Key Points
Recurrent mutations in FOXO1 affect the DNA binding domain and the T24 phosphorylation site, which disrupt interactions with 14-3-3.
Presence of FOXO1 mutations is associated with decreased OS, particularly in DLBCL patients of the low-risk R-IPI categories.
Abstract
Diffuse large B-cell lymphoma (DLBCL) accounts for 30% to 40% of newly diagnosed lymphomas and has an overall cure rate of approximately 60%. Previously, we observed FOXO1 mutations in non-Hodgkin lymphoma patient samples. To explore the effects of FOXO1 mutations, we assessed FOXO1 status in 279 DLBCL patient samples and 22 DLBCL-derived cell lines. FOXO1 mutations were found in 8.6% (24/279) of DLBCL cases: 92.3% (24/26) of mutations were in the first exon, 46.2% (12/26) were recurrent mutations affecting the N-terminal region, and another 38.5% (10/26) affected the Forkhead DNA binding domain. Recurrent mutations in the N-terminal region resulted in diminished T24 phosphorylation, loss of interaction with 14-3-3, and nuclear retention. FOXO1 mutation was associated with decreased overall survival in patients treated with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (P = .037), independent of cell of origin (COO) and the revised International Prognostic Index (R-IPI). This association was particularly evident (P = .003) in patients in the low-risk R-IPI categories. The independent relationship of mutations in FOXO1 to survival, transcending the prognostic influence of the R-IPI and COO, indicates that FOXO1 mutation is a novel prognostic factor that plays an important role in DLBCL pathogenesis.
Introduction
FOXO proteins comprise a family of transcription factors involved in several diseases including cancers, where they may act as tumor suppressors. Misregulated FOXOs have been observed in breast cancer, prostate cancer, colon carcinoma, ovarian cancer, multiple myeloma, B-chronic lymphocytic leukemia, and chronic myelogenous leukemia.1-4 The tumor suppressive roles of FOXO proteins owe, in part, to their regulation of a large subset of genes involved in DNA repair, cell cycle regulation, and apoptosis.1-5 Somatic deletion of Foxo1, Foxo3, and Foxo4 in mice results in thymic lymphomas and hemangiomas.6 FOXO1 expression was found to be reduced in classical Hodgkin lymphoma and lymphocyte-predominant Hodgkin lymphoma relative to non-Hodgkin lymphomas and normal germinal center B cells, and when FOXO1 was ectopically reexpressed in classical Hodgkin lymphoma cell lines, apoptosis was induced.7
Studies in myeloid leukemias have demonstrated a potential oncogenic role of FOXO factors, suggesting their roles in cancers are likely dependent on the cell of origin (COO).8,9 The maintenance of leukemia-initiating cells requires inhibition of maturation and differentiation programs, which requires activated protein kinase B (PKB/AKT) and/or the presence of activated/nuclear FOXO protein.9 Modulation of leukemia-initiating cell activity is achieved through FOXO3 expression, which has been localized to the nucleus in primary acute myeloid leukemias.9
B-cell commitment is a multistage process involving cell survival, proliferation, gene rearrangements, class-switch recombination, and terminal differentiation.10 Transcriptional regulation plays a critical role in this process. During B-cell commitment and differentiation, survival signals are transmitted in part by stimulation of the B-cell receptor and other cell surface receptors that can activate phosphoinositide 3-kinase and, as a result, AKT.11 Subsequent to its activation, AKT phosphorylates FOXO1, which results in cytoplasmic sequestration by 14-3-3 and suppression of FOXO1 transcriptional activity.12
FOXO1 has diverse roles among different cell types1,4 and distinct functions at the various stages of B-cell development.10,13-15 Conditional deletion of FOXO1 resulted in partial blockade of the pre-pro B to early pro-B transition, as a consequence of decreased interleukin-7 receptor signaling and diminished transcription of RAG genes, leading to impairment of the V(D)J rearrangement.10 FOXO1 also has roles in the maturation of peripheral B cells and class-switch recombination through transcriptional regulation of activation-induced cytidine deaminase.10
Diffuse large B-cell lymphoma (DLBCL) is an aggressive cancer associated with recently described somatic driver mutations.16,17 There are two molecular subtypes of DLBCL that are defined by distinct gene expression profiles indicative of the COO, the activated B-cell (ABC) and germinal center B-cell (GCB) types, which respond differently to the current treatment standard (rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone [R-CHOP]).18-22 ABC type DLBCL is associated with less favorable outcomes compared with GCB DLBCL. This molecular classification adds prognostic value to the widely used International Prognostic Index (IPI) and revised IPI (R-IPI) that constitute the current gold standard for identifying patients with higher likelihood of poor prognosis.23,24 Although gene expression signatures and single gene mutation (or expression)–based prognosticators have been described, most of these molecular features, with the exception of TP53 mutation and MYC expression, are surrogates for either the R-IPI or COO subgroups.22
Using genome and transcriptome sequencing, we identified FOXO1 as a recurrent target of somatic mutation in DLBCL.16 Here, we analyze the pattern of recurrent mutations affecting the N-terminal region corresponding to the T24 phosphorylation site of FOXO1 and identify a cluster of mutations affecting the Forkhead DNA binding domain. We demonstrate an association between the presence of FOXO1 mutation and decreased overall survival (OS) in DLBCL patients, independent of COO and the R-IPI. We further demonstrate that the N-terminal hot-spot mutations diminish FOXO1 phosphorylation and result in loss of interaction with 14-3-3, which suggests aberrant nuclear localization of FOXO1.
Methods
Patients and samples
FOXO1 mutations were initially reported in Morin et al.16 Here, we examined FOXO1 mutational status in 279 DLBCL tumor samples and 22 cell lines. One hundred ninety-three patients were uniformly treated with R-CHOP and were used for survival analysis (supplemental Figure 1 on the Blood website). The median follow-up for living patients in this 193 patient cohort was 5.6 years. The study was approved by the University of British Columbia-British Columbia Cancer Agency Research Ethics Board and conducted in accordance with the Declaration of Helsinki (UBC BCCA REB# H05-60103).
Targeted exon resequencing of FOXO1 and mutation validation
Using a polymerase chain reaction approach (supplemental Methods; supplemental Table 1), the coding exons of FOXO1 were amplified in 189 de novo DLBCL patient samples and Illumina libraries generated for sequencing on an Illumina HiSeq 2000. Targeted exon resequencing was also used to reanalyze 69 DLBCL samples previously analyzed using RNA-Seq.16 Sanger sequencing of FOXO1 exons was used to confirm the somatic status of FOXO1 mutations in 5 of these patient samples in which both the tumor and germ-line DNA were available. Sanger sequencing was also used to confirm FOXO1 mutation status in the 22 DLBCL cell lines.
Statistical analysis
Group comparisons were performed using the Fisher exact test, χ2 test, and Student t test. OS was measured from the time of initial diagnosis to death from any cause. Cox proportional-hazards models and time-to-event analyses with the use of the Kaplan-Meier method were performed using SPSS software, version 14.0. Two-sided P < .05 indicated statistical significance.
Plasmid constructs
Full-length human FOXO1 (GC-F0954-CF) was purchased from GeneCoepoeia (Rockville, MD), and site-directed mutagenesis was conducted with the use of the QuikChange II Site Directed Mutagenesis kit (Stratagene, Santa Clara, CA) to convert the clone to match with the reference genome (Hg18; supplemental Table 2). FOXO1 mutants were generated (M1L, R19Q, R21C, and T24A), and the presence of the desired mutations was confirmed by DNA sequencing. Wild-type and mutant FOXO1 was then subcloned into the pcDNA-DEST47 and pLenti6.2/V5-DEST constructs (Invitrogen, Grand Island, NY) using the LR Clonase II enzyme mix (Invitrogen).
Cell culture
HEK-293 cells were maintained in Dulbecco’s modified Eagle medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen) in a 37°C incubator with a 5% CO2 humidified atmosphere. Human wild-type or mutant FOXO1 constructs were transiently transfected into HEK-293 using TurboFect (ThermoScientific, Waltham, MA) transfection reagent.
DB, DOHH2, Karpas422, Nu-DHL-1, Nu-DUL-1, Su-DHL-6, and WSU-DLCL2 are cell lines obtained from DSMZ. Pfeiffer and Toledo were obtained from ATCC, and all OCI-Ly lines (1, 3, 7, 10, and 19) were obtained from Louis Staudt (National Institutes of Health, Bethesda, MD). The cell lines MD903 and Su-DHL-9 were obtained from Martin Dyer (University of Leicester). DLBCL cells were maintained at 37°C in RPMI 1640 (Invitrogen) media supplemented with 10% (v/v) fetal bovine serum and 1% penicillin/streptomycin (Invitrogen) in a 37°C incubator with a 5% CO2 humidified atmosphere.
Recombinant human and insulin growth factor-1 (IGF-1; PeproTech, Rocky Hill, NJ) was added to cells at a final concentration of 100 ng/mL from a stock of 100 µg/mL in water.
Lentiviral transduction
Replication-defective lentivirus was produced by cotransfection of FOXO1 lentiviral constructs (pLenti6.2) with packaging plasmids (CMV DeltaR8.91 and pMD2 VSV-G envelope) into 293T cells using TransIT-LT1 transfection reagent (Mirus Bio, Madison, WI). Viral supernatant was collected 48 and 72 hours after transfection, passed through a 0.45-μm filter, and concentrated.
HEK-293 cells were transduced with the viral supernatant and Polybrene (Sigma) in their respective growth media and allowed to culture overnight. FOXO1-expressing HEK-293 stable cells were generated through selection with 5 µg/mL Blasticidin (Invitrogen).
Immunoblotting
Total cell extracts were prepared by lysis using radio-immunoprecipitation assay buffer (Santa Cruz Biotechnologies, Santa Cruz, CA) supplemented with Complete protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktails A and B (Santa Cruz Biotechnologies). Total protein was quantified with the BCA Reagent Kit (Pierce, Rockford, IL) using bovine serum albumin as a standard. Equal amounts of protein (10-20 µg) were size-fractionated by 4% to 12% Bis-Tris gels (Invitrogen), transferred to polyvinylidene difluoride (Millipore, Billerica, MA) membranes, and blocked with 5% skim milk in phosphate buffered saline with 0.01% tween 20. Membranes were probed overnight with primary antibody (supplemental Table 3) at 1:1000 dilution in 5% skim milk in phosphate buffered saline with 0.01% tween 20 and probed with their corresponding secondary antibody at 1:5000 dilution for 1 hour, and chemiluminescence was detected.
Immunoprecipitations
HEK-293 cells were transfected with wild-type or mutant FOXO1 constructs, and 48 hours following transfection, were harvested and lysed in NP40 lysis buffer supplemented with Complete protease inhibitor cocktail. Generally, 500 µg of cell lysate was immunoprecipitated with green fluorescent protein (GFP)-Trap magnetic beads (ChromoTek) or blocked magnetic beads for control for 2 hours at 4°C. Immunoprecipitates were washed extensively with phosphate-buffered saline and eluted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer for western blot analysis.
Subcellular fractionation
Nuclear and cytoplasmic extracts were prepared with an isotonic lysis buffer (150 mM NaCl, 1.5 mM MgCl2, 10 mM Tris-HCl, at pH 7.5, 0.5% NP-40; Complete protease inhibitor cocktail). Cells were harvested in ice-cold lysis buffer and centrifuged at 700g for 7 minutes at 4°C. The supernatant was further clarified by centrifugation at 13 000g for 15 minutes at 4°C, representing the cytoplasmic fraction. The pellet was suspended in ice-cold nuclear lysis buffer (250 mM NaCl, 20 mM sodium phosphate at pH 7.0, 30 mM sodium pyrophosphate, 5 mM EDTA, 10 mM NaF, 0.1% NP-40, 10% glycerol, 1 mM dithiothreitol, Complete protease inhibitor cocktail), and sonicated 3 times for 15 seconds. The supernatant was clarified by centrifugation at 13 000g for 15 minutes at 4°C, representing the nuclear fraction. Separation of the nuclear and cytoplasmic fractions was verified by blotting with the cytosolic protein β-tubulin and the nuclear protein TBP.
Results
FOXO1 mutations in DLBCL
In an exploratory RNA-Seq analysis, we identified FOXO1 mutations in a cohort of 110 NHL cases, including both DLBCL and follicular lymphoma (FL).16 A total of 10 missense mutations were found in 8 cases including 7 of 110 NHL patients and 1 of 10 DLBCL cell lines. Two codons in the FOXO1 gene were mutated in >1 patient, namely those encoding M1 (2 patients and 1 cell line) and T24 (2 patients).16 Two of the DLBCL patients had FOXO1 mutations, and despite treatment with R-CHOP, both exhibited significantly reduced OS compared with the other patients without FOXO1 mutations (data not shown). On the basis of this observation and the apparent recurrence of FOXO1 mutations, we sequenced FOXO1 exons in a larger cohort of 279 DLBCL cases to more accurately estimate FOXO1 mutation frequency and to assess the association between mutations and OS (supplemental Figure 1; supplemental Methods). The cohort contained 193 patients who received uniform treatment with R-CHOP, and these were considered in our survival analysis. We found a total of 26 mutations of FOXO1 in 24 of 279 cases including the 3 mutations in 3 cases already noted,16 plus 21 additional single nucleotide variants and 2 deletions in 21 cases (2 cases had 2 mutations; 4 additional single nucleotide variants and 1 deletion were found using targeted sequencing in the 69 overlapped RNA-Seq cases), yielding a mutation frequency in primary DLBCL cases of 8.6% (24/279; Figure 1A; Table 1). More than half (16/26 mutations) of all detected mutations affected 6 amino acids (Table 1). Nearly all mutations were found within the first exon (24/26 mutations; 92.3%) of FOXO1, which encodes the entire N-terminal region and part of the Forkhead box domain (Figure 1A). With the exception of the M1 mutation in the OCI-Ly1 cell line, which is hemizygous, the mutations in FOXO1 were heterozygous (Table 1; data not shown). The somatic status for each unique mutation was determined by sequencing the matched germ-line DNA from ≥1 case, if available (Figure 1A; Table 1). Fifty percent (13/26) of the mutations clustered near the beginning of the coding sequence. These included recurrently mutated sites affecting M1 (6/26 mutations; 23.1%), R19 (2/26 mutations; 7.7%), R21 (2/26 mutations; 7.7%), and T24 (2/26 mutations; 7.7%) (Figure 1A-B; Table 1).
Mutations in FOXO1 affecting phosphorylation sites and the DNA binding domain. (A) Mutations affecting FOXO1 in relation to the exons, protein domains, and phosphoinositide 3-kinase/AKT phosphorylation sites. Sequencing of FOXO1 revealed mainly missense mutations (green circles) with two deletions (orange triangles) located primarily in the first exon of FOXO1. A majority of the recurrent hot-spot mutations affect the consensus [RxRxxT] PKB/AKT recognition motif surrounding the T24 phosphorylation site. A cluster of mutations were also found in the Forkhead DNA binding domain. •Sites in which the mutation has been confirmed to be somatic in ≥1 patient sample. (B) Cross-species sequence conservation of the N-terminal RxRxxT site in FOXO1 proteins. *Sites of recurrent mutations in DLBCL samples. (C) Ribbon representation of the FOXO1 DBD-DNA complex (green). The sites of DBD mutations are marked with the wild-type amino acid structures (blue) and mutated amino acid structures (red). Many of the FOXO1 mutations in the Forkhead domain are found within the helical structures.
Mutations in FOXO1 affecting phosphorylation sites and the DNA binding domain. (A) Mutations affecting FOXO1 in relation to the exons, protein domains, and phosphoinositide 3-kinase/AKT phosphorylation sites. Sequencing of FOXO1 revealed mainly missense mutations (green circles) with two deletions (orange triangles) located primarily in the first exon of FOXO1. A majority of the recurrent hot-spot mutations affect the consensus [RxRxxT] PKB/AKT recognition motif surrounding the T24 phosphorylation site. A cluster of mutations were also found in the Forkhead DNA binding domain. •Sites in which the mutation has been confirmed to be somatic in ≥1 patient sample. (B) Cross-species sequence conservation of the N-terminal RxRxxT site in FOXO1 proteins. *Sites of recurrent mutations in DLBCL samples. (C) Ribbon representation of the FOXO1 DBD-DNA complex (green). The sites of DBD mutations are marked with the wild-type amino acid structures (blue) and mutated amino acid structures (red). Many of the FOXO1 mutations in the Forkhead domain are found within the helical structures.
FOXO1 mutations in DLBCL patients and cell lines
| Sample . | Coordinate . | Nucleotide change . | Amino acid substitution . | Verification result . | Zygosity . | Mutation frequency* . | Variant frequency† . |
|---|---|---|---|---|---|---|---|
| 05-30349 | chr13:40138349 | T>A | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 95-16931 | chr13:40138349 | T>C | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-20828 | chr13:40138349 | T>C | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-31713 | chr13:40138349 | T>C | M1V | Somatic | Heterozygous | 2.2% (6/279) | 23.1% (6/26) |
| 03-23488 | chr13:40138349 | T>A | M1L | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-31974 | chr13:40138349 | T>G | M1L | No germ line–validated mutation | Heterozygous | 2.2% (6/279) | 23.1% (6/26) |
| OCI-Ly1 | chr13:40138349 | T>A | M1L | Cell line–validated mutation | Hemizygous | ||
| Su-DHL-5 | chr13:40138349 | T>A | M1L | Cell line–validated mutation | Heterozygous | ||
| 95-12860 | chr13:40138279 | G>A | T24I | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| 05-17793 | chr13:40138280 | T>C | T24A | Somatic | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| 06-19659 | chr13:40138288 | C>A | R21L | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| 01-15178 | chr13:40138288 | C>G | R21P | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| Nu-DUL-1 | chr13:40138288 | C>G | R21P | Cell line–validated mutation | Heterozygous | . | |
| Karpas422 | chr13:40138289 | G>A | R21C | Cell line–validated mutation | Heterozygous | ||
| 04-11156 | chr13:40138294 | C>G | R19P | No germ line–validated mutation | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| 06-18547 | chr13:40138295 | G>A | R19W | Somatic | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| DoHH2 | chr13:40138295 | G>A | R19W | Cell line–validated mutation | Heterozygous | ||
| 06-27347 | chr13:40137853 | G>A | A166V | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 00-18839 | chr13:40137853 | G>A | A166V | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 02-16987 | chr13:40137853 | G>C | A166G | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 75-50777 | chr13:40137736 | C>T | S205N | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| Su-DHL-5 | chr13:40137736 | C>G | S205T | Cell line–validated mutation | Heterozygous | ||
| OCI-Ly1 | chr13:40138322 | T>C | I10V | Cell line–validated mutation | Hemizygous | ||
| 06-28900 | chr13:40138312 | deletion | D11 | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 06-23907 | chr13:40138068 | deletion | V94 (-VAAA) | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 05-17793 | chr13:40137894 | G>T | S152R | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 99-25549 | chr13:40137891 | G>C | S153R | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 95-11015 | chr13:40137848 | G>C | L168V | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 03-23488 | chr13:40137839 | T>C | K171E | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 05-18695 | chr13:40137802 | A>G | L183P | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 04-11650 | chr13:40137793 | A>G | I186T | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 05-30239 | chr13:40032490 | G>A | L380F | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 03-25415 | chr13:40032313 | T>C | I439V | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 8.6% (24/279) | 9.3% (26/279) | 100.00% |
| Sample . | Coordinate . | Nucleotide change . | Amino acid substitution . | Verification result . | Zygosity . | Mutation frequency* . | Variant frequency† . |
|---|---|---|---|---|---|---|---|
| 05-30349 | chr13:40138349 | T>A | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 95-16931 | chr13:40138349 | T>C | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-20828 | chr13:40138349 | T>C | M1V | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-31713 | chr13:40138349 | T>C | M1V | Somatic | Heterozygous | 2.2% (6/279) | 23.1% (6/26) |
| 03-23488 | chr13:40138349 | T>A | M1L | Heterozygous | 2.2% (6/279) | 23.1% (6/26) | |
| 03-31974 | chr13:40138349 | T>G | M1L | No germ line–validated mutation | Heterozygous | 2.2% (6/279) | 23.1% (6/26) |
| OCI-Ly1 | chr13:40138349 | T>A | M1L | Cell line–validated mutation | Hemizygous | ||
| Su-DHL-5 | chr13:40138349 | T>A | M1L | Cell line–validated mutation | Heterozygous | ||
| 95-12860 | chr13:40138279 | G>A | T24I | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| 05-17793 | chr13:40138280 | T>C | T24A | Somatic | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| 06-19659 | chr13:40138288 | C>A | R21L | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| 01-15178 | chr13:40138288 | C>G | R21P | Heterozygous | 0.72% (2/279) | 7.7% (2/26) | |
| Nu-DUL-1 | chr13:40138288 | C>G | R21P | Cell line–validated mutation | Heterozygous | . | |
| Karpas422 | chr13:40138289 | G>A | R21C | Cell line–validated mutation | Heterozygous | ||
| 04-11156 | chr13:40138294 | C>G | R19P | No germ line–validated mutation | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| 06-18547 | chr13:40138295 | G>A | R19W | Somatic | Heterozygous | 0.72% (2/279) | 7.7% (2/26) |
| DoHH2 | chr13:40138295 | G>A | R19W | Cell line–validated mutation | Heterozygous | ||
| 06-27347 | chr13:40137853 | G>A | A166V | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 00-18839 | chr13:40137853 | G>A | A166V | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 02-16987 | chr13:40137853 | G>C | A166G | Heterozygous | 1.1% (3/279) | 11.5% (3/26) | |
| 75-50777 | chr13:40137736 | C>T | S205N | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| Su-DHL-5 | chr13:40137736 | C>G | S205T | Cell line–validated mutation | Heterozygous | ||
| OCI-Ly1 | chr13:40138322 | T>C | I10V | Cell line–validated mutation | Hemizygous | ||
| 06-28900 | chr13:40138312 | deletion | D11 | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 06-23907 | chr13:40138068 | deletion | V94 (-VAAA) | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 05-17793 | chr13:40137894 | G>T | S152R | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 99-25549 | chr13:40137891 | G>C | S153R | Somatic | Heterozygous | 0.36% (1/279) | 3.8% (1/26) |
| 95-11015 | chr13:40137848 | G>C | L168V | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 03-23488 | chr13:40137839 | T>C | K171E | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 05-18695 | chr13:40137802 | A>G | L183P | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 04-11650 | chr13:40137793 | A>G | I186T | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 05-30239 | chr13:40032490 | G>A | L380F | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 03-25415 | chr13:40032313 | T>C | I439V | Heterozygous | 0.36% (1/279) | 3.8% (1/26) | |
| 8.6% (24/279) | 9.3% (26/279) | 100.00% |
Mutation frequency is the percentage of mutations calculated relative to all DLBCL patient samples.
Variant frequency, the percentage of mutations affecting that specific amino acid relative to all other mutations, is calculated based on the mutations found in DLBCL patients samples.
We observed a second cluster of mutations affecting the region of the gene encoding the Forkhead domain (Figure 1A,C; Table 1). The Forkhead domain is a 110 amino acid winged helix DNA binding domain that folds into a 3-dimensional structure consisting of 3 α-helices (H1; H2; and H3), 3 β-strands (S1; S2; and S3), and 2 wing-like loops (W1 and W2) arranged in the order of H1-S1-H2-H3-S2-W1-S3-W2.25 The 5 amino acids N-terminal to helix H3 are unique to FOXO proteins and may lie in a region that specifies the regulation of a distinct set of target genes.26 A majority of FOXO1 mutations in the Forkhead domain are located in the helical structures, with a few mutations in these 5 amino acids (Figure 1C). Two recurrently mutated sites were found in the Forkhead box domain: A166 (3/26 mutations; 11.5%) and S205 (1/26 mutations; 3.8%; also observed in 1 DLBCL cell line and 2 FL samples; Figure 1; Table 1; data not shown).
FOXO1 mutations were also observed in 5 of 22 DLBCL cell lines. We found a hemizygous M1L mutation in the DLBCL cell line OCI-Ly1, a heterozygous M1L mutation in Su-DHL-5, and heterozygous R19W, R21C, R21P, and S205T mutations in the DoHH2, Karpas422, Nu-DUL-1, and Su-DHL-5 cell lines, respectively (Table 1; supplemental Figure 2).
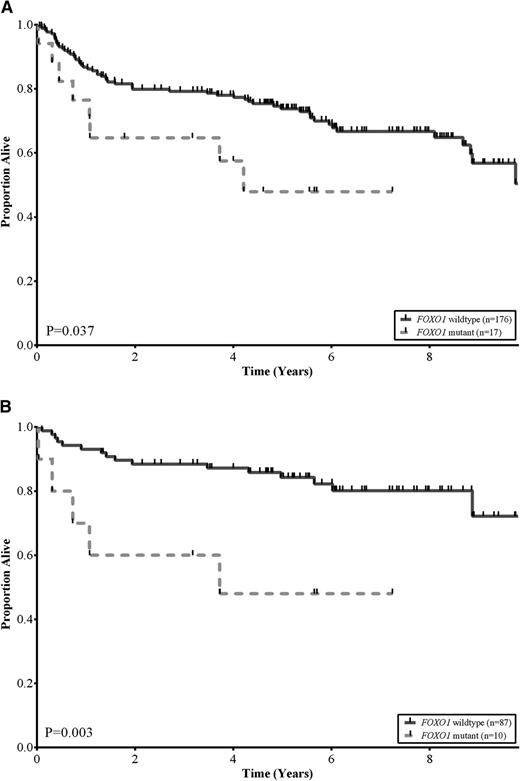
No statistically significant differences were observed among the baseline clinical characteristics of patients with and without FOXO1 mutations, including COO subtype (P = .42) and R-IPI categories (P = .84; Table 2). This was also the case when these comparisons were restricted to those patients treated with R-CHOP (supplemental Table 4). We assessed the association between FOXO1 mutation status and OS in the 193 DLBCL patients that had been treated with R-CHOP. Patients with tumors that harbored FOXO1 mutations had a significantly inferior OS compared with patients with wild-type FOXO1 (log-rank, P = .037; Figure 2A). The estimated 5-year OS of patients with FOXO1 mutations was 48% compared with 74% for patients with no mutations detected. In multivariate analyses, using outcomes from the 125 patients where all variables were available, the FOXO1 mutational status was associated with inferior OS independent of both the COO and R-IPI categories (supplemental Table 5). Interestingly, the differences in OS were particularly evident in patients in the low-risk R-IPI categories (very good and good; IPI scores 0-2). In this group, the estimated 5-year OS of patients with FOXO1 mutations was 48% compared with 84% for patients with no mutations detected (log-rank, P = .003; Figure 2B).
Clinical characteristics of the 279 patients with de novo DLBCL
| Demographic or clinical characteristic . | FOXO1 mutated (24 patients) . | FOXO1 wild type(255 patients) . | P* . |
|---|---|---|---|
| Male, % | 67 | 64 | 0.78 |
| Age, median (range) | 66 (45-90) | 64 (16-92) | 0.27 |
| Stage, n (%) | |||
| I/II | 10 (45) | 115 (48) | 0.84 |
| III/IV | 12 (55) | 126 (52) | 0.84 |
| NA | 2 | 14 | 0.84 |
| Lactate dehydrogenase > ULN, n (%) | |||
| No | 7 (37) | 104 (48) | 0.34 |
| Yes | 12 (63) | 112 (52) | 0.34 |
| NA | 5 | 39 | 0.34 |
| ECOG performance status, n (%) | |||
| 0-1 | 13 (62) | 170 (71) | 0.37 |
| ≥2 | 8 (38) | 69 (29) | 0.37 |
| NA | 3 | 16 | 0.37 |
| Extranodal sites, n (%) | |||
| 0-1 | 20 (95) | 187 (82) | 0.14 |
| >1 | 1 (5) | 42 (18) | 0.14 |
| NA | 3 | 26 | 0.14 |
| R-IPI,†n (%) | |||
| Very good and good (0-2) | 12 (63) | 132 (61) | 0.84 |
| Poor (3-5) | 7 (37) | 85 (39) | 0.84 |
| NA | 5 | 38 | 0.84 |
| COO,‡n (%) | |||
| GCB | 11 (58) | 87 (48) | 0.42 |
| Non-GCB | 8 (42) | 94 (52) | 0.42 |
| NA | 5 | 74 | 0.42 |
| BCL2 FISH break-apart,§n (%) | |||
| Positive | 4 (33) | 43 (35) | 1.0 |
| Negative | 8 (67) | 81 (65) | 1.0 |
| NA | 12 | 131 | 1.0 |
| BCL6 FISH break-apart,§n (%) | |||
| Positive | 1 (8) | 26 (21) | 0.46 |
| Negative | 11 (92) | 95 (79) | 0.46 |
| NA | 12 | 134 | 0.46 |
| MYC FISH break-apart,§n (%) | |||
| Positive | 2 (18) | 9 (7) | 0.22 |
| Negative | 9 (82) | 115 (93) | 0.22 |
| NA | 13 | 131 | 0.22 |
| B symptoms, n (%) | |||
| Absent | 14 (67) | 157 (66) | 0.92 |
| Present | 7 (33) | 82 (37) | 0.92 |
| NA | 3 | 16 | 0.92 |
| Demographic or clinical characteristic . | FOXO1 mutated (24 patients) . | FOXO1 wild type(255 patients) . | P* . |
|---|---|---|---|
| Male, % | 67 | 64 | 0.78 |
| Age, median (range) | 66 (45-90) | 64 (16-92) | 0.27 |
| Stage, n (%) | |||
| I/II | 10 (45) | 115 (48) | 0.84 |
| III/IV | 12 (55) | 126 (52) | 0.84 |
| NA | 2 | 14 | 0.84 |
| Lactate dehydrogenase > ULN, n (%) | |||
| No | 7 (37) | 104 (48) | 0.34 |
| Yes | 12 (63) | 112 (52) | 0.34 |
| NA | 5 | 39 | 0.34 |
| ECOG performance status, n (%) | |||
| 0-1 | 13 (62) | 170 (71) | 0.37 |
| ≥2 | 8 (38) | 69 (29) | 0.37 |
| NA | 3 | 16 | 0.37 |
| Extranodal sites, n (%) | |||
| 0-1 | 20 (95) | 187 (82) | 0.14 |
| >1 | 1 (5) | 42 (18) | 0.14 |
| NA | 3 | 26 | 0.14 |
| R-IPI,†n (%) | |||
| Very good and good (0-2) | 12 (63) | 132 (61) | 0.84 |
| Poor (3-5) | 7 (37) | 85 (39) | 0.84 |
| NA | 5 | 38 | 0.84 |
| COO,‡n (%) | |||
| GCB | 11 (58) | 87 (48) | 0.42 |
| Non-GCB | 8 (42) | 94 (52) | 0.42 |
| NA | 5 | 74 | 0.42 |
| BCL2 FISH break-apart,§n (%) | |||
| Positive | 4 (33) | 43 (35) | 1.0 |
| Negative | 8 (67) | 81 (65) | 1.0 |
| NA | 12 | 131 | 1.0 |
| BCL6 FISH break-apart,§n (%) | |||
| Positive | 1 (8) | 26 (21) | 0.46 |
| Negative | 11 (92) | 95 (79) | 0.46 |
| NA | 12 | 134 | 0.46 |
| MYC FISH break-apart,§n (%) | |||
| Positive | 2 (18) | 9 (7) | 0.22 |
| Negative | 9 (82) | 115 (93) | 0.22 |
| NA | 13 | 131 | 0.22 |
| B symptoms, n (%) | |||
| Absent | 14 (67) | 157 (66) | 0.92 |
| Present | 7 (33) | 82 (37) | 0.92 |
| NA | 3 | 16 | 0.92 |
ECOG, Eastern Cooperative Oncology Group; GCB, germinal center B-cell like; NA, not available; ULN, upper limit of normal.
P values are for comparisons between the FOXO1 mutated and wild-type groups excluding patients where the variable is not available.
COO was determined by gene expression profiling in 140 cases and by immunochemistry using the Hans Classifier in 60 cases.18-20
The presence of translocations was determined using commercial dual color “break-apart” probes from Abbott Molecular on a tissue microarray using the method described in Chin et al.41
FOXO1 mutations correlate with reduced survival in DLBCL. (A) Kaplan-Meier curves of OS in a cohort of 193 de novo DLBCL patients uniformly treated with R-CHOP, comparing FOXO1 wild-type (solid black) vs FOXO1 mutant (dotted grey) subgroups. Patients with a mutation in FOXO1 had lower OS rates relative to patients without a mutation (log-rank, P = .037). (B) Kaplan-Meier curves of OS in a cohort of 97 low-risk R-IPI categories (very good and good; IPI scores 0-2) DLBCL patients uniformly treated with R-CHOP, comparing FOXO1 wild-type (solid black) vs FOXO1 mutant (dotted grey) subgroups. Patients with a mutation in FOXO1 had lower OS rates relative to patients without a mutation (log-rank, P = .003).
FOXO1 mutations correlate with reduced survival in DLBCL. (A) Kaplan-Meier curves of OS in a cohort of 193 de novo DLBCL patients uniformly treated with R-CHOP, comparing FOXO1 wild-type (solid black) vs FOXO1 mutant (dotted grey) subgroups. Patients with a mutation in FOXO1 had lower OS rates relative to patients without a mutation (log-rank, P = .037). (B) Kaplan-Meier curves of OS in a cohort of 97 low-risk R-IPI categories (very good and good; IPI scores 0-2) DLBCL patients uniformly treated with R-CHOP, comparing FOXO1 wild-type (solid black) vs FOXO1 mutant (dotted grey) subgroups. Patients with a mutation in FOXO1 had lower OS rates relative to patients without a mutation (log-rank, P = .003).
Effect of FOXO1 mutations on phosphorylation and protein interactions
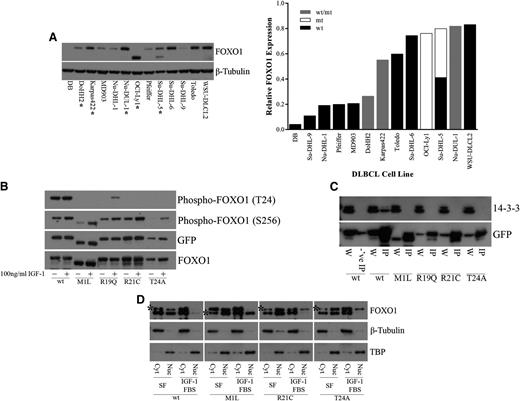
In the nucleus, FOXO1 functions as a transcription factor to regulate genes involved in differentiation, apoptosis, cell cycle regulation, DNA damage repair, oxidative stress, and glucose metabolism.1-5 Regulation of FOXO1 activity results from a balance between signaling events that regulate posttranslational modifications and the subcellular distribution of FOXO1 proteins.27-34 Inactivation of FOXO1 occurs through the relocalization of FOXO1 to the cytoplasm.31 FOXO1 phosphorylation by AKT can occur at three different sites, T24, S256, and S319, owing to the AKT recognition motif (RxRxxS/T).32 R19, R21, and T24, all of which were observed to be mutated in DLBCL samples, correspond to the 3 fixed positions of the AKT recognition motif at the N-terminal FOXO1 phosphorylation site (Figure 1B). We found that another mutated site, M1, also affects this N-terminal phosphorylation site. The M1 mutation results in a shift of the transcription initiation site to the next in-frame methionine, located 70 amino acids downstream. This results in a shortened FOXO1 protein that lacks the T24 residue. This mutation is found in the OCI-Ly1 and Su-DHL-5 cell lines (Figure 3A). The presence of mutations did not appear to alter the expression of FOXO1 at the protein level, which varied among the different DLBCL cell lines (Figure 3A) or at the RNA level in DLBCL patient samples and DLBCL cell lines for which RNA-Seq was performed (supplemental Figure 3).
Recurrent FOXO1 mutations alter phosphoinositide 3-kinase/AKT phosphorylation and protein-protein interactions. (A) Whole cell extracts from DLBCL cells were resolved on 4% to 12% Bis-Tris gels, following by immunoblotting (left) with anti-FOXO1 antibody. Total protein level was assayed by immunoblot using anti–β-tubulin antibody. *Mutant FOXO1 cell lines. The smaller molecular weight band in the OCI-Ly1 and Su-DHL-5 cell lines, hemizygous and heterozygous for the M1 mutation, respectively, indicates a shift in translation initiation to the next methionine. Relative FOXO1 protein expression (normalized to β-tubulin) is displayed in increasing expression on the right. Where possible, the expression from the individual alleles was determined; (wt/mt) indicates an unknown proportion of expression from the wild-type and mutant protein. (B) HEK-293 cells were transiently transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with GFP. Twenty-four hours after transfection, cells were serum starved for 24 hours and treated with 10% fetal bovine serum and 100 ng/mL recombinant IGF-1 for 1 hour. Whole cell extracts were resolved on 4% to 12% Bis-Tris gels and immunoblotted with phospho-specific antibodies directed against FOXO1. The total level of tagged protein was assayed by immunoblot using the anti-GFP and anti-FOXO1 antibodies. A reduction in phosphorylation at T24 was observed in the N-terminal mutants but not in the wild type. (C) HEK-293 cells were transiently transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with GFP. Whole cell extracts were immunoprecipitated with GFP-Trap, resolved on 4% to 12% Bis-Tris gels, followed by immunoblotting with anti-GFP antibody. Blocked magnetic beads were used as a negative control. A loss of interaction with 14-3-3 was observed with the N-terminal mutants but not in the wild type. W, whole cell extract; IP, immunoprecipitation; -‘ve IP, negative control IP. (D) HEK-293 cells were stably transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with V5. Cells were serum starved for 24 hours and treated with 10% fetal bovine serum and 100 ng/mL recombinant IGF-1 for 1 hour. Biochemical fractionation was performed, and cytoplasmic and nuclear fractions were resolved on 4% to 12% Bis-Tris gels and immunoblotted with anti-FOXO1 antibody. *V5-tagged FOXO1. Note: wt-V5, R21C-V5, and T24A-V5 are the upper bands, and M1L-V5 is the lower band. Purity of the cytoplasmic and nuclear fractions was assayed by immunoblot using anti–β-tubulin and anti-TBP antibodies, respectively. An inability to be exported from the nucleus to the cytoplasm following IGF-1 treatment was observed in our mutants but not for wild-type FOXO1. Cyt, cytoplasmic fraction; Nuc, nuclear fraction; SF, serum-free media.
Recurrent FOXO1 mutations alter phosphoinositide 3-kinase/AKT phosphorylation and protein-protein interactions. (A) Whole cell extracts from DLBCL cells were resolved on 4% to 12% Bis-Tris gels, following by immunoblotting (left) with anti-FOXO1 antibody. Total protein level was assayed by immunoblot using anti–β-tubulin antibody. *Mutant FOXO1 cell lines. The smaller molecular weight band in the OCI-Ly1 and Su-DHL-5 cell lines, hemizygous and heterozygous for the M1 mutation, respectively, indicates a shift in translation initiation to the next methionine. Relative FOXO1 protein expression (normalized to β-tubulin) is displayed in increasing expression on the right. Where possible, the expression from the individual alleles was determined; (wt/mt) indicates an unknown proportion of expression from the wild-type and mutant protein. (B) HEK-293 cells were transiently transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with GFP. Twenty-four hours after transfection, cells were serum starved for 24 hours and treated with 10% fetal bovine serum and 100 ng/mL recombinant IGF-1 for 1 hour. Whole cell extracts were resolved on 4% to 12% Bis-Tris gels and immunoblotted with phospho-specific antibodies directed against FOXO1. The total level of tagged protein was assayed by immunoblot using the anti-GFP and anti-FOXO1 antibodies. A reduction in phosphorylation at T24 was observed in the N-terminal mutants but not in the wild type. (C) HEK-293 cells were transiently transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with GFP. Whole cell extracts were immunoprecipitated with GFP-Trap, resolved on 4% to 12% Bis-Tris gels, followed by immunoblotting with anti-GFP antibody. Blocked magnetic beads were used as a negative control. A loss of interaction with 14-3-3 was observed with the N-terminal mutants but not in the wild type. W, whole cell extract; IP, immunoprecipitation; -‘ve IP, negative control IP. (D) HEK-293 cells were stably transfected with constructs encoding FOXO1 (wild type or mutants) C-terminally tagged with V5. Cells were serum starved for 24 hours and treated with 10% fetal bovine serum and 100 ng/mL recombinant IGF-1 for 1 hour. Biochemical fractionation was performed, and cytoplasmic and nuclear fractions were resolved on 4% to 12% Bis-Tris gels and immunoblotted with anti-FOXO1 antibody. *V5-tagged FOXO1. Note: wt-V5, R21C-V5, and T24A-V5 are the upper bands, and M1L-V5 is the lower band. Purity of the cytoplasmic and nuclear fractions was assayed by immunoblot using anti–β-tubulin and anti-TBP antibodies, respectively. An inability to be exported from the nucleus to the cytoplasm following IGF-1 treatment was observed in our mutants but not for wild-type FOXO1. Cyt, cytoplasmic fraction; Nuc, nuclear fraction; SF, serum-free media.
To investigate the effects of recurrent mutations on FOXO1 function, we created C-terminal GFP- or V5-tagged wild-type and mutant constructs of FOXO1 (“Methods”). On the basis of the prevalence of mutations in our cohort, we constructed clones representing each of M1L, R19Q, R21C, and T24A. Insulin receptor and IGF-1 receptor signaling plays a major role in activating phosphoinositide 3-kinase/AKT signaling, which can induce phosphorylation of FOXO1.28,29,32 Therefore, we tested whether these mutations altered phosphorylation at the N-terminal T24 and at the Forkhead S256 phosphorylation sites following treatment with IGF-1 in FOXO1-transfected HEK-293 cells. Mutations in M1, R19, R21, and T24 appeared to greatly diminish phosphorylation at T24 with no effect on phosphorylation at S256 (Figure 3B). Phosphorylation of T24 by AKT generates a recognition motif for the binding of the 14-3-3 protein.27,28,30,31 Thus, we assessed whether interaction with 14-3-3 was affected by this reduction or loss in phosphorylation at T24. Immunoprecipitation of FOXO1-transfected HEK-293 cells demonstrated a consistent loss of interaction between each of the tested FOXO1 mutants and 14-3-3 (Figure 3C). Nuclear to cytoplasmic shuttling of FOXO1 requires both the N-terminal AKT and the Forkhead AKT binding sites for 14-3-3 binding and subsequent dimerization.27,28,31 Binding alters access to the nuclear localization sequence and induces subsequent nuclear export and cytoplasmic sequestration of FOXO1.27,30,33,34 We chose to stably express our FOXO1 constructs to obtain a level of expression of FOXO1 similar to that of endogenous FOXO1 in HEK-293 cells to prevent dominant negative inhibition of PKB/AKT.28 Cellular fractionation of FOXO1 stably transfected HEK-293 cells demonstrated a shift of both endogenous FOXO1 and wild-type tagged FOXO1 from the nucleus to the cytoplasm; however, the M1, R21, and T24 tagged mutants remained in the nucleus despite IGF-1 treatment (Figure 3D).
Discussion
FOXO1 was one of 109 genes found to be a recurrent target of somatic mutation in a cohort of B-cell NHLs enriched for DLBCL cases.16 To explore the prevalence of FOXO1 mutations in DLBCL and to assess the impact of FOXO1 mutations on OS, we sequenced the coding exons of FOXO1 in a cohort of DLBCL patients and cell lines. The overall prevalence of FOXO1 mutations in DLBCL was 8.6% (24/279 cases), and half (16/26 mutations) of all detected mutations affected 6 amino acids. Four of these recurrently mutated positions (M1 [6/26], R19 [2/26], R21 [2/26], and T24 [2/26]) were in the N-terminal region. Two others (A166 [3/26] and S205 [1/26; also found in 1 DLBCL cell line and 2 FL samples]), were in the Forkhead DNA binding domain. FOXO1’s function as a transcription factor is dependent on its nuclear localization and the function of the Forkhead box DNA binding domain, which mediates protein-DNA contacts primarily in α-helix H3 and the first and second loops.3,25 The ability of FOXO1 to regulate transcription may be altered by the presence of a cluster of mutations in this latter domain.
To our knowledge, the presence of FOXO1 point mutations has only been previously reported in DLBCL by our group16 ; however, chromosomal rearrangements involving FOXOs have been reported to generate fusion proteins in other cancers. Recurrent FOXO1 fusion proteins with PAX3 and PAX7 have been reported in alveolar rhabdomyosarcomas as a result of the chromosomal translocations t(2;13)(q35;q14) and t(1;13)(p36;q14), resulting in the fusion of the DNA binding domain of PAX proteins to the transactivation domain of FOXO1.35 In addition, FOXO3-MLL and FOXO4-MLL fusion proteins have been reported in acute leukemias as a result of the chromosomal translocations t(6;11)(q21;q23) and t(X;11)(q21;q23), respectively.15,36
The expression of FOXO1 protein in DLBCL cell lines was found to be variable in our western blots (Figure 3A), and no visible difference in expression was observed in cell lines possessing a mutation in FOXO1 compared with cell lines that were wild type for FOXO1. This is consistent with the analysis reported by Xie et al,7 which also demonstrated variable expression levels of FOXO1 that were comparable to normal B cells.
Mutations in the N-terminal region of FOXO1 results in the loss of phosphorylation of the consensus RxRxxT phosphorylation site, likely through complete ablation of the site or loss of recognition of the consensus motif.30,32 Mutation of T24 in FOXO1 has previously been shown to ablate a single phosphorylation site, resulting in increased nuclear localization and transcriptional activity.34 Phosphorylation alone, however, is unable to alter the state of the protein. This requires the association of another protein induced by phosphorylation: often a role occupied by 14-3-3, which binds to the T24 phosphorylation site.27,28,30,31 The binding of 14-3-3 to FOXO1 may result in the masking or exposing of target sequences, altering cellular localization and sequestration of the FOXO1 protein.27,30,33,34 The loss of interaction of FOXO members with 14-3-3 has been previously observed with the introduction of a mutation of the N-terminal AKT phosphorylation and 14-3-3 recognition site in both DAF-16 (T54)37 and FOXO3 (T32).27 Nuclear retention of FOXO1 as a result of mutation at T24 has previously been observed with HA-tagged and myc epitope-tagged FOXO1 constructs.29,34 Our observation of diminished phosphorylation of T24 and loss of interaction with 14-3-3 (Figure 3) in our recurrent N-terminal FOXO1 mutants (M1L, R19Q, R21C, and T24A) suggests altered mechanisms regulating cellular localization and persistence of mutant FOXO1 in the nucleus, where it may remain transcriptionally active.
Phosphorylated FOXO1 has been demonstrated to be a prognostic factor for survival in patients with soft tissue sarcoma38 and gastric cancer,39 and FOXO1 expression correlates with longer disease-free survival in breast cancer.40 Our study presents a novel finding linking mutations in FOXO1 to decreased OS in DLBCL patients uniformly treated with R-CHOP independent of COO or R-IPI. The independent relationship of mutations in FOXO1 to survival, transcending the prognostic influence of the R-IPI and COO, indicates that FOXO1 mutations play a critical role in DLBCL pathogenesis. Hence, determining FOXO1 mutation status may serve as a novel prognostic indicator in patients treated with R-CHOP and with otherwise good prognosis, allowing for improved risk stratification and identification of patients for which alternative treatment options should be explored.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr David Huntsman for helpful discussions during the preparation of the manuscript; A. Troussard, S. Chittaranjan, S. Chan, E. Lim, R. Huff, and J. Pon for constructive feedback and assistance in the laboratory; the Library Construction, Biospecimen, Sequencing, and Bioinformatics teams at Canada’s Michael Smith Genome Sciences Centre for expert technical assistance; Suman Singh for assistance with the British Columbia (BC) Cancer Agency Lymphoid Cancer Database; and all the patients and their families for their support of this study.
R.D.M. and J.M.C. are supported by the BC Cancer Foundation. D.W.S. is supported by a Postdoctoral Fellowship from the Canadian Institutes of Health Research. M.M.-L. is supported by a Michael Smith Foundation for Health Research (MSFHR) research trainee award. C.S. is supported by a Career Investigator Award from the MSFHR. S.J.M.J. is a Senior Scholar of the MSFHR. M.A.M. is University of British Columbia Canada Research Chair in Genome Science. The BC Cancer Agency Genome Sciences Centre gratefully acknowledges funding support from the BC Cancer Foundation, Genome Canada, Genome British Columbia, the Cancer Research Society, and the Leukemia and Lymphoma Society of Canada. The research reported here was funded in part by The Terry Fox Foundation (grant 019001). This project has been funded in whole or in part with Federal Funds from the National Cancer Institute, National Institutes of Health, under contract NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Authorship
Contribution: D.L.T., D.W.S., R.D.M, and M.A.M. designed and performed the research, analyzed and interpreted data, and wrote the paper; M.M.-L. helped perform functional experiments and analyzed and interpreted data; J.A. and S.J.M.J. produced Figure 1C; A.J.M. and Y.Z. performed high-throughput sequencing; J.S. provided DNA from patient samples; and C.S., J.M.C., and R.D.G. participated in the original design of the project, reviewed the manuscript, and provided editorial input.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marco Marra, Genome Sciences Centre, BC Cancer Agency, 675 West 10th Avenue, Vancouver, BC, Canada V5Z 1L3; e-mail: mmarra@bcgsc.ca.
References
Author notes
D.W.S. and R.D.M. contributed equally to this work.

![Figure 1. Mutations in FOXO1 affecting phosphorylation sites and the DNA binding domain. (A) Mutations affecting FOXO1 in relation to the exons, protein domains, and phosphoinositide 3-kinase/AKT phosphorylation sites. Sequencing of FOXO1 revealed mainly missense mutations (green circles) with two deletions (orange triangles) located primarily in the first exon of FOXO1. A majority of the recurrent hot-spot mutations affect the consensus [RxRxxT] PKB/AKT recognition motif surrounding the T24 phosphorylation site. A cluster of mutations were also found in the Forkhead DNA binding domain. •Sites in which the mutation has been confirmed to be somatic in ≥1 patient sample. (B) Cross-species sequence conservation of the N-terminal RxRxxT site in FOXO1 proteins. *Sites of recurrent mutations in DLBCL samples. (C) Ribbon representation of the FOXO1 DBD-DNA complex (green). The sites of DBD mutations are marked with the wild-type amino acid structures (blue) and mutated amino acid structures (red). Many of the FOXO1 mutations in the Forkhead domain are found within the helical structures.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/18/10.1182_blood-2013-01-479865/4/m_3666f1.jpeg?Expires=1769092852&Signature=YQ9GxlByYPCKgYeSgA4Eq1YGd91Gpmbb53mj37tjJGddafl42n00rgwbRFWOobnTzriUYMVh6Z1Vz8ky2s5S7p8yVREmdZBJeIdThcQ7UIIBh8evZeU3AJxg3MIwo7GlZZigp5NYogUvT~r6srbLUSTppEVKsbI6A7Kb~QgNu72tjmpyBqiWbJ5c93nV8ZEmtMIpzY1qa6QaFVpp7MmjS3Xy-ri7Q9Kti~jyhxqSIYRNE61y13uhb4zia2sG8pfcJMz6fU4-pYoRsKilaCyKG9qz2eRos5M3JcWaioHDI0L6MUlbWuoJjg19okYNYlXa-E1sQ208qkkGbYgTlqQOhA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal