) cells detected by western blot using anti-HA antibody after lentiviral infection and puromycin selection. α-Tubulin is used as a loading control. (D) Cell morphologies of sequential EB formation from H9 cells at day 5 and 11 under spin EB differentiation condition. Hemato-endothelial structure could be observed in RUNX1a transduced EBs since day 5 EBs. Scale bars, 50 μm. (E) Percentage of EBs containing typical sac (cystic) structure at spin EB day 7 and 11. Error bars represent SDs of 3 independent experiments. *P = .0158, **P = .0347.
Enforced expression of RUNX1a in hESC (H9) and sequential hematopoietic differentiation through spin EBs. (A) Schematic representation of lentiviral constructs used to express RUNX1a. EGFP, enhanced green fluorescent protein; HA, human influenza hemagglutinin; MSCV, murine stem cell virus; PGK, phosphoglycerate kinase; PURO, puromycin. (B) Expression of transduced RUNX1a in hESC (H9) cells detected by RT-qPCR after lentiviral infection and puromycin selection. GAPDH is used as an internal control. Results from parental cells (WT) that represent endogenous RUNX1a are set as 1. Bar chart represents RUNX1a expression in vector or RUNX1a transduced cells relative to parental cells. Error bars represent SDs of 3 independent experiments. (C) Expression of transduced RUNX1a in hESC (H9) cells detected by western blot using anti-HA antibody after lentiviral infection and puromycin selection. α-Tubulin is used as a loading control. (D) Cell morphologies of sequential EB formation from H9 cells at day 5 and 11 under spin EB differentiation condition. Hemato-endothelial structure could be observed in RUNX1a transduced EBs since day 5 EBs. Scale bars, 50 μm. (E) Percentage of EBs containing typical sac (cystic) structure at spin EB day 7 and 11. Error bars represent SDs of 3 independent experiments. *P = .0158, **P = .0347.
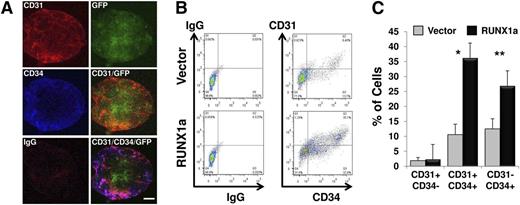
Precursors of definitive blood cells are from CD31+CD34+CD45− hemogenic endothelium.26,27 To address whether RUNX1a expression enhances hemato-endothelial precursor formation, H9 EBs expressing RUNX1a were subjected to immunostaining. Expression of CD34 and CD31 was clearly detected at day 6 of EB differentiation (Figure 3A). With FACS analysis, we observed a significantly higher percentage of CD31+CD34+ or total CD34+ cells in RUNX1a EBs than that in control EBs at day 7 (Figure 3B-C). In agreement with this finding, we observed enrichment of the CD31+CD34+ population in EBs derived from RUNX1a iCB5 cells (data not shown). In addition, we also checked whether RUNX1a affected expression of transcription factors critical for mesoderm development and hematopoiesis, including Brachyury, KDR, SCL, GATA2, and PU.1, in EBs at different time points. For mesoderm differentiation-related factors, expression of Brachyury and KDR was up-regulated in the presence of RUNX1a (supplemental Figure 5). For hematopoietic differentiation-related factors, expression of SCL, GATA2, and PU.1 showed a two- to fivefold increase in hESCs on RUNX1a expression (supplemental Figure 6, day 0). As differentiation progressed, the increase of PU.1 expression in RUNX1a cells stayed steady. However, the relative expressions of transcription factors GATA2 and SCL were clearly enhanced (supplemental Figure 6). Collectively, our gene expression analysis suggests that RUNX1a not only pushed the lineage commitment of mesoderm but also specifically enhanced the hemogenic differentiation.
Efficient and enhanced mesoderm and hematopoietic differentiation on RUNX1a overexpression in hESC (H9) cells. (A) Confocal fluorescence images of day 6 EBs derived from RUNX1a-transduced H9 cells show expression of GFP, CD31, and CD34. Scale bars, 200 µm. (B) Representative FACS analysis of CD31/CD34 expression on day 7 EBs derived from vector or RUNX1a transduced H9 cells. (C) Summary of FACS analysis from panel B. Percentages of 3 populations are shown: CD31+CD34−, CD31+CD34+, and CD31−CD34+. Error bars represent SDs of 3 independent experiments.*P = .0001, **P = .02.
Efficient and enhanced mesoderm and hematopoietic differentiation on RUNX1a overexpression in hESC (H9) cells. (A) Confocal fluorescence images of day 6 EBs derived from RUNX1a-transduced H9 cells show expression of GFP, CD31, and CD34. Scale bars, 200 µm. (B) Representative FACS analysis of CD31/CD34 expression on day 7 EBs derived from vector or RUNX1a transduced H9 cells. (C) Summary of FACS analysis from panel B. Percentages of 3 populations are shown: CD31+CD34−, CD31+CD34+, and CD31−CD34+. Error bars represent SDs of 3 independent experiments.*P = .0001, **P = .02.
To further prove that RUNX1a enhanced hemogenic endothelial commitment during hPSC differentiation, we sorted CD31+CD34+CD45− cells and assayed for the emergence of cells with both endothelial and hematopoietic potentials. As expected, these cells gave rise to endothelial morphology and phenotype when cultured under endothelial condition. Further analysis demonstrated that these cells expressed von Willebrand factor and were able to uptake acetylated LDL (supplemental Figure 7A-C). Likewise, when cultured under hematopoietic differentiation conditions, these cells differentiated into CD45+ hematopoietic cells (supplemental Figure 7D). Our results asserted that endothelial and hematopoietic bi-potentiality can be attributed to a population of CD31+CD34+CD45− cells.
Enforced expression of RUNX1a augmented hematopoietic progenitor cells from pluripotent stem cells
We next addressed whether the enforced expression of RUNX1a increased hematopoietic specification of hPSCs. Differentiated EBs derived from vector or RUNX1a-transduced H9 cells were analyzed for expression of CD34, CD43, and CD45 (surface markers for HPCs) at days 9, 11, and 13 by flow cytometry. At day 9, we demonstrated significantly higher CD43+, CD43+CD45+, and CD34+CD45+ populations in RUNX1a cells (Figure 4A-B). As differentiation progressed further, a similar trend was also observed at days 11 and 13 (Figure 4C-D). Furthermore, CD34+CD43+ cells were also increased (supplemental Figure 8). Importantly, we found the same enrichment of HPCs in hiPSCs (iCB5 and BC1) expressing RUNX1a (supplemental Figure 9). Taken together, our results clearly indicate that ectopic RUNX1a expression induced a robust increase in the population of hematopoietic stem and progenitor cells. Notably, CD34+ cells from RUNX1a H9 EBs also showed more potential for myeloid development. After 14-day culture in semisolid myeloid conditions, >80% of these CD34+ cells became CD45+ cells (in contrast to 55% from CD34+ cells of vector H9 EBs). Among these CD45+ cells, significantly more CD33+ and fewer CD11b+ cells were found in H9 EBs expressing RUNX1a, suggesting that more myeloid progenitors cells were generated in the presence of RUNX1a (supplemental Figure 10).
Robust enhancement of CD34+/CD43+/CD45+ HPCs in EBs derived from RUNX1a-transduced H9 cells. (A) Representative FACS analysis of CD34/CD45 or CD43/CD45 expression on day 9 EBs derived from vector or RUNX1a-transduced H9 cells. (B) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 9 EBs derived from vector or RUNX1a-transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .0002, **P = .0078. (C) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 11 EB derived from vector or RUNX1a-transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .04, **P = .03. (D) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 13 EBs derived from vector or RUNX1a transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .0003, **P = .001.
Robust enhancement of CD34+/CD43+/CD45+ HPCs in EBs derived from RUNX1a-transduced H9 cells. (A) Representative FACS analysis of CD34/CD45 or CD43/CD45 expression on day 9 EBs derived from vector or RUNX1a-transduced H9 cells. (B) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 9 EBs derived from vector or RUNX1a-transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .0002, **P = .0078. (C) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 11 EB derived from vector or RUNX1a-transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .04, **P = .03. (D) Summary of FACS analysis of CD34/CD45 or CD43/CD45 expression on day 13 EBs derived from vector or RUNX1a transduced H9 cells. Error bars represent SDs of 3 independent experiments. *P = .0003, **P = .001.
Enforced expression of RUNX1a in hPSCs expands the hematopoietic cells that have the ability to differentiate into multilineage cells
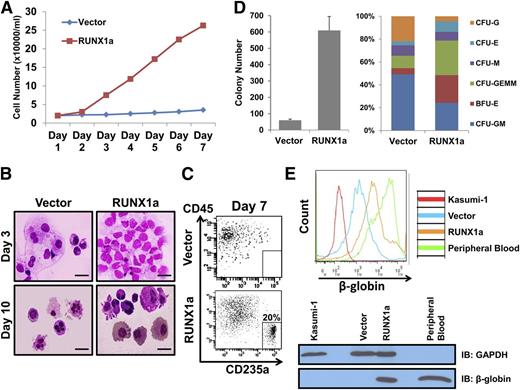
To investigate the function of RUNX1a-expressing hematopoietic cells derived from hPSCs, we expanded them in cytokine-driven ex vivo culture conditions. In contrast to control CD34+CD45+ cells, RUNX1a-expressing CD34+CD45+ cells expanded with significantly higher efficiency within 7 days of ex vivo expansion (Figure 5A; supplemental Figure 11A), indicating the increased ex vivo expansion capacity of RUNX1a-expressing HSPCs derived from hPSCs. The same conclusion was drawn when we subjected CD34+CD45+ cells from RUNX1a-BC1 EBs to colony-forming unit (CFU) and replating assays (supplemental Figure 12). We also performed Wright-Giemsa staining on progenitor cells from liquid culture on days 3 and 10. Our results suggest that, even with higher ex vivo expansion capability, RUNX1a progenitor cells were still capable of differentiating into multiple hematopoietic lineages (Figure 5B). We also analyzed adult red blood cell development. RUNX1a-transduced hematopoietic progenitors had a high percentage of CD235a+CD45− (20%) (Figure 5C), suggesting that expression of RUNX1a in HPCs enhanced definitive hematopoiesis.1 In addition, CD34+CD45+ progenitor cells from H9 day 13 EBs were collected and subjected to CFU assay. The number of CFUs generated from CD34+CD45+ progenitor cells of vector only or RUNX1a-enhanced EBs was quantified (Figure 5D, left). We observed a significant increase in CFCs with RUNX1a over that without RUNX1a. Further characterization revealed differences in composition of CFU subtypes (Figure 5D, right). Supplemental Figure 11B shows typical images of CFU subtypes derived from RUNX1a EBs. Notably, we observed much higher percentages of CFU-E, CFU-GEMM, and BFU-E cells from RUNX1a EBs, hinting at the increase of erythropoiesis potential.
Long-term ex vivo–expanded RUNX1a progenitor cells are capable of differentiating into multiple lineages. (A) Progenitor cells (CD34+CD45+) from vector or RUNX1a-transduced H9 cells are expanded ex vivo for 7 days in liquid culture. Cell numbers are monitored every day. (B) Wright-Giemsa staining of 3- or 10-day ex vivo–expanded progenitor cells from vector or RUNX1a-transduced H9 cells. Scale bar, 50 μm. (C) Representative FACS analysis of CD235a/CD45 expression on ex vivo–expanded progenitor CD34+CD45+ cells from vector or RUNX1a-transduced H9 cells in liquid culture. (D) CFU assay on progenitor cells from vector or RUNX1a-transduced H9 cells. (Left) Numbers of colonies formed from 10 000 cells. (Right) Scoring of CFUs reveals differences in CFU subtypes. BFU, burst-forming unit; E, erythroid; G, granulocyte; GEMM, granulocyte, erythrocyte, monocyte, megakaryocyte; GM, granulocyte-macrophage; M, macrophage. (E) Expression of β-globin is detected in HPCs differentiated from RUNX1a EBs after 14-day expansion by FACS analysis or western blot. Kasumi-1 is used as a negative control. Peripheral blood is used as a positive control. GAPDH is used in western blot as a loading control.
Long-term ex vivo–expanded RUNX1a progenitor cells are capable of differentiating into multiple lineages. (A) Progenitor cells (CD34+CD45+) from vector or RUNX1a-transduced H9 cells are expanded ex vivo for 7 days in liquid culture. Cell numbers are monitored every day. (B) Wright-Giemsa staining of 3- or 10-day ex vivo–expanded progenitor cells from vector or RUNX1a-transduced H9 cells. Scale bar, 50 μm. (C) Representative FACS analysis of CD235a/CD45 expression on ex vivo–expanded progenitor CD34+CD45+ cells from vector or RUNX1a-transduced H9 cells in liquid culture. (D) CFU assay on progenitor cells from vector or RUNX1a-transduced H9 cells. (Left) Numbers of colonies formed from 10 000 cells. (Right) Scoring of CFUs reveals differences in CFU subtypes. BFU, burst-forming unit; E, erythroid; G, granulocyte; GEMM, granulocyte, erythrocyte, monocyte, megakaryocyte; GM, granulocyte-macrophage; M, macrophage. (E) Expression of β-globin is detected in HPCs differentiated from RUNX1a EBs after 14-day expansion by FACS analysis or western blot. Kasumi-1 is used as a negative control. Peripheral blood is used as a positive control. GAPDH is used in western blot as a loading control.
To confirm the increased definitive hematopoiesis of RUNX1a EBs, we examined adult β-globin expression in cells collected from the 14-day culture of H9 HPCs in semisolid serum-free media supplemented with erythropoietin. We found that HPC cells from RUNX1a-expressing EBs gave much higher β-globin expression than vector EBs, demonstrated by both flow cytometry and western blot (Figure 5E). It is worth noting that, in addition to definitive adult hemoglobin (β-globin), we also observed elevated transcripts of primitive (ε-globin and ζ-globin) and fetal (γ-globin) hemoglobins (supplemental Figure 13). Together, our data demonstrate that RUNX1a positively regulates hematopoiesis specification and blood cell generation from human pluripotent stem cells.
Hematopoietic progenitors derived from RUNX1a transduced hESCs have long-term expansion ability through interaction with surrogate niche
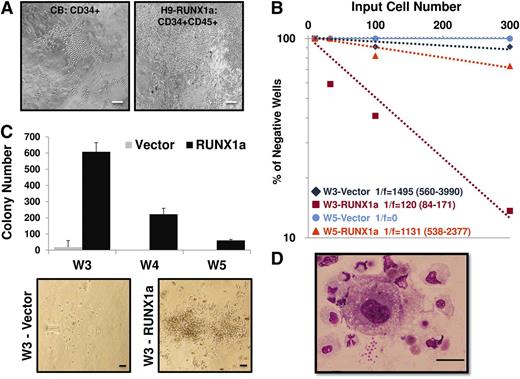
The LTC-IC assay serves the purpose of quantification of putative HPSCs in a given population to be cultured for an extended period.28,29 To investigate whether RUNX1a contributed to HPC activity, we also cultured hESC-derived HPCs in the presence of stromal cells. Like CD34+ cells from cord blood, we observed that the RUNX1a-expressing HPCs were expanded through direct interaction with stromal cells in 2 independent experiments (Figure 6A).21 These results indicate that RUNX1a-transduced hESC-derived hematopoietic progenitors possessed long-term expansion ability through interaction with surrogate niche.22 We also performed limiting dilution assays on 3- or 5-week expanded HPCs to measure their stem cell frequencies. Results from this assay revealed that the frequency of LTC-IC was higher when RUNX1a was present (Figure 6B). Moreover, in secondary CFU in culture assays conducted after 3- to 5-week culture under in vitro LTC-IC conditions, we found significant increases in colony numbers in RUNX1a HPCs in 2 independent experiments (Figure 6C). The morphology of these colony cells was evaluated by Wright-Giemsa staining. Colonies from RUNX1a HPCs showed multilineage morphologies, including differentiated macrophages, monocytes, neutrophils, and some nucleated erythroid cells (Figure 6D). Collectively, these results demonstrate the positive effects of RUNX1a on the potential of hematopoietic stem cell and progenitor function.
Hematopoietic progenitors derived from RUNX1a hESCs can be expanded through interaction with surrogate niche. (A) Morphology of colonies from bulk long-term culture of CD34+ core blood cells or RUNX1a H9–derived HPCs (CD34+CD45+) on M2-10B4 stromal cells at week 5. Scale bars, 200 μm. (B) Limiting dilution assay on 3- or 5-week expanded CD34+CD45+ cells from vector or RUNX1a-transduced H9 EBs. (C) (Top) Numbers of CFU generated from 40 000 CD34+CD45+ cells of vector or RUNX1a EBs derived from H9 cells after 3- to 5-week culture under LTC conditions followed by 2-week expansion in semisolid methylcellulose media. (Bottom) Representative CFU pictures at week 3. Scale bars, 100 μm. (D) Wright-Giemsa staining of RUNX1a H9–derived progenitor cells at week 5 in bulk LTC conditions and 2-week expansion in semisolid methylcellulose media. Scale bar, 100 μm.
Hematopoietic progenitors derived from RUNX1a hESCs can be expanded through interaction with surrogate niche. (A) Morphology of colonies from bulk long-term culture of CD34+ core blood cells or RUNX1a H9–derived HPCs (CD34+CD45+) on M2-10B4 stromal cells at week 5. Scale bars, 200 μm. (B) Limiting dilution assay on 3- or 5-week expanded CD34+CD45+ cells from vector or RUNX1a-transduced H9 EBs. (C) (Top) Numbers of CFU generated from 40 000 CD34+CD45+ cells of vector or RUNX1a EBs derived from H9 cells after 3- to 5-week culture under LTC conditions followed by 2-week expansion in semisolid methylcellulose media. (Bottom) Representative CFU pictures at week 3. Scale bars, 100 μm. (D) Wright-Giemsa staining of RUNX1a H9–derived progenitor cells at week 5 in bulk LTC conditions and 2-week expansion in semisolid methylcellulose media. Scale bar, 100 μm.
Reconstitution of NSG mice engraftment
Last, we performed IBMT to evaluate the hematopoietic engraftment ability of RUNX1a-derived progenitor cells (CD34+CD45+). A total of 15 NSG mice were injected following the IBMT method. Four mice encountered early death (within 2 weeks), possibly because of irradiation and infection. At week 9 after transplantation, human hematopoietic engraftment was evaluated by measuring the percentage of hCD45+ cells in both injected and contra-injected bone marrow of all surviving recipient mice (n = 5; Figure 7A). The high mortality most likely came from pulmonary emboli formation in hESC-derived cells as reported previously,23 independent of RUNX1a. We also examined the hematopoietic multilineage reconstitution, including myeloid (CD45+CD33+), lymphoid (CD45+CD19+), and erythroid (CD36+MHC-1+) hematopoietic lineages in surviving mice (>9 weeks; Figure 7B). The success of human chimerism in bone marrow cells taken from injected femurs was confirmed by qRT-PCR using human RUNX1a-specific primers (Figure 7C). Our results provide direct evidence that hematopoietic cells derived from RUNX1a HSPCs possess the developmental potential to generate multilineages in vivo.
Reconstitution of NSG mice engraftment. Approximately 1.5 × 105 to 3 × 105 RUNX1a progenitor cells (CD34+CD45+) were injected into the femur of irradiated NSG mice (n = 15). (Ai) Representative histogram analysis of human CD45 (hCD45) expression in recipient mice 9 weeks after transplantation. (Aii) Comparative analysis of hCD45 expression level as human chimerism in injected femur and contralateral femur in recipient mice (n = 5). (B) hCD45+ cells in recipient bone marrow 9 weeks after transplantation were measured and analyzed by flow cytometry. hCD45+ cells were further gated and analyzed for myeloid (hCD33+) and B-lymphoid (hCD19+) populations. MHC-I+ was gated for erythroid cells (CD36+) compositions. (Bi) Representative results from FACS analysis. (Bii) Summary of FACS analysis on 5 surviving recipient mice. (C) Representative RT-qPCR analysis of human RUNX1a in bone marrow of recipient mice after 6 weeks of survival.
Reconstitution of NSG mice engraftment. Approximately 1.5 × 105 to 3 × 105 RUNX1a progenitor cells (CD34+CD45+) were injected into the femur of irradiated NSG mice (n = 15). (Ai) Representative histogram analysis of human CD45 (hCD45) expression in recipient mice 9 weeks after transplantation. (Aii) Comparative analysis of hCD45 expression level as human chimerism in injected femur and contralateral femur in recipient mice (n = 5). (B) hCD45+ cells in recipient bone marrow 9 weeks after transplantation were measured and analyzed by flow cytometry. hCD45+ cells were further gated and analyzed for myeloid (hCD33+) and B-lymphoid (hCD19+) populations. MHC-I+ was gated for erythroid cells (CD36+) compositions. (Bi) Representative results from FACS analysis. (Bii) Summary of FACS analysis on 5 surviving recipient mice. (C) Representative RT-qPCR analysis of human RUNX1a in bone marrow of recipient mice after 6 weeks of survival.
Discussion
Many experimental results have demonstrated that growth- and environmental factor–induced signal transduction and transcription factor–regulated gene expression are 2 major approaches to control PSC maintenance, expansion, and differentiation.25,30,31 One very elusive cell type that has been proven to be difficult to differentiate and expand from hPSCs is HSCs capable of long-term multilineage engraftment. It has been reported that RUNX1 serves as a master regulator in hematopoiesis and in various blood-related disorders. We and others have reported previously that expression of RUNX1 is hardly detectable in both human and mouse embryonic stem cells but is dramatically increased during hematopoiesis.1,32 Interestingly, it has been shown that human cord blood cells expressing lentivirus-transduced RUNX1a contain significantly more LTC-ICs and cobblestone area–forming cells.20 In addition, overexpression of human RUNX1a in mouse bone marrow progenitor cells can potentiate engraftment ability on competitive transplantation.20 In this study, we successfully cultured and demonstrated the developmental impact of RUNX1a on human embryonic and hematopoietic development, harnessing the availability of hPSCs as a working model. Therefore, this report provides the first evidence that RUNX1a is a valuable factor to facilitate therapeutic production of human blood cells from both embryonic stem cells and induced pluripotent cells.
Several studies have demonstrated that hemogenic endothelial (HE) cells are the direct precursors of HSCs. The most compelling evidence comes from tracing hematopoietic cell emergence in the aorta-gonad-mesonephros region of both zebrafish and mouse.33 During hESC specification, a specific population of hemogenic endothelium cells (CD31+CD34+CD45−) is involved in both hematopoietic and endothelial development.23,34 With a defined spin EB differentiation system,35 we showed that RUNX1a enhances and highlights the entire process of hematopoietic differentiation of hPSCs and recapitulates the main stages of early hematopoietic development, including (1) mesoderm commitment of hPSCs; (2) specification of HE cells expressing CD31 from the early mesoderm cells; (3) emergence of CD31+CD34+CD45− cells from HE cells; and (4) emergence of CD43+CD34+ and CD43+CD45+/CD34+CD45+ HPCs. Previous publications have demonstrated that RUNX1a can influence human CD34+ cord blood cells, leading to enhanced self-renewal activity and HSPCs expansion ex vivo.20 Our work takes a step further to establish that RUNX1a expression promotes ex vivo expansion of HPCs derived from PSCs and promotes multilineage differentiation and definitive hematopoiesis.
In zebrafish, where the transcriptional regulation of RUNX1 using 2 promoters is conserved, transgenic lines that express fluorescently labeled RUNX1 isoforms specific for each of the 2 promoters show that the distal isoform is expressed in areas where erythromyeloid progenitors arise, whereas the proximal isoform originates where definitive HSCs develop.36 More recent studies using mouse knock-in models to label the expression patterns of the RUNX1 distal and proximal promoters have also shown that the proximal promoter may be important for hemogenic endothelium development, whereas the distal promoter is active in more mature progenitors for the initial development of definitive hematopoietic cells.19 In human cells, the RUNX1a isoform is driven by the P2 promoter (proximal). We showed that enforced RUNX1a expression pushed the development of hemogenic endothelium ancestors along the embryonic differentiation. However, it remains to be addressed whether promoter selection for RUNX1 gene determines differentiation specificity.
Transcription factor RUNX1 is expressed in the hemangioblast, the multipotent precursor cells that can differentiate into both hematopoietic and endothelial cells, during embryonic development.37 Although target genes of RUNX1 have been well documented,38,39 direct targets critical for the onset of hematopoiesis remain to be identified. Our study on RUNX1a at the onset of definitive hematopoiesis has firmly established its role as a pivotal player in HSC emergence. It will be of great interest to decipher the gene regulatory network of RUNX1a on the initiation of and during hematopoiesis. Information obtained from this genome-wide analysis on RUNX1a function will not only provide an important mechanistic framework of HSC emergence but also benefit future studies on modulating stem cell maintenance and/or de novo generation. Additionally, combinatorial transcriptional control may be crucial in HSPCs. Besides RUNX1, transcription factors HOXB4 and SCL are also known to facilitate hematopoietic specification in ESCs.27,40-42 We showed that the capability for long-term expansion in RUNX1a-expressing HPCs is not as strong as in CD34+ cord blood cells ex vivo. It has been suggested that multiple transcription factors work together to specify differentiation of progenitors into distinct lineages.43-45 More importantly, integrated analysis of 10 key hematopoietic transcriptional regulators, including RUNX1, in hematopoietic progenitor cells also hints at the close collaboration between HSPC-associated transcription factors for determining cell fate.46 It is therefore reasonable to speculate that RUNX1a may collaborate with other factor(s) for direct programming of hematopoietic differentiation.
In this study, we delivered RUNX1a into hPSCs via lentiviral transduction. One major concern with this approach is that random virus integration may disrupt genome integrity and ultimately enhance oncogenic potential. The use of cell-penetrating transcription factors by direct addition to culture medium has been successfully demonstrated to provide sufficient signals for iPSC generation,47,48 therefore presenting an alternative method to evaluate the function of RUNX1a in directing hematopoietic differentiation. It is intriguing to see whether cell-penetrating RUNX1 proteins can safely and efficiently promote blood cell formation from hPSCs.
In summary, we identified that RUNX1a enhances the establishment of definitive HSCs from human PSCs. RUNX1a is also critical for the maintenance, expansion, and multilineage differentiation of these HSCs. Our study not only provides advanced understanding of stem cell biology but also useful tools to enhance the specificity and efficiency of generating functional blood stem cells and other types of blood cells from PSCs through transcription factor gene regulation. Applications of our findings will expedite the future use of human pluripotent stem cells as a therapy for blood disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dennis J. Young (Flow Cytometry Shared Resource at Moores UCSD Cancer Center) for flow cytometer expertise, Laura Bendzick (D.A.K.’s laboratory) and Sarah Dowey (L.C.’s laboratory) for excellent technical assistance, Russell DeKelver and Kentson Lam for editing the manuscript, and all other members of D.-E.Z.’s laboratory for valuable discussions.
This work was supported in part by California Institute of Regenerative Medicine grant RB2-01645 (D.-E.Z.), National Institutes of Health grants R01-HL073781 (L.C.), R01-HL067828 (D.S.K.), and T32 Medical Scientist Training Program Grant GM008244 (D.A.K.), Maryland Stem Cell Research Fund grant 2011-MSCRFE-0087 (Z.Y.), the Leukemia Research Fund of the University of Minnesota Cancer Center (D.S.K.), the William Lawrence and Blanche Hughes Foundation (D.S.K.), Stem Cell Biology training grant T32HD060536 (D.A.K.), and Developmental Biology training grant 5T32HD007480 (P.I.F.).
Authorship
Contribution: D.R. and W.-J.S. designed and performed the experiments, analyzed the data, and wrote the manuscript; M.-C.L. and J.-B.F. designed and performed experiments; D.A.K., P.I.F., and Z.Y. provided experimental assistance and data analysis; M.Y. performed experiments and analyzed the data; L.C. and D.S.K. provided critical experimental advices and valuable discussion; and D.-E.Z. supervised experimental design, data analysis, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong-Er Zhang, Mail Drop 0815, Moores Cancer Center, University of California at San Diego, 3855 Health Sciences Drive, La Jolla, CA 92093; e-mail: d7zhang@ucsd.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal