Key Points
Biallelic inheritance of a telomerase T-motif mutation selectively impairs repeat addition processivity and results in severe disease.
Computational algorithms commonly used to predict the impact of variants on protein function have limited sensitivity with regard to hTERT.
Abstract
Hoyeraal Hreidarsson syndrome (HHS) is a form of dyskeratosis congenita (DC) characterized by bone marrow failure, intrauterine growth retardation, developmental delay, microcephaly, cerebellar hypoplasia, immunodeficiency, and extremely short telomeres. As with DC, mutations in genes encoding factors required for telomere maintenance, such as telomerase reverse transcriptase (TERT), have been found in patients with HHS. We describe 2 sibling HHS cases caused by a homozygous mutation (p.T567M) within the TERT T motif. This mutation resulted in a marked reduction in the capacity of telomerase to processively synthesize telomeric repeats, indicating a role for the T motif in this unique aspect of telomerase function. We support this finding by demonstrating defective processivity in the previously reported p.K570N T-motif mutation. The consanguineous, heterozygous p.T567M parents exhibited telomere lengths around the first percentile and no evidence of a DC phenotype. Although heterozygous processivity defects have been associated with familial, adult-onset pulmonary fibrosis, these cases demonstrate the severe clinical and functional impact of biallelic processivity mutations. Thus, despite retaining the capacity to add short stretches of telomeric repeats onto the shortest telomeres, sole expression of telomerase processivity mutants can lead to a profound failure of telomere maintenance and early-onset multisystem disease.
Introduction
Telomeres, composed of repetitive TTAGGG hexamers and associated factors localized to chromosome ends, serve to maintain chromosome integrity by preventing end-to-end fusions, potentially deleterious recombination events, and loss of terminal coding regions during DNA replication.1 Maintenance of telomeric DNA is crucial to cell survival, as critically short telomeres are recognized as DNA damage and provoke cellular senescence or apoptosis. In the absence of this response, such dysfunctional telomeres give rise to genomic instability and have the potential to promote malignant transformation. The hematopoietic consequences of defective telomere maintenance may manifest as bone marrow failure (BMF), myelodysplastic syndrome (MDS), or acute myeloid leukemia (AML).
Telomerase, a specialized reverse transcriptase, maintains the length of the telomeric repeats in select cell populations in which it is expressed (eg, stem cells and germ cells). The catalytic protein subunit of telomerase, hTERT, is encoded by telomerase reverse transcriptase (TERT), and the integral RNA subunit, hTR, is encoded by telomerase RNA component (TERC).2 ,3 At present, 34 disease-associated sequence variants have been described within TERT, the majority of which are missense changes.4 Markedly short germ-line telomeres that result from such variants characterize disorders of telomere biology, the prototype of which is dyskeratosis congenita (DC). DC includes a 90% lifetime risk for BMF; predisposition to MDS, AML, and other malignancies; skin and mucosal abnormalities; pulmonary fibrosis; liver disease; and additional clinical features.5,6 Cells are very sensitive to a reduction in levels of telomerase such that even heterozygous mutations in TERT or TERC may result in short telomeres and a clinical phenotype via haploinsufficiency.7,8 Variants in DKC1, TINF2, NHP2, NOP10, WRAP53, CTC1, and RTEL1 have also been associated with reduced telomere length and a clinical phenotype consistent with DC.9-11
The telomere biology disorders represent a clinical spectrum.12 Mutations in DKC1, TERT, TERC, and TINF2 have been reported not only in patients with classic DC but also in individuals with a single predominant phenotype, such as BMF, MDS, liver disease, or idiopathic pulmonary fibrosis.13,14 On the severe end of the spectrum is Hoyeraal Hreidarsson syndrome (HHS), a multisystem disorder characterized by intrauterine growth retardation and cerebellar hypoplasia in addition to features of DC.15,16 Correlating with severe clinical features, patients with HHS are noted to have the most extremely shortened telomeres among the telomere biology disorder spectrum of patients. HHS has been associated with mutations in DKC1, TERT, and TINF2,17-19 as well as a splice variant of DCLRE1B.20 As with classic DC, a substantial proportion of patients with HHS remain genetically uncharacterized.
In the HHS cases described here, the mutation impacting telomerase function was found to lie within TERT. The hTERT protein consists of 4 structural domains: the telomerase essential N-terminal domain, the telomerase RNA binding domain, the catalytic reverse transcriptase (RT) domain, and the C-terminal extension.21 The RT domain is responsible for the addition of nucleotides onto the 3′ end of the chromosome by reverse transcribing the telomeric complementary template contained within hTR.22 In addition, telomerases from many species, including human, have the ability to repetitively copy the template after a single binding event, resulting in the synthesis of multiple telomeric repeats prior to telomerase’s release from the DNA substrate. This property of repeat addition processivity (referred to hereafter as processivity) requires 3 successive steps: (1) alignment of the 3′ end of the telomeric DNA with the 3′ portion of the template region of hTR, (2) reverse transcription of the RNA template adding nucleotides to the DNA 3′ end until the 5′ boundary of the template is reached, and (3) template translocation, which involves DNA/RNA strand separation and realignment of DNA to the 3′ portion of template, followed by repositioning of the DNA/RNA hybrid within the enzymatic active site.21,23 All 4 structural domains of hTERT contribute to processivity.24-26 Moreover, processivity can be enhanced by interactions with TPP1-POT1 via the telomerase essential N-terminal and C-terminal extension domains.27,28
The T motif, which lies within the telomerase RNA binding domain, was the first region of hTERT recognized as being both highly conserved and telomerase specific,2 as well as the first non-RT motif shown to be required for telomerase activity.29 The T motif is encoded by amino acids 547–594, within which lies a near-universally conserved sequence motif, FYXTE. Targeted mutagenesis of residues within this motif result in varying degrees of reduced telomerase activity,29 as well as significantly increased telomere extension rates, without affecting either hTR binding or measured processivity.30 Only 1 naturally occurring mutant in the T motif has been described to date; it was found in a multigenerational family manifesting blood diseases from macrocytosis to AML but without other clinical features of DC.31 Use of the telomerase repeat amplification protocol (TRAP) to functionally analyze the mutated residue (p.K570N) demonstrated a drastic reduction in telomerase enzymatic activity, correlating with severely shortened telomere length.31
Recently, 2 families with an ancestral TERT mutation were reported as having a predominantly adult-onset pulmonary fibrosis phenotype over multiple generations, with only 4 of 18 mutation carriers developing other characteristics of a telomere biology disorder, such as subclinical cytopenias, liver function abnormalities, and AML.32 This was striking because a notable feature of both heterozygous TERT and TERC mutations is disease anticipation, resulting in an increasingly severe phenotype and earlier onset across successive generations, correlating with progressive telomere shortening.7,33 In the reported families, however, although telomere length was consistently below the 10th percentile, with the majority falling below the first percentile, there was no evidence for evolution to a more severe DC clinical presentation with this ancestral mutation; the absence of the classical mucocutaneous features of DC was notable. Disease phenotypes in these families tracked with a double heterozygous mutation in the hTERT RT domain, p.V791I/V867M, that, when reconstituted in vitro at physiologic nucleotide concentrations, had little impact on telomerase catalytic activity but demonstrated a significant decrease in processivity. A second study revealed that the hTERT RT domain mutation p.R865H, also associated with familial adult-onset pulmonary fibrosis and telomere shortening,34 impacted processivity without affecting telomerase catalytic activity.35 These findings have led to the proposal that a selective defect in processivity, while limiting the extent of telomere lengthening, allows for sufficient replenishment of telomeric repeats on the shortest telomeres, thereby preventing severe, early-onset telomere-related disease.32
Here, we describe 2 cases of an inherited DC-spectrum disease resulting from a sequence variant within the hTERT T motif, just C terminal of the critical FYXTE region. Functional analysis revealed this variant to be associated with significantly reduced processivity. Notably, this patient and his similarly affected sister carried a homozygous mutation and manifested a phenotype consistent with HHS. Thus, we show that reduced processivity alone may drastically impair telomere maintenance and result in a severe clinical phenotype.
Materials and methods
Human subjects
The Baylor College of Medicine Institutional Review Board approved this study. Informed consent was obtained from the participants in accordance with the Declaration of Helsinki.
Telomerase genotyping
Telomerase genotyping was performed by bidirectional polymerase chain reaction (PCR)-based double-stranded automated sequencing of the exons and flanking intronic regions (Ambry Genetics).
Telomere length
Telomere flow fluorescence in situ hybridization (FISH) was carried out as part of clinical testing as previously described.36 Telomere length analysis on the proband was performed by the Universitat Bern (Bern, Switzerland) and on the parents of the proband at Repeat Diagnostics (Vancouver, British Columbia).
Telomerase in vivo reconstitution
In vivo reconstitution of telomerase was carried out in 293FT cells (Invitrogen) by cotransfection of pcDNA-hTERT and pBS-U1-hTR vectors. Substitutions K570N and T567M were introduced into pcDNA-hTERT by site-direct mutagenesis using overlapping PCR.37 Cell transfection and lysate preparation were carried out as previously described.23
Northern blotting
Total RNA was isolated from transfected 293FT cells using TRI Reagent (Molecular Research Center) following the manufacturer’s instructions. Five micrograms of total RNA was resolved on a 4% polyacrylamide/8M urea denaturing gel, followed by electroblotting to a Hybond-XL membrane (GE Healthcare) and UV crosslinking. The membrane was hybridized with riboprobes targeting 5S rRNA and hTR as described previously.38
Western blotting
Thirty micrograms of protein for each lysate was heated at 95°C for 5 minutes in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer, resolved on an 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, and electrotransferred onto a polyvinylidene difluoride membrane. The membrane was blocked in 5% nonfat milk/1× TTBS buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.05% Tween-20), followed by incubation with anti-hTERT goat polyclonal antibody L-20 (Santa Cruz Biotechnology) or anti–glyceraldehyde-3-phosphate dehydrogenase mouse monoclonal antibody 6C5 (Life Technologies) in 5% nonfat milk/1× TTBS for 1 hour. After washing 3 times with 1× TTBS buffer, the membrane was incubated with the HRP-conjugated donkey–anti-goat antibody (Santa Cruz Biotechnology) or goat–anti-mouse antibody (Bio-Rad) in 5% nonfat milk/1× TTBS for 1 hour at room temperature. Following 3 washes with 1× TTBS, the membrane was developed and analyzed as previously described.23
Telomerase direct primer-extension assay
The direct primer-extension assay was performed using 1 μL of transfected 293FT cell lysate at 1.5 μg protein/μL as previously described.23 Telomerase activity was quantitated and normalized to the loading control and the expression level of hTR, which is limiting in the in vivo reconstitution system. Telomerase processivity was quantitated as previously described.38
TRAP assay
A 2-tube TRAP assay was performed using 1 μL of diluted cell lysate with 3 ng/μL and 0.6 ng/μL of protein concentrations as previously described, with minor modifications.39 Cell lysate was mixed with 0.5 μM of TS primer (5′-AATCCGTCGAGCAGAGTT-3′) in 1× primer extension buffer (50 mM Tris-HCl, pH 8.3, 2 mM DTT, 0.5 mM magnesium chloride, and 1 mM spermidine) containing 20 μM dATP, 20 μM dTTP, and 10 μM dGTP, followed by 1 hour incubation at 30°C. The TS primer was purified by phenol/chloroform extraction and ethanol precipitation. The telomerase-extended products were PCR amplified in a 25-μL reaction consisting of 1× Taq buffer (NEB), 50 μM of dNTP, 0.4 μM of 32P end-labeled TS primer, 0.4 μM of ACX primer (5′-GCGCGGCTTACCCTTACCCTTACCCTAACC-3′), 0.4 μM of NT primer (5′-ATCGCTTCTCGGCCTTTT-3′), 2 × 10−13 M of TSNT primer (5′-CAATCCGTCGAGCAGAGTTAAAAGGCCGAGAAGCGATC-3′), and 1 unit of Taq (NEB). Samples were denatured at 94°C for 2 minutes, followed by 25 cycles at 94°C for 25 seconds and at 59°C for 30 seconds. PCR products were resolved on a 10% polyacrylamide/2% glycerol gel. The gel was dried, exposed to a phosphor storage screen, and analyzed with a Bio-Rad FX-Pro molecular imager. Total activity was quantitated and normalized to the intensity of an internal control band.
TERT variant analysis
Data for the TERT variant table (Table 1) were compiled using dbNSFP version light 1.3 (December 2011), incorporating recent updates to both PolyPhen-2 and SIFT. Of the 34 TERT mutations listed in the Telomerase Database (http://telomerase.asu.edu/), the 6 insertions or deletions were excluded because they could not be evaluated by the algorithms used by dbNSFP. An additional 3 mutations were excluded because the specific change was not included in the dbNSFP database. Because the H412Y variant has reported telomerase activity of between 50% and 91%, it was excluded for ease of interpretation. Therefore, 24 previously described mutations were included (Table 1), in addition to the novel variant described here. The “total predicted damaging” score was calculated by assigning 1 point to each of the 4 algorithms for prediction of either “damaging” or “deleterious.” A half-point was added for a prediction of “possibly damaging” from PolyPhen-2. Therefore, the scores ranged from 0, indicating prediction as benign or nondeleterious by all 4 algorithms, to 4, indicating prediction as deleterious or damaging by all 4 algorithms. “Telomerase activity” and “Present in ≥ relatives” columns were summarized from published literature describing these variants (see Reference column) in terms of functional analysis and presence in at least 2 members of a pedigree.
Naturally occurring TERT variants resulting in a missense change associated with results from 4 algorithms predicting functional impact, telomerase activity, and presence or absence in a family pedigree
| AA position . | Reference nucleotide . | Alternative nucleotide . | Reference AA . | Alternative AA . | PhyloP . | SIFT . | PolyPhen 2 . | LRT . | MutationTaster . | Total score . | Telomerase activity, % . | Present in ≥2 relatives . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 567 | G | A | T | M | C | T | P | N | NDe | 0.5 | 60 | Yes | This study |
| 33 | G | A | P | S | NC | T | B | N | NDe | 0 | 80 | Yes | 34 |
| 55 | A | T | L | Q | C | D | P | N | De | 2.5 | 35–40 | Yes | 13,44 |
| 144 | C | T | V | M | NC | D | D | N | De | 3 | 75 | Yes | 34 |
| 202 | C | T | A | T | NC | T | B | N | NDe | 0 | <1 | Yes | 14,45 |
| 279 | C | T | A | T | NC | T | P | N | NDe | 0.5 | 81–100 | No | 31,44,46,47 |
| 412 | G | A | H | Y | C | T | B | N | De | 1 | 50–91 | Yes | 14,44 |
| 486 | G | A | R | C | C | T | D | N | De | 2 | 45 | Yes | 34 |
| 570 | C | G | K | N | C | D | B | De | De | 3 | <1 | No | 31 |
| 682 | C | T | G | D | C | T | P | De | De | 2.5 | <1 | No | 31,48 |
| 694 | C | T | V | M | NC | D | D | N | De | 3 | <1 | Yes | 14 |
| 704 | G | A | P | S | C | T | P | N | De | 1.5 | 13 | No | 46,49 |
| 716 | G | A | A | V | C | D | P | De | De | 3.5 | 14 | No | 46 |
| 721 | G | C | P | R | C | D | D | De | De | 4 | 100 | Yes | 31,47 |
| 726 | G | A | T | M | NC | T | B | N | NDe | 0 | 100 | No | 31,48 |
| 772 | T | C | Y | C | C | T | D | N | De | 2 | <1 | Yes | 14 |
| 811 | G | A | R | C | C | D | D | N | NDe | 2 | <50 | Yes | 19 |
| 846 | T | C | Y | C | NC | D | D | De | De | 4 | 10 | No | 49 |
| 865 | C | T | R | H | C | D | D | De | De | 4 | 28 | Yes | 34 |
| 876 | G | C | H | Q | C | T | P | De | NDe | 1.5 | 10 | No | 49 |
| 901 | G | A | R | W | NC | D | B | N | NDe | 1 | <25 | Yes | 19 |
| 902 | C | G | K | N | C | D | P | De | De | 3.5 | <1 | Yes | 7,31 |
| 979 | G | A | R | W | NC | D | P | N | De | 2.5 | 100 | Yes | 31,45 |
| 1015 | A | G | C | R | C | T | P | De | De | 2.5 | 9 | No | 46 |
| 1062 | C | T | A | T | NC | T | B | N | NDe | 0 | 100 | No | 44,46 |
| 1090 | C | T | V | M | NC | D | B | N | NDe | 1 | <1 | No | 14 |
| AA position . | Reference nucleotide . | Alternative nucleotide . | Reference AA . | Alternative AA . | PhyloP . | SIFT . | PolyPhen 2 . | LRT . | MutationTaster . | Total score . | Telomerase activity, % . | Present in ≥2 relatives . | Reference . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 567 | G | A | T | M | C | T | P | N | NDe | 0.5 | 60 | Yes | This study |
| 33 | G | A | P | S | NC | T | B | N | NDe | 0 | 80 | Yes | 34 |
| 55 | A | T | L | Q | C | D | P | N | De | 2.5 | 35–40 | Yes | 13,44 |
| 144 | C | T | V | M | NC | D | D | N | De | 3 | 75 | Yes | 34 |
| 202 | C | T | A | T | NC | T | B | N | NDe | 0 | <1 | Yes | 14,45 |
| 279 | C | T | A | T | NC | T | P | N | NDe | 0.5 | 81–100 | No | 31,44,46,47 |
| 412 | G | A | H | Y | C | T | B | N | De | 1 | 50–91 | Yes | 14,44 |
| 486 | G | A | R | C | C | T | D | N | De | 2 | 45 | Yes | 34 |
| 570 | C | G | K | N | C | D | B | De | De | 3 | <1 | No | 31 |
| 682 | C | T | G | D | C | T | P | De | De | 2.5 | <1 | No | 31,48 |
| 694 | C | T | V | M | NC | D | D | N | De | 3 | <1 | Yes | 14 |
| 704 | G | A | P | S | C | T | P | N | De | 1.5 | 13 | No | 46,49 |
| 716 | G | A | A | V | C | D | P | De | De | 3.5 | 14 | No | 46 |
| 721 | G | C | P | R | C | D | D | De | De | 4 | 100 | Yes | 31,47 |
| 726 | G | A | T | M | NC | T | B | N | NDe | 0 | 100 | No | 31,48 |
| 772 | T | C | Y | C | C | T | D | N | De | 2 | <1 | Yes | 14 |
| 811 | G | A | R | C | C | D | D | N | NDe | 2 | <50 | Yes | 19 |
| 846 | T | C | Y | C | NC | D | D | De | De | 4 | 10 | No | 49 |
| 865 | C | T | R | H | C | D | D | De | De | 4 | 28 | Yes | 34 |
| 876 | G | C | H | Q | C | T | P | De | NDe | 1.5 | 10 | No | 49 |
| 901 | G | A | R | W | NC | D | B | N | NDe | 1 | <25 | Yes | 19 |
| 902 | C | G | K | N | C | D | P | De | De | 3.5 | <1 | Yes | 7,31 |
| 979 | G | A | R | W | NC | D | P | N | De | 2.5 | 100 | Yes | 31,45 |
| 1015 | A | G | C | R | C | T | P | De | De | 2.5 | 9 | No | 46 |
| 1062 | C | T | A | T | NC | T | B | N | NDe | 0 | 100 | No | 44,46 |
| 1090 | C | T | V | M | NC | D | B | N | NDe | 1 | <1 | No | 14 |
B, benign; C, conserved; D, damaging; De, deleterious; N, neutral; NC, nonconserved; NDe, nondeleterious; P, possibly damaging; T, tolerated.
Results
An HHS phenotype is associated with a homozygous TERT p.T567M mutation within the T motif
The male patient presented at age 30 months as a referral from the Middle East, with pancytopenia, developmental delay, ataxia, and cerebellar hypoplasia. Pancytopenia, with 50% bone marrow cellularity, was first noted at age 20 months, following a viral illness. He was born at term with evidence of intrauterine growth retardation (birth weight 1.9 kg) and hospitalized for the first 2 weeks of life with respiratory and swallowing difficulties. His weight was in the fifth percentile and his height was below the third percentile. His exam was notable for oral leukoplakia, ataxia, hypotonia, and decreased reflexes. No pigmentation abnormalities were evident, and nails were thin but not dystrophic. His speech and gross and fine motor development were significantly delayed, with inability to walk independently and to dress or feed himself. He spoke no intelligible words, but was able to follow simple 1-step commands.
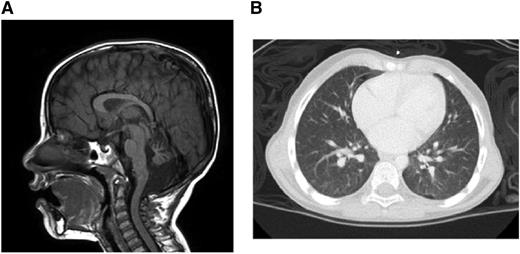
Brain magnetic resonance imaging demonstrated multiple findings including profound atrophy of the cerebellar vermis (Figure 1A). A chest computed tomography revealed diffuse, ill-defined ground-glass nodules and bilateral pleural blebs (Figure 1B). Liver function test results were within normal limits.
Clinical manifestations. (A) Brain magnetic resonance imaging. (B) Chest computed tomography.
Clinical manifestations. (A) Brain magnetic resonance imaging. (B) Chest computed tomography.
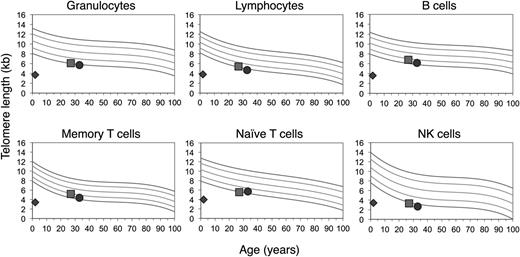
Given his clinical findings, telomere length testing was performed at age 28 months using the telomere flow FISH assay (University Hospital, Bern).36 His telomere lengths were well below the first percentile in all white blood cell (WBC) subsets (Figure 2). Sequencing of TINF2, NHP2, and NOP10 revealed no variants (Ambry Genetics). Sequencing of DKC1 and TERC was not performed as these were found to be without variants in his similarly affected sibling (see below). TERT sequencing revealed a homozygous variant of unknown significance, p.T567M (c.1700C>T), in exon 3, mapping to the T motif (Figure 3).
Homozygosity of TERT p.T567M results in dramatic telomere shortening. Telomere lengths determined by telomere flow FISH analysis (courtesy of University Hospital, Bern and Repeat Diagnostics, Vancouver). Reference percentiles indicated in the top left graph obtained from the determination of telomere length in 400 normal controls.14 The proband’s telomere length is indicated by the diamond, the mother’s by the square, and the father’s by the circle.
Homozygosity of TERT p.T567M results in dramatic telomere shortening. Telomere lengths determined by telomere flow FISH analysis (courtesy of University Hospital, Bern and Repeat Diagnostics, Vancouver). Reference percentiles indicated in the top left graph obtained from the determination of telomere length in 400 normal controls.14 The proband’s telomere length is indicated by the diamond, the mother’s by the square, and the father’s by the circle.
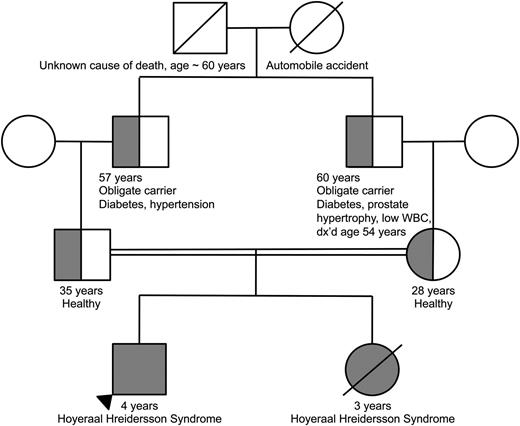
Family pedigree demonstrating consanguinity between the proband’s parents. The homozygous proband and deceased sibling are indicated by the completely filled circle/square. The heterozygous parents as well as obligate heterozygous grandfathers are indicated by the half-filled circles/ half-filled squares.
Family pedigree demonstrating consanguinity between the proband’s parents. The homozygous proband and deceased sibling are indicated by the completely filled circle/square. The heterozygous parents as well as obligate heterozygous grandfathers are indicated by the half-filled circles/ half-filled squares.
The patient was diagnosed with HHS. Given his progressive, severe cytopenias and bone marrow hypocellularity (<5% to 10% to 20%), he received a bone marrow transplant from a 9/10 matched unrelated donor. The combination of fludarabine, melphalan, and alemtuzumab was used as a conditioning regimen, and cyclosporine and prednisone were used as graft-versus-host-disease prophylaxis. He engrafted on day 14, and subsequent chimerism studies revealed 100% donor cells. His immediate post transplant course was complicated by transaminitis, engraftment syndrome, and adenovirus enteritis. At the time of manuscript preparation, he continues to be in remission from his marrow failure.
Biallelic inheritance of TERT p.T567M (c.1700C>T) results in severe disease and dramatic telomere shortening, whereas heterozygosity for the variant has minimal clinical consequence
The patient’s sister had clinical features that were similar to those of her brother, also presenting at an early age with ataxia and BMF. At age 3 years, she underwent matched unrelated cord blood transplantation. However, a complicated post transplant course, including idiopathic pulmonary syndrome and persistent cytopenias, resulted in her death 1 month following transplant. She did not receive telomere length testing; however, post-mortem sequencing confirmed homozygosity for TERT p.T567M (GeneDx).
The parents, who were first cousins, were found to be heterozygous for TERT p.T567M (Figure 3). They were healthy and reported no clinical manifestations consistent with a telomere biology disorder. Their complete blood counts and indices were normal. Telomere flow FISH analysis revealed their telomere lengths near or just below the first percentile for all leukocyte subsets (Figure 2). Pedigree analysis indicated that the paternal and maternal grandfathers, who were brothers, were obligate carriers, with the mutation being inherited from one of their parents. The maternal grandfather reported having a low WBC count for the past 6 to 7 years. Otherwise, there was no reported family history of other blood disorders, cancers, or hepatic or pulmonary disease.
hTERT p.T567M is associated with reduced telomerase processivity
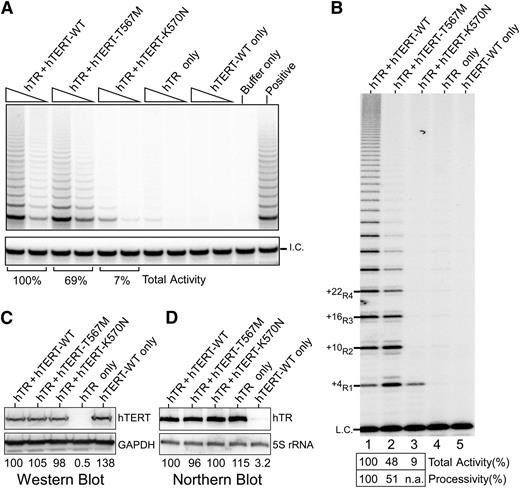
Given the disease severity in the proband, we explored the impact of this mutation on telomerase activity and processivity. We analyzed the effect of the mutation on in vivo reconstituted telomerase using the TRAP assay. We found total activity was only modestly reduced to 69% of wild-type (WT) levels (Figure 4A), which was surprising given the extreme telomere shortening in the proband. As the TRAP assay does not directly measure processivity, we then analyzed the same lysates using a direct primer extension assay, which allows quantitative assessment of processivity.21 In this assay, the T567M-mutant telomerase exhibited a marked decrease in processivity, specifically showing a dramatic decrease of the longer length products corresponding to +22R4 and above bands relative to WT (Figure 4B). These differences could not be accounted for by differences in hTERT or hTR levels (Figure 4C-D). A similar processivity defect was observed using telomerase reconstituted in vitro (supplemental Figure 1), thus indicating an inherent defect in telomerase processivity. Although the total activity was reduced (48% of WT), this was accounted for mostly by the loss of the multiple repeat addition products, ie, defective processivity.
Telomerase activity analyses of TERT p.K570N and p.T567M mutants. (A) Telomerase TRAP assay of TERT mutants. Two concentrations of lysate (3 ng and 0.6 ng of protein) are analyzed in a 2-step TRAP reaction. Control reactions with CHAPS lysis buffer in place of lysate (buffer only) and a telomerase positive cell lysate (positive) are included. (B) Telomerase direct assay of TERT p.K570N and p.T567M mutant telomerase reconstituted in vivo. Cell lysates (1.5 μg of protein) of 293FT cells transfected with TERC and TERT-WT (lane 1), T567M (lane 2), or K570N (lane 3) are analyzed by direct primer extension assays. Transfections of TERC (lane 4) or TERT-WT (lane 5) alone are included as controls. A 32P end-labeled 18-mer oligonucleotide is included as the loading control (L.C.). (C) Western blot of ectopic TERT expression. The glyceraldehyde-3-phosphate dehydrogenase protein is used as the internal control. (D) Northern blot analysis of ectopic TERC expression. The endogenous 5S rRNA is probed to ensure equal loadings.
Telomerase activity analyses of TERT p.K570N and p.T567M mutants. (A) Telomerase TRAP assay of TERT mutants. Two concentrations of lysate (3 ng and 0.6 ng of protein) are analyzed in a 2-step TRAP reaction. Control reactions with CHAPS lysis buffer in place of lysate (buffer only) and a telomerase positive cell lysate (positive) are included. (B) Telomerase direct assay of TERT p.K570N and p.T567M mutant telomerase reconstituted in vivo. Cell lysates (1.5 μg of protein) of 293FT cells transfected with TERC and TERT-WT (lane 1), T567M (lane 2), or K570N (lane 3) are analyzed by direct primer extension assays. Transfections of TERC (lane 4) or TERT-WT (lane 5) alone are included as controls. A 32P end-labeled 18-mer oligonucleotide is included as the loading control (L.C.). (C) Western blot of ectopic TERT expression. The glyceraldehyde-3-phosphate dehydrogenase protein is used as the internal control. (D) Northern blot analysis of ectopic TERC expression. The endogenous 5S rRNA is probed to ensure equal loadings.
Given the impact of the TERT p.T567M mutation on telomerase processivity, we addressed whether the other disease-causing motif T mutation K570N31 had a similar impact. Remarkably, in the direct primer extension assay, the in vivo reconstituted K570N-mutant telomerase was nonprocessive, producing only the +4R1 product (Figure 4B). In the TRAP assay, the K570N-mutant telomerase showed 7% of WT activity (Figure 4A). However, since the PCR amplification step of TRAP does not favor the single-repeat product, the activity in the TRAP assay does not correctly represent the actual activity or processivity of this mutant. Thus, as revealed by the direct assay, the effect of this mutation in vivo is likely a consequence of defective processivity.
Position 567 in hTERT is not an evolutionarily conserved residue
As noted in Figure 5, although a threonine is present at the position aligning with human 567 in 6 of the 14 orthologs examined, the remainder contains either a methionine or valine at this position. This was surprising, as the substitution in our case, which resulted in a profound defect in processivity, was a methionine. Moreover, among the species with a methionine at this position, Gallus gallus and Takifugu rubripes have telomerases that are processive in vitro,40,41 whereas Xenopus laevis telomerase is known to be nonprocessive, producing at most 2 repeats when analyzed by conventional assays.42 No reports of telomerase activity and processivity in Coturnix japonica were found. Thus, while human telomerase contains threonine at position 567 for an optimal processivity, other species adapt other amino acids for maintaining processivity or simply tolerate a lower processivity.
Sequence alignment of the TERT T motif across species. Colors represent amino acids with similar properties. Residue 567 is outlined in blue.
Sequence alignment of the TERT T motif across species. Colors represent amino acids with similar properties. Residue 567 is outlined in blue.
Computational predictions of effect of variants on protein function are inconsistent with measured telomerase activity and/or observed clinical phenotype
Several algorithms have been developed to predict the impact of an amino acid substitution on protein function. Among the commonly used algorithms, SIFT (http://sift.jcvi.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), MutationTaster (http://www.mutationtaster.org), and LRT43 predicted no effect of the hTERT p.T567M substitution on protein function, reflecting, in part, the lack of evolutionary conservation of T567. However, as the proband’s phenotype was so severe and the functional analyses demonstrated definitive impact on processivity, we hypothesized that these algorithms might be limited in their capacity for predicting the effect of missense changes on telomerase function. To investigate this hypothesis, we extracted missense changes associated with a clinical phenotype found in the Telomerase Database4 and correlated each with its reported telomerase activity, as well as whether the mutation was observed in more than 1 family member with a telomere biology disorder phenotype. In addition, we used dbNSFP (https://sites.google.com/site/jpopgen/dbNSFP) to compile predictions for each of these mutations and generated a cumulative predictive score for deleterious effect across all 4 algorithms. As outlined in Table 1, the predictions generated by these programs were often inconsistent with both clinical observation and measured telomerase function. Table 2 summarizes these findings in terms of the number of variants with cumulative predictive scores of 0 to 1, 1.5 to 2, 2.5 to 3, and 3.5 to 4, categorized by telomerase functional analysis (≤80% or >80%) and presence of the variant in 2 or more affected individuals within a pedigree. By dividing this table into 4 quadrants, we found that of the 12 variants predicted to be deleterious (cumulative score of 2.5 to 4), only 2 were associated with normal telomerase activity in vitro. Conversely, of the 12 variants predicted to be benign (cumulative score of 0 to 2), 9 were associated with significantly reduced telomerase activity in vitro. Thus, these algorithms exhibited a poor sensitivity for telomerase defects.
Association between cumulative scores predicting consequence of a missense change and telomerase functional analysis as well as association with a hereditary clinical phenotype
| Variant characteristics . | Score 0 to 1 . | Score 1.5 to 2 . | Sum of variants with scores of 0 to 2 . | Score 2.5 to 3 . | Score 3.5 to 4 . | Sum of variants with scores of 2.5 to 4 . |
|---|---|---|---|---|---|---|
| Reported in pedigree (2 or more relatives) | 3 | 3 | 3 | 2 | ||
| Telomerase activity no higher than 80% | 1 | 2 | 3 | 2 | ||
| Number of variants reported in pedigree and with reduced telomerase activity | 4 | 5 | 9 | 6 | 4 | 10 |
| Not reported in pedigree | 0 | 0 | 1 | 1 | ||
| Telomerase activity more than 80% | 3 | 0 | 0 | 0 | ||
| Number of variants not reported in a pedigree and with normal telomerase activity | 3 | 0 | 3 | 1 | 1 | 2 |
| Variant characteristics . | Score 0 to 1 . | Score 1.5 to 2 . | Sum of variants with scores of 0 to 2 . | Score 2.5 to 3 . | Score 3.5 to 4 . | Sum of variants with scores of 2.5 to 4 . |
|---|---|---|---|---|---|---|
| Reported in pedigree (2 or more relatives) | 3 | 3 | 3 | 2 | ||
| Telomerase activity no higher than 80% | 1 | 2 | 3 | 2 | ||
| Number of variants reported in pedigree and with reduced telomerase activity | 4 | 5 | 9 | 6 | 4 | 10 |
| Not reported in pedigree | 0 | 0 | 1 | 1 | ||
| Telomerase activity more than 80% | 3 | 0 | 0 | 0 | ||
| Number of variants not reported in a pedigree and with normal telomerase activity | 3 | 0 | 3 | 1 | 1 | 2 |
Discussion
Here we report a novel TERT variant with marked effects on processivity, resulting in significantly shortened telomere length and severe clinical manifestations when inherited as a homozygous mutation. This mutation represents the second naturally occurring mutation reported within the T motif of hTERT. This motif, although both highly conserved and known to be required for normal telomerase catalytic activity, had thus far not been implicated in promoting processivity. We demonstrate that both of the disease-associated T-motif variants, T567M and K570N, impact processivity, underscoring the T motif’s role in this aspect of telomerase function.
Within this pedigree, we noted the absence of a telomere biology disorder phenotype in the proband’s parents, both heterozygous for the mutation described, despite having telomere lengths near the first percentile in all leukocyte subsets. The proband’s grandfathers, who were obligate carriers, lacked a significant history as well, except for the maternal grandfather who had a several-year history of low WBC count. However, this mutation was associated with marked telomere shortening and a severe HHS phenotype when both alleles were affected in the parents’ offspring. Telomere lengths on the sister were not measured; however, the proband’s telomere lengths fell well below the first percentile for his age, as would be expected in this severe form of DC. As has been noted in familial pulmonary fibrosis cohorts, individuals with heterozygous mutations affecting processivity may benefit from telomerase’s natural ability to preferentially elongate the shortest telomeres.32 These heterozygous individuals, though incapable of producing telomeres of normal length, are able to maintain lengths above the critical threshold for senescence, thus minimizing the potential for manifestation of a telomere biology disorder. However, in the absence of a functional copy of TERT, the moderate processivity defect of the T567M mutation is able to result in a severe phenotype.
The direct primer-extension assay shows that the T567M mutation moderately reduces processivity, whereas the K570N mutation abolishes telomerase processivity (Figure 4B). Telomerase processivity mutants have higher probability of enzyme dissociation from the DNA products and could, thus, increase enzyme turnover in the reaction.38 This effect is observed with the p.T567M mutant, which generates more initial telomere repeat products, ie, the +4R1 and +10R2, than WT, presumably due to enzyme turnover. In comparison, the nonprocessive p.K570N mutant generates the +4R1 repeat product at the WT level, suggesting a low enzyme turnover. However, telomerase processivity likely plays a more important role than enzyme turnover in telomere lengthening in cells where the telomere substrate and telomerase concentration are low. Notably, the heterozygous TERT p.K570N mutation tracked with various blood disorders across a 4-generation pedigree.31 As in the familial pulmonary fibrosis cohort, classical manifestations of DC were not observed, again contrasting the effects of inheritance of a heterozygous vs homozygous processivity mutant.
We also note that the hTERT p.T567M substitution does not occur within an evolutionarily conserved residue. In addition, species with a naturally occurring methionine at the position cognate to human TERT 567 do not consistently demonstrate reduced telomerase processivity, suggesting the presence of alternative factors that are influencing telomerase processivity, eg, coevolution of TERT and TERC. Last, our survey of known disease-associated TERT mutations demonstrates that algorithms commonly used to predict functional impact are unreliable in many cases, particularly when a missense change is predicted to be benign. A possible explanation for this discrepancy is failure of the algorithm to consider the effect of a residue substitution on RNA–protein interactions. Therefore, we recommend reliance on measured in vitro functional analysis, including processivity assays, when determining the impact of TERT variants on telomerase function, including rare variants predicted to be benign.
Presented in abstract form at the 54th annual meeting of the American Society of Hematology, Atlanta, GA, December 8-11, 2012.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the described family for their participation. The authors also thank Vinayaka Prasad, Albert Einstein School of Medicine, and Joachim Lingner, Swiss Institute for Experimental Cancer Research, for the pNFLAG-hTERT, and pcDNA-hTERT and pBS-U1-hTR plasmids, respectively.
This work was supported by grants from the National Institutes of Health (5K12CA090433-10 to M.M.G. and R01GM094450 to J.J.-L.C.) and from the Cancer Prevention Research Institute of Texas (RP120076 to A.A.B.).
Authorship
Contribution: A.A.B. and J.J.-L.C. were the principal investigators and take primary responsibility for the paper; M.M.G. drafted the manuscript and performed the research related to the computational algorithms; X.Q. performed the telomerase functional analyses; G.S.S. and A.A.B. were involved directly in the patient’s care, obtained consents for the research study, and collected samples and clinical information; and all authors analyzed the data, critically revised the manuscript, and agreed upon the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alison A Bertuch, Baylor College of Medicine, 1102 Bates St, Suite 1200, Houston, TX 77030; e-mail: abertuch@bcm.edu.
References
Author notes
M.M.G. and X.Q. contributed equally to this study, and J.J.-L.C. and A.A.B. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal