Key Points
A comprehensive study of 19 gene mutations and their cooperation, including the first report of ASXL1 and TET2 mutations in pediatric AML.
The development of pediatric AML requires fewer gene mutations than adult AML.
Abstract
Gene mutations involving epigenetic regulators recently have been described in adult acute myeloid leukemia (AML). Similar studies are limited in children. We analyzed gene mutations and cooperation in pediatric AML with special reference on mutated epigenetic regulators. Nineteen gene mutations, including 8 class I genes, 4 class II genes, WT1 and TP53 (class III), and 5 epigenetic regulator genes (class IV), were analyzed in 206 children with de novo AML. Mutational analysis was performed with polymerase chain reaction−based assay followed by direct sequencing. One hundred seventeen of 206 patients (56.8%) had at least one mutation: 51% class I, 13% class II, 6.8% class III, and 5.6% class IV. FLT3-internal tandem duplication was most frequent, and 29% of patients had more than one gene mutation. Two patients carried ASXL1 mutations, both with t(8;21), 2 had DNMT3A mutations, 2 had IDH1 mutations, 1 had IDH2 mutation, and 3 had TET2 mutations. Both patients with IDH1 mutations had AML-M0 subtype and MLL-partial tandem duplication. Cooperating mutations with mutated epigenetic regulators were observed in 8 of 10 patients. We conclude that mutated epigenetic regulators were much less than those in adult AML but with frequent cooperating mutations. ASXL1, TET2, and IDH1 mutations were associated with specific genetic subtypes.
Introduction
Comprehensive analyses in de novo childhood acute myeloid leukemia (AML) of gene mutations involving epigenetic regulators have been limited. The ASXL1 (additional sex comb-like 1) gene mapping to chromosome 20q acts as a cofactor of retinoic acid receptor via binding to steroid receptor coactivation-1 and belongs to enhancer of trithorax and polycomb genes that can both activate and repress the HOX gene.1,2 Very recently, it has been demonstrated that ASXL1 loss-of-function mutations result in the loss of polycomb repressive complex 2−mediated histone H3 lysine 27 trimethylation, which promotes myeloid leukemia transformation.3 ASXL1 mutations conferred a poor outcome in adult AML,4,5 but there have been no reports of ASXL1 mutations in childhood AML. TET proteins encode α-ketoglutarate-dependent oxygenases, which are involved in the conversion of 5-methylcytosine to 5-hydroxymethylcytosine.6 TET2 protein is important for normal myelopoiesis, and disruption of TET2 enzymatic activity results in altered DNA methylation and favors myeloid neoplasm transformation.7 IDH1 and IDH2 mutations convert α-ketoglutarate to 2-hydroxyglutarate, which disrupts TET2 function.8,9
Somatic mutations of TET2 were identified with microdeletion at 4q24 in myeloid neoplasms by the use of high-resolution single-nucleotide polymorphism microarrays.10,11 TET2 mutations were detected in adult AML with a frequency ranging from 7% to 23%, but the prognostic relevance of these results was controversial.12 There have been no published reports on TET2 mutations in pediatric AML except in abstract form.13 Mutation of the codon 132 of IDH1 gene was identified first in an adult AML patient with normal karyotype by the use of whole-genome sequencing.14 IDH2 mutations in codons R140 and R172 were later reported in adult AML.15 IDH1 or IDH2 mutations also have occurred rarely in pediatric patients with AML.16-18 Mutation of DNMT3A, which encodes a DNA methyltransferase, was first identified by whole-genome sequencing in an AML patient with normal karyotype and detected in 22% of adult patients with de novo AML, especially those in the intermediate-risk cytogenetic group.19 DNMT3A mutations have been rare in pediatric patients with AML: in one study that included 180 cases, no patients were found,20 and only 1.0% and 2.1% of the patients in the other two studies were reported.21,22 In this study of 206 pediatric patients with de novo AML, we systematically analyzed known mutated genes, examined more patients and genes than we have reported previously,23 and included 5 recently identified genes that encode epigenetic modifiers. We also sought to determine the presence of multiple mutated gene combinations within a single individual.
Materials and methods
Patients and samples
Two hundred six consecutive children with de novo AML who were diagnosed at Chang Gung Memorial Hospital, Taoyuan, and Mackay Memorial Hospital, Taipei, between December 1995 and June 2011 were enrolled. The study was approved by the institutional review board of Mackay Memorial Hospital and was performed in compliance with the Declaration of Helsinki. The morphologic subtypes were classified according to the French-American-British (FAB) classification system. Immunophenotyping and cytogenetic/genetic analyses were performed at initial diagnosis as has been described previously.23,24 MLL gene rearrangement was screened by cytogenetics, Southern blot analysis, or fluorescent in situ hybridization followed by reverse transcriptase-polymerase chain reaction (PCR) assays or panhandle PCR to detect the MLL fusion transcripts as previously described.25
The earlier cohort of patients with acute promyelocytic leukemia (APL) was treated with the Taiwan Pediatric Oncology Group (TPOG)-APL-97 protocol, which consisted of all-trans retinoic acid followed by idarubicin and cytarabine. The postremission therapy consisted of 6 courses of idarubicin and cytarabine.26 Since 2001, patients with APL have been treated with the TPOG-APL-2001 protocol, which was modified from the Programa para el Estudio de la Terape´utica en Hemopatı´a Maligna (PETHEMA) protocol.27 The non-APL patients were randomized to TPOG-AML-97A protocol26 or AML-97B protocol, which was modified from Medical Research Council (MRC) AML 10.
Cell fractionation, DNA and RNA extraction, and cDNA preparation
Mononuclear cells were obtained from bone marrow aspirates at diagnosis and cryopreserved until testing. Genomic DNA (gDNA) and RNA were extracted from freshly frozen cells. RNA was reversely transcribed to complementary DNA (cDNA) with the Superscript II RNase H2 reverse transcriptase kit (Invitrogen Corporation, Carlsbad, CA), as described previously.28
Mutational analysis
Detections of gene mutations, including FLT3-internal tandem duplication (ITD), FLT3-tyrosine kinase domain (TKD), C-FMS (exons 6-22), C-KIT (exons 7-21), exons 2 and 3 of NRAS and KRAS, MLL-partial tandem duplication (PTD), and the entire coding sequences of CEBPα and RUNX1 were performed as previously described,23,25,29-31 and the results were updated in the present study.
Detection of ASXL1 mutation
We performed mutational analysis of ASXL1 exon 13 (original exon 12) by using the method described by Gelsi-Boyer et al.32
Detection of TET2 mutations
Mutational analysis of TET2 was performed with a gDNA PCR assay to amplify the whole coding sequences (exons 3-11) of TET2. The PCR products were subjected to direct sequencing. gDNA-PCR was performed with primers described by Delhommeau et al10 or Langemeijer et al11 with some modifications (supplemental Table 1; see the Blood Web site). TET2 missense mutations reported before or occurring in the two conserved regions (amino acids 1134-1444 and 1842-1921) were deemed somatic TET2 mutations. Otherwise, missense mutations outside the conserved regions were considered polymorphisms if they were present in complete remission (CR) samples.
Detection of DNMT3A mutation
Mutational analysis of DNMT3A was performed by PCR assay to amplify the entire coding sequence (exons 2-23) of DNMT3A. The PCR products were first screened by denaturing high-performance liquid chromatography (DHPLC; WAVE Transgenomic, Omaha, NE), to which guanine and cytosine clamps were added to the primers to facilitate the detection of mutations, as described previously.33 Samples with an abnormal DHPLC profile were sequenced directly on both directions. The sequences of primers used for PCR-based analysis are shown in supplemental Tables 2 and 3. DHPLC sensitivity was determined by mixing various quantities of sequence-confirmed mutants with the wild type (5%-50%). The detection limit was 5% in our assay system (supplemental Figure 1), which was more sensitive than that of direct sequencing.
Detection of IDH1 and IDH2 mutations
Mutational analyses of exon 4 of IDH1 and IDH2 were conducted with gDNA-PCR followed by direct sequencing. The primers used are shown in supplemental Table 4, which cover the coding sequences of exon 4 of IDH1 and IDH2 containing the mutational hot spot codons R132, R140, and R172.
Detection of additional gene mutations
The mutational analyses of exons 1-3 (supplemental Table 4) and 7-9 of WT1,34 TP53 (exons 5-9),35 JAK2V617F,36 and NPM1 (exon 11, original exon 12)37 were performed according to the previously described methods of other investigators with some modifications. Mutational analysis of the entire PTPN11 coding regions (exons 1-15) was performed using cDNA-PCR assay with primers shown in supplemental Table 4. For patients without available RNA samples, a gDNA PCR assay was performed according to the method of Tartaglia et al.38 For all the mutational analyses, the detected mutations were confirmed in a second independent analysis, and/or by the use of cDNA samples, and/or by the use of different primers to confirm mutations.
Statistical analysis
The Fisher exact test, the χ2 analysis, and Wilcoxon rank-sum test were used whenever appropriate to make comparisons between groups. A Kaplan-Meier estimation was used to plot the overall survival (OS) and event-free survival (EFS) of each subgroup. Comparisons of estimated survival curves were analyzed by the log-rank test. A P value < .05 was considered as statistically significant. SPSS version 17 software (SPSS Inc) was used to perform the statistical analyses.
Results
Frequency and distribution of 19 gene mutations in pediatric de novo AML
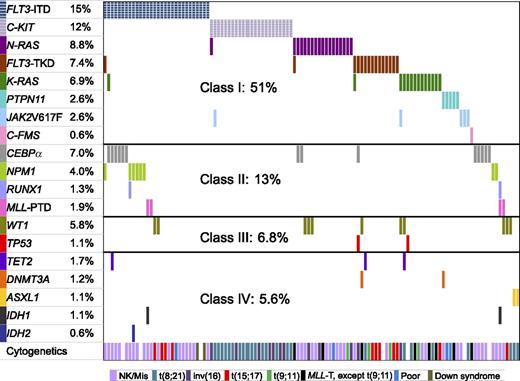
We divided the 19 mutated genes into 4 functional classes: class I included gene mutations involving signaling transduction and RAS pathways, class II included hematopoietic transcription factor genes, TP53 and WT1 comprised class III, and class IV comprised epigenetic regulator genes. The frequencies and distribution of mutated genes and their cooperation are shown in Figure 1. Approximately one-half of the patients had class I gene mutations, 13% had class II mutations, 6.8% of patients had TP53 and WT1 mutations, and 5.6% of patients had class IV gene mutations. The occurrence of classes I, II, and III mutations in the whole cohort of children with AML according to the order of frequency was as follows: class I (FLT3-ITD, C-KIT, NRAS, FLT3-TKD, KRAS, PTPN11, JAK2V617F, C-FMS); class II (CEBPα, NPM1, RUNX1, MLL-PTD); class III (WT1, TP53). Class IV mutations occurred rarely: TET2 mutations in 1.7%, DNMT3A in 1.2%, ASXL1 in 1.1%, IDH1 in 1.1%, and IDH2 in 0.6% of patients; there was no overlap in involvement among the 5 genes. Taken together, 56.8% of patients had at least one mutation among the genes we examined.
The frequencies and distribution of 19 gene mutations and their cooperativity. Each column represents one individual patient with at least one mutated gene(s) shown by different colored bars. The top 8 genes belong to class I, the next 4 genes class II, WT1 and TP53 class III, and last 5 genes class IV. The last raw represents the cytogenetics for each patient.
The frequencies and distribution of 19 gene mutations and their cooperativity. Each column represents one individual patient with at least one mutated gene(s) shown by different colored bars. The top 8 genes belong to class I, the next 4 genes class II, WT1 and TP53 class III, and last 5 genes class IV. The last raw represents the cytogenetics for each patient.
Characteristics of patients with gene mutations involving epigenetic regulators and their cooperating mutations
In total, 10 patients harbored 1 of the 5 gene mutations involving the epigenetic modifiers. Of the 8 patients who achieved CR, 6 had CR samples available for analysis, and no mutation was detected in any of the 6 CR samples examined, including the 1 patient with missense mutation of TET2 (F760Y) located outside the conserved regions; this finding indicated that they were somatic mutations.
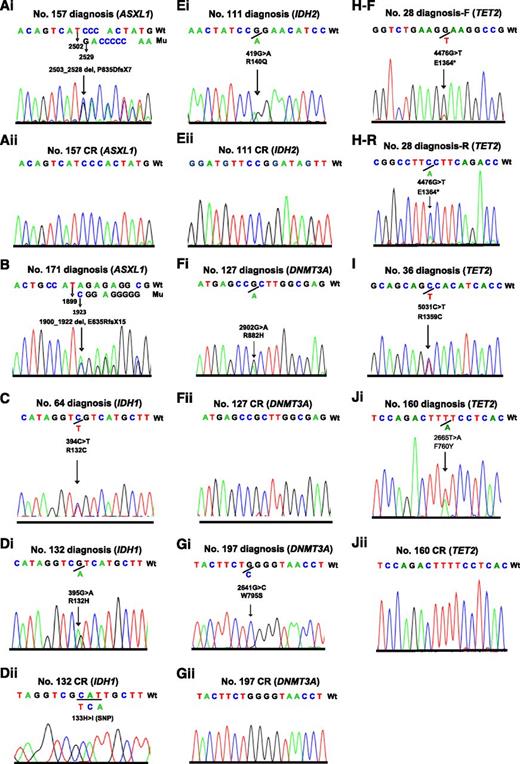
One patient with a frameshift mutation of ASXL1 (E635Rfs*15) and another one patient with nonsense mutation of TET2 (E1364*) that resulted in truncated protein who were expected to have loss of function mutations did not achieve CR. In addition, one patient each with IDH1 (R132C) and TET2 (R1359C located in highly conserved region and reported previously39 ) did not have a CR sample available, and they were considered as somatic mutations. The electropherograms of the 5 gene mutations in the 10 cases at diagnosis and the 6 samples in CR are shown in Figure 2. The clinicohematological features and characteristics of mutated genes of epigenetic modifiers as well as their coexisted mutations in the 10 patients are summarized in Table 1. The 2 patients with ASXL1 mutations had t(8;21)/RUNX1-RUNX1T1 and FAB M2 subtypes. The frequency of ASXL1 mutations in pediatric t(8;21) AML was 5.6% (2/36) compared with none of the 141 patients with non-t(8;21) AML (P = .04).
Electropherograms of the 10 pediatric patients with AML carrying 5 mutated genes of epigenetic regulators. Two patients with ASXL1 mutations (Ai and B), 2 patients with IDH1 mutations (C and Di), 1 patient with IDH2 mutation (Ei), 2 patients with DNMT3A mutations (Fi and Gi), and 3 patients with TET2 mutations (H-F reads forward, H-R reads reversely, I, and Ji) at diagnosis, and wild types of the 5 genes in complete remission of the corresponding patients (Aii, Dii, Eii, Fii, Gii, and Jii).
Electropherograms of the 10 pediatric patients with AML carrying 5 mutated genes of epigenetic regulators. Two patients with ASXL1 mutations (Ai and B), 2 patients with IDH1 mutations (C and Di), 1 patient with IDH2 mutation (Ei), 2 patients with DNMT3A mutations (Fi and Gi), and 3 patients with TET2 mutations (H-F reads forward, H-R reads reversely, I, and Ji) at diagnosis, and wild types of the 5 genes in complete remission of the corresponding patients (Aii, Dii, Eii, Fii, Gii, and Jii).
Clinicohematological features and characteristics of the mutated genes of epigenetic modifiers and their coexisted mutations in 10 pediatric AML patients
| Patient no. . | Age, y/sex . | FAB . | Cytogenetics/genetic subclass . | Mutated gene . | Amino acid change . | Cooperative mutations . | Treatment protocol . | Event-free survival, mo . |
|---|---|---|---|---|---|---|---|---|
| 157 | 10/M | M2 | 45×,-Y,t(8;21) | ASXL1 | P835DfsX7 | Nil | 97A | 68+ |
| 171 | 2/F | M2 | 46XX,t(8;12;21) | ASXL1 | E635RfsX15 | Nil | 97A | 17 |
| 64 | 5/F | M0 | 47XX,+ 21 | IDH1 | R132C | MLL-PTD | 97B | 14 |
| RUNX1 (V333Dfs*242) | ||||||||
| 132 | 11/M | M0 | 46XY | IDH1 | R132H | FLT3-ITD | 97A | 87+ |
| MLL-PTD | ||||||||
| 111 | 10/M | M2 | 46XY | IDH2 | R140Q | FLT3-ITD | 97B | 110+ |
| NPM1 (type D) | ||||||||
| 127 | 11/M | M5 | 48XY,+11,+20 | DNMT3A | R882H | PTPN11 (E76K) | 97A | 93+ |
| 197 | 14/M | M4 | 46XY,MLL translocation | DNMT3A | W795S | FLT3-TKD (D835E) | 97A | 10 |
| WT1 (A382Gfs*3) | ||||||||
| 28 | 13/M | M2 | 45×,-Y,t(8;21) | TET2 | E1364* | FLT3-TKD (D835V) | 97B | 0 |
| 36 | 14/M | M1 | 46XY | TET2 | R1359C | FLT3- ITD, CEBPα [P46H fs*115 (;) K304_Q305 insL] | 97B | 167+ |
| 160 | 16/M | M2 | 45×,-Y,t(8;21) | TET2 | F760Y | KRAS (G13R), WT1 (R434Hfs*86) | 97B | 8.9 |
| Patient no. . | Age, y/sex . | FAB . | Cytogenetics/genetic subclass . | Mutated gene . | Amino acid change . | Cooperative mutations . | Treatment protocol . | Event-free survival, mo . |
|---|---|---|---|---|---|---|---|---|
| 157 | 10/M | M2 | 45×,-Y,t(8;21) | ASXL1 | P835DfsX7 | Nil | 97A | 68+ |
| 171 | 2/F | M2 | 46XX,t(8;12;21) | ASXL1 | E635RfsX15 | Nil | 97A | 17 |
| 64 | 5/F | M0 | 47XX,+ 21 | IDH1 | R132C | MLL-PTD | 97B | 14 |
| RUNX1 (V333Dfs*242) | ||||||||
| 132 | 11/M | M0 | 46XY | IDH1 | R132H | FLT3-ITD | 97A | 87+ |
| MLL-PTD | ||||||||
| 111 | 10/M | M2 | 46XY | IDH2 | R140Q | FLT3-ITD | 97B | 110+ |
| NPM1 (type D) | ||||||||
| 127 | 11/M | M5 | 48XY,+11,+20 | DNMT3A | R882H | PTPN11 (E76K) | 97A | 93+ |
| 197 | 14/M | M4 | 46XY,MLL translocation | DNMT3A | W795S | FLT3-TKD (D835E) | 97A | 10 |
| WT1 (A382Gfs*3) | ||||||||
| 28 | 13/M | M2 | 45×,-Y,t(8;21) | TET2 | E1364* | FLT3-TKD (D835V) | 97B | 0 |
| 36 | 14/M | M1 | 46XY | TET2 | R1359C | FLT3- ITD, CEBPα [P46H fs*115 (;) K304_Q305 insL] | 97B | 167+ |
| 160 | 16/M | M2 | 45×,-Y,t(8;21) | TET2 | F760Y | KRAS (G13R), WT1 (R434Hfs*86) | 97B | 8.9 |
AML indicates acute myeloid leukemia; FAB, French-American-British classification; ITD, internal tandem duplication; PTD, partial tandem duplication of MLL; and TKD, tyrosine kinase domain.
In the present series, 4 patients had MLL-PTD, and 2 of them, both with FAB M0 subtype, had IDH1 mutations (one each of R132C and R132H). There was a strong association of MLL-PTD with IDH1 mutations compared with none of the 174 patients with non-MLL-PTD AML (P < .0001). Likewise, an association of IDH1 mutation with M0 subtype also was statistically significant (P = .003). Another one patient with FAB M2 subtype and a normal karyotype harbored the IDH2 mutation (R140Q). Both patients with DNMT3A mutations (R882H and W795S) were older than 10 years of age and had FAB M4 or M5, and 1 had an MLL translocation (with unknown partner gene) that was detected by fluorescence in situ hybridization analysis but not by conventional cytogenetics. All 3 patients with TET2 mutations were older than 10 years of age and had FAB M1 or M2; 2 had t(8;21).
Correlations between other gene mutations and clinicohematological features
Of the 19 mutated genes examined, apart from the 5 mutated genes of epigenetic modifiers, 11 of the remaining 14 gene mutations were detected in more than 3 patients; we analyzed the correlation between their clinicohematological parameters and mutation status. Only 6 mutated genes were found to have clinical correlations as shown in supplemental Table 5. FLT3-ITD and CEBPα mutations were significantly associated with older age and greater white blood cell counts. Patients with K-RAS or CEBPα mutations had a significant lower platelet counts. The NPM1 mutation was borderline significantly associated with older age. At least one mutated gene was detected throughout all FAB subtypes and cytogenetic risk groups (supplemental Table 6); however, their frequencies varied among different subgroups, with AML-M6 or M7 and patients with MLL translocations or unfavorable cytogenetic risk groups being less frequent than other subgroups.
Impact of gene mutations on outcomes in pediatric AML
Because the number in each subgroup of patients with individual gene mutation was very small, we combined the subgroups on the basis of the functional classes for the outcome analysis. We excluded patients with APL from the analysis. There were no significant differences in the EFS or OS according to the mutational status of class I, II, or III in non-APL patients (supplemental Figures 2A-4B), although a trend of inferior outcome was observed in patients with class I mutation (P = .072 for EFS and P = .088 for OS). The 10 patients with class IV gene mutations had a 5-year EFS of 50.0 ± 15.8% compared with 44.7 ± 4.4% for other non-APL patients without the mutations (P = .668, supplemental Figure 5A); and also no difference in OS was observed (50.0 ± 15.8% vs 51.0 ± 4.5%, P = .904, supplemental Figure 5B).
We further analyzed whether FLT3-ITD had an effect on outcome and found no difference (P = .268 for EFS and P = .196 for OS). The presence of C-KIT mutations in core-binding factor-AML did not affect the 5-year EFS [P = .726 for t(8;21) and P = .486 for inv(16)]. We found that the patients with MLL-PTD had a 5-year EFS of only 33.3 ± 27.2%. The 2 patients with TP53 mutations had very grave outcomes, both with OS of <4 months.
We observed that 3 patients harboring 3 mutations across classes I and II had 5-year EFS of 6, 8, and 60+ months, respectively. One patient with a combination of 3 classes I, II, and III mutations had no EFS and survived only for 1 month. Three patients harboring class IV mutations that coexisted with class I and class II mutations had EFS of 87+, 110+, and 167+ months, respectively, suggesting that mutations of epigenetic modifiers did not adversely affect outcome in patients carrying other mutated genes.
Coexistence of gene mutations in pediatric AML
Among the 117 patients with at least one mutated gene, the coexistence of more than one gene mutation was detected in 34 patients (29%). The occurrence of gene mutations within the same functional classes was rare. They were mutually exclusive in the 13 patients carrying TP53 and WT1 mutations and in the 10 patients harboring mutated genes of epigenetic regulators. Only 2 of 26 patients carrying class II mutations and 7 of 104 patients carrying class I mutations had gene mutations within the same class. As shown in Figure 2, cooperating mutations across different classes were observed in 30 patients. Combination of classes I and II was detected in 13 patients, I and III in 7 patients, I and IV in 2 patients, II and III in 1, and II and IV in 1. Six patients had multiple mutations, 1 had class I plus II plus III, 2 had class I plus III plus IV, and an additional 3 had class I plus II and IV mutations. Cooperating mutations of epigenetic regulator genes with other genes were common and present in 8 patients (Table 1), especially with class I mutations in 7. Of the 3 patients with TET2 mutations, one each coexisted with FLT3-TKD (D835V), FLT3-ITD plus CEBPα mutations, and KRAS (G13R) plus WT1 (R434Hfs*86). In addition to MLL-PTD, the 2 patients carrying IDH1 mutants had coexisting FLT3-ITD and RUNX1 (V333Dfs*242) mutations, respectively. The patient with IDH2 mutation also harbored FLT3-ITD and NPM1 (CTCG duplication, type D) mutations. Of the 2 patients with DNMT3A mutations, one cooperated with the PTPN11 (E76K) mutation, and the other coexisted with FLT3-TKD (D835E) and WT1 (A382Gfs*3) mutations. The 2 patients carrying ASXL1 mutations did not have concurrent mutations with other 18 genes analyzed; however, both had a RUNX1-RUNX1T1 transcript.
Discussion
Compared with adult AML, there were fewer studies of gene mutations in childhood AML. We previously reported a decrease in the frequencies of FLT3-ITD, FLT3-TKD, and CEBPα in childhood AML compared with those in adult AML.29,31,40 The frequencies of FLT3 and CEBPα mutations that differed considerably between children and adults were later confirmed by the Children’s Oncology Group.41,42 Other investigators also found that the frequency of NPM1 mutations was 4 times greater in adult AML compared with those of pediatric AML.43 All these findings suggested a different ontogeny between childhood and adult AML.
In the present study, we extended our previous study and systematically analyzed 19 known gene mutations involved in adult myeloid neoplasms in a large cohort of children with de novo AML. If we took the functional groups of gene mutations into consideration, mutations occurring most frequently in childhood AML was class I mutations, which involve receptor tyrosine kinases and RAS signaling pathway. Together, they accounted for one-half of our patients, with FLT3-ITD being the most frequent. The frequency of mutations that block hematopoietic differentiation was 13%, and the frequency of mutations involving apoptosis or tumor suppressor genes, ie, WT1 and TP53, was 6.9%, with the latter mutation being rare. The mutation frequencies of class I and II were similar to the report by Radtke et al,44 they used single-nucleotide polymorphism array and resequencing of candidate cancer genes in a cohort of 111 children with de novo AML. The results of our extended study on pediatric patients again confirm a great difference in the frequency of gene mutations between childhood and adult AML.
The present study showed that the recently identified gene mutations involving the DNA methylation and histone modification were very rare, accounting for approximately one tenth of those reported in adult AML. ASXL1 mutations have not been reported in childhood AML; the 2 mutations we detected were both frameshift mutations that caused a truncation of C-terminal plant homeodomain of ASXL1 and leukemia transformation.3 Our result showed a low frequency (1.1%) of ASXL1 mutations in pediatric AML, a finding in sharp contrast with those described in adult AML studies, in which the frequencies are 5- to 10-fold greater.4,5 TET2 mutations in pediatric AML only have been reported in an abstract.13 We detected a frequency of 1.7%, which was much lower than the frequency of 12% to 22% reported in adult AML.12
The very low frequencies of IDH1, IDH2, and DNMT3A mutations detected in the present series were in line with the recent reports of childhood AML16-18,20 but were in contrast to the findings in adult AML with a reported frequencies of 15% to 33% for IDH12 and 12% to 22% for DNMT3A.12,19 The authors of a recent study from Germany found 1.0% (2/195) of childhood AML had DNMT3A mutations.21 We also screened the entire coding sequences and found that both mutants were located in the C-terminal methyltransferase domain, where the greatest number of previously reported mutations in adult series were located.19,45 The mutations of ASXL1, TET2, IDH1, IDH2, and DNMT3A were mutually exclusive in our patients. Of the 10 patients with mutated genes of epigenetic regulators, apart from the one with ASXL1 E635Rfs*15, which had been reported in adult patients with AML,4,5 and another with the hot spot of R132C IDH mutation, the remaining 6 patients with IDH1(R132H), IDH2 (R140Q), ASXL1 (P835Dfs*7), DNMT3A missense mutations (W795S and R882H), or TET2 (F760Y) at diagnosis did not have mutations detected in the CR samples. Our results indicated that these mutations were somatic acquired and leukemia-associated.
Although mutations in epigenetic modifiers are particularly rare in pediatric AML, many known oncogenes and tumor suppressors might contribute, at least in part, to leukemia transformation through direct or indirect alterations in the epigenetic state. Therefore, additional molecular alterations of disordered DNA methylation, such as promoter hypermethylation with gene silencing, overexpression of DNMT, or MLL translocation, may contribute to the leukemogenesis.46 Global methylation signature or more extensive genome-wide epigenomic research are valuable tools to further uncover the molecular pathway of epigenetic abnormalities, which in turn will provide a rationale molecular basis for therapeutic reversal strategies with dehypomethylating agents or histone deacetylase inhibitors.
We found that the 5 mutated genes of epigenetic modifiers were associated with some interesting clinical characteristics in our 10 patients. Notably, both patients with DNMT3A mutations had AML-M4 or M5, and one of them had an MLL translocation that was not present in the 2 reported pediatric patients with DNMT3A mutations.21,22 In adult AML, the DNMT3A mutation also was reported to be associated with acute monocytic leukemia.47 All of our 3 patients with TET2 mutations and 2 patients with DNMT3A mutations were older than 10 years. Both patients harboring ASXL1 mutations and 2 of the 3 patients carrying TET2 mutations had t(8;21) AML. The frequency of ASXL1 mutations in our t(8;21) AML seemed to be greater than those without the mutations. This association was not observed in one adult AML study in which 8% of t(8;21)AML had ASXL1 mutations compared with 5.3% of entire AML cohort.5 Whether ASXL1 mutations are associated with t(8;21) AML remains to be examined in a larger number of t(8;21) patients.
We also observed a strong association between the IDH1 mutation and MLL-PTD or AML-M0 subtype. In one of the recent reported pediatric series on IDH mutations, 2 of the 7 IDH1-mutated patients and 1 of 9 patients with IDH2 mutations harbored MLL-PTD.18 One of 3 children with IDH1 mutations was associated with AML-M0 in another study.17 The patient with IDH2 mutation had a coexisting NPM1 mutation. We correlated the mutation status of individual gene of other functional classes with age, sex, complete blood counts, and percentage of blasts in bone marrow. Patients with FLT3-ITD and CEBPα mutations were older and had greater white blood cell counts, whereas KRAS and CEBPα mutations are associated with lower platelet counts. We observed gene mutations occurred less frequently in AML-M6 or M7 as well as in unfavorable cytogenetic risk groups. Similar to adult AML, FLT3-ITD and NPM1 mutations were present more frequently in normal karyotypes, and C-KIT mutations were strongly associated with core-binding factor AML in children.
It had been hypothesized that the development of AML is associated with at least a two-hit process with the cooperation of activation mutations of signaling pathway (class I mutations) and mutations of hematopoietic transcription factors which block differentiation (class II mutations).48 With the subsequent discovery of the increasing number of mutated genes and their cooperating mutations, a mechanism of multistep leukemogenesis was suggested.49 In the present study, we observed that cooperation of mutated genes of epigenetic regulators with other known gene mutations was very common, occurring in 8 of 10 pediatric AML patients, 7 with class I (5 FLT3, 1 KRAS, and 1 PTPN11) and 4 with class II (2 MLL-PTD, 1 CEBPα, and one each for RUNX1 and NPM1) mutations, and 2 with WT1 mutations. The 2 patients with ASXL1 mutations did not have other coexisted mutated genes, but both carried RUNX1/RUNX1T1 with disruption of RUNX1 at C-terminal region, which, like RUNX1 mutations, would result in a dysregulated hematopoietic transcription factor and impair differentiation, a finding functionally confirmed in mouse models.50
Regarding the prognostic significance of gene mutations in pediatric AML, we and others found that FLT3-ITD was most common among the mutated genes examined.20,44 We failed to find a significant difference in outcome between patients with and without FLT3-ITD. The number of other individual mutated genes in class I, II, or III was too small, and the distribution among subgroups was very heterogeneous, which precluded a meaningful analysis of the prognostic significance. We thus analyzed any of class I, II, or III genes versus no mutation in each class of genes and found no significant difference except a trend of adverse outcome in patients carrying FLT3-ITD. Class IV mutations in adult AML conferred an inferior outcome.4,5,12,39,45 One half of our patients with class IV mutations experienced long-term EFS, although there were no significant differences in OS and EFS between those with and without mutations because of the small number of patients carrying these mutations. Together, the frequency of gene mutations was much less in pediatric AML compared with those in adult patients. The clinical and prognostic relevance of gene mutations on childhood AML remains to be determined by a larger cohort of pediatric patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Ting-Yu Huang for secretarial assistance.
This study was supported by grants from Mackay Memorial Hospital, Taipei, Taiwan (MMH-E-99009 and MMH-E100009), grants from the National Science Council, Taiwan (NSC-94-2314-B-195-003 and NSC-96-2314-B-195-006-MY3), and grants from National Health Research Institute (NHRI-EX96-9434SI) and Department of Health, Taiwan (DOH100-TD-C-111-006).
Authorship
Contribution: D.-C.L. and L.-Y.S. designed the experiment; D.-C.L., L.-Y.S., and H.-C.L. analyzed the data and wrote the paper; Y.-J.H., Y.-S.S., Y.-H.H., and T.-H.L. performed the experiments; and D.-C.L., H.-C.L., C.-P.Y., T.-H.J., I.-J.H., T.-C.Y., S.-H.C. and J.-Y.H. provided patient samples and clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lee-Yung Shih, Division of Hematology-Oncology, Department of Internal Medicine, Chang Gung Memorial Hospital,199 Tung-Hwa North Road, Taipei 105, Taiwan; e-mail: sly7012@adm.cgmh.org.tw.