Abstract
Precursor mRNA splicing is catalyzed by the spliceosome, a macromolecule composed of small nuclear RNAs associated with proteins. The SF3B1 gene encodes subunit 1 of the splicing factor 3b, which is important for anchoring the spliceosome to precursor mRNA. In 2011, whole-exome sequencing studies showed recurrent somatic mutations of SF3B1 and other genes of the RNA splicing machinery in patients with myelodysplastic syndrome or myelodysplastic/myeloproliferative neoplasm. SF3B1 mutations had a particularly high frequency among conditions characterized by ring sideroblasts, which is consistent with a causal relationship. SF3B1 mutants were also detected at a lower frequency in a variety of other tumor types. In chronic lymphocytic leukemia, SF3B1 was found to be the second most frequently mutated gene. In myelodysplastic syndromes, SF3B1 mutations appear to be founding genetic lesions and are associated with a low risk of leukemic evolution. In contrast, SF3B1 mutations have a lower incidence in early stages of chronic lymphocytic leukemia, are more common in advanced disease, and tend to be associated with poor prognosis, suggesting that they occur during clonal evolution of the disease. The assessment of SF3B1 mutation status may become innovative diagnostic and prognostic tools and the availability of spliceosome modulators opens novel therapeutic prospects.
Introduction
Until recently, the molecular basis of myelodysplastic syndromes (MDS) has remained essentially elusive. In the last few years, however, there have been major advances in this field and it has become apparent that somatic mutations of genes encoding core components of the RNA splicing machinery play a major role in the pathophysiology of MDS and other hematologic malignancies.1-4 In this perspective article, we examine the biologic and clinical significance of mutations in SF3B1, a gene that encodes subunit 1 of the splicing factor 3b protein complex, in myeloid and lymphoid neoplasms.
Basic concepts of RNA splicing
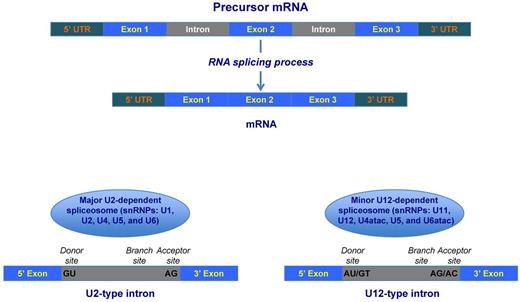
Most eukaryotic genes have coding sequences (exon) and noncoding (intron) sequences and are initially expressed as precursor messenger RNA (pre-mRNA) that includes all of these sequences (Figure 1).5 The term mRNA splicing or maturation defines the process by which introns are excised from pre-mRNA and flanking exons are ligated together.5 In higher eukaryotes, most genes undergo different patterns of splicing, generating alternative mRNA isoforms that have different functions and contribute to biologic complexity.
Schematic representation of precursor mRNA (pre-mRNA) splicing. Top: Human genes have coding (exon) and noncoding (intron) sequences and are initially expressed as pre-mRNA that includes all of these sequences. In the process of precursor mRNA splicing or maturation, introns are excised and flanking exons are joined into the mature sequence of RNA. Bottom: Excision and ligation processes of pre-mRNA splicing are catalyzed by the spliceosome, a macromolecular structure composed of 5 snRNAs associated with proteins to form particles termed snRNPs. Each U1, U2, and U5 snRNP contains a single snRNA and several proteins, whereas U4 and U6 snRNPs contains 2 snRNAs and several proteins. In most eukaryotes, there are 2 classes of introns: the common U2 type (representing more than 99% of human introns) and the rare U12 type; these 2 types differ in their consensus splice-site sequences (indicated in black at the 5′ and 3′ end of the intron). The removal of these 2 types of introns is catalyzed by 2 different spliceosomes, called U2- and U12-dependent spliceosomes, respectively. The 2 spliceosomes differ in their snRNA composition, whereas most of the proteins are shared. The assembly of the spliceosome occurs anew on each pre-mRNA, which contains specific sequence elements that drive this process. The crucial signal sequences are the splice donor site (5′ end of the intron), the branch site (near the 3′ end), and the splice acceptor site (3′ end). In the major U2-dependent spliceosome, the 5′ donor site is recognized by the U1 snRNP, whereas the branch site is recognized by the U2 snRNP.
Schematic representation of precursor mRNA (pre-mRNA) splicing. Top: Human genes have coding (exon) and noncoding (intron) sequences and are initially expressed as pre-mRNA that includes all of these sequences. In the process of precursor mRNA splicing or maturation, introns are excised and flanking exons are joined into the mature sequence of RNA. Bottom: Excision and ligation processes of pre-mRNA splicing are catalyzed by the spliceosome, a macromolecular structure composed of 5 snRNAs associated with proteins to form particles termed snRNPs. Each U1, U2, and U5 snRNP contains a single snRNA and several proteins, whereas U4 and U6 snRNPs contains 2 snRNAs and several proteins. In most eukaryotes, there are 2 classes of introns: the common U2 type (representing more than 99% of human introns) and the rare U12 type; these 2 types differ in their consensus splice-site sequences (indicated in black at the 5′ and 3′ end of the intron). The removal of these 2 types of introns is catalyzed by 2 different spliceosomes, called U2- and U12-dependent spliceosomes, respectively. The 2 spliceosomes differ in their snRNA composition, whereas most of the proteins are shared. The assembly of the spliceosome occurs anew on each pre-mRNA, which contains specific sequence elements that drive this process. The crucial signal sequences are the splice donor site (5′ end of the intron), the branch site (near the 3′ end), and the splice acceptor site (3′ end). In the major U2-dependent spliceosome, the 5′ donor site is recognized by the U1 snRNP, whereas the branch site is recognized by the U2 snRNP.
Pre-mRNA splicing is catalyzed by the spliceosome, a macromolecule composed of 5 small nuclear RNAs (snRNAs) associated with proteins to form particles termed small nuclear ribonucleoproteins (snRNPs; Figure 1).6,7 In most eukaryotes, including humans, there are 2 classes of introns: the common U2 type, representing more than 99% of human introns, and the rare U12 type, with the 2 differing in their consensus splice-site sequences. The removal of these 2 types of introns is catalyzed by 2 different spliceosomes called the U2- and U12-dependent spliceosomes, respectively. The 2 spliceosomes differ in their snRNA composition (Figure 1) but share most of the proteins.
The assembly of the spliceosome occurs anew on each pre-mRNA, which contains specific sequences that drive and regulate this process. The crucial signal sequences are the splice donor site (5′ end), the branch site (near the 3′ end), and the splice acceptor site (3′ end of the intron; Figure 1). In the major U2-dependent spliceosome, the donor site is recognized by the U1 snRNP, whereas the branch site is recognized by the U2 snRNP.
Several proteins interact with snRNAs to form snRNPs within the spliceosome.5 Those interacting with U1 and U2 snRNAs in the early phase of intron recognition and spliceosome formation include SRSF2, U2AF1, ZRSR2, U2AF65, SF1, SF3B1, SF3A1, and PRPF40B (Table 1).6 The SF3B1 protein is a core component of the U2 snRNP, which binds to the branch site, thereby base-pairing with the intron RNA.6
Genes encoding proteins that are core components of U1 and U2 snRNPs of the spliceosome, and their mutation status in human malignancies
| Gene (symbol, full official name, and chromosomal location) . | Encoded protein . | Human malignancies in which somatic mutations have been detected . |
|---|---|---|
| SRSF2 (serine/arginine-rich splicing factor 2; 17q25.1) | The encoded protein is a member of the serine/arginine (SR)–rich family of pre-mRNA splicing factors. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. SRSF2 enhances the splicing process both at 5′ and 3′ and is also involved in mRNA export from the nucleus and in translation. | Myeloid neoplasms with a particular enrichment in chronic myelomonocytic leukemia |
| SF1 (splicing factor 1; 11q13) | The encoded protein specifically recognizes the intron branch point sequence and is required for the early stages of spliceosome assembly. | Myeloid neoplasms |
| SF3B1 (splicing factor 3b, subunit 1, 155kDa; 2q33.1) | The encoded protein is subunit 1 of the splicing factor 3b protein complex. Splicing factor 3b, together with splicing factor 3a and a 12S RNA unit, forms the U2 snRNP. The splicing factor 3b/3a complex binds pre-mRNA upstream of the intron's branch site in a sequence-independent manner and may anchor the U2 snRNP to the pre-mRNA. | Myeloid neoplasms with a specific enrichment in conditions characterized by ring sideroblasts Chronic lymphocytic leukemia Various tumors |
| SF3A1 (splicing factor 3a, subunit 1, 120kDa; 22q12.2) | The encoded protein is subunit 1 of the splicing factor 3a protein complex. | Myeloid neoplasms |
| U2AF1 (U2 small nuclear RNA auxiliary factor 1; 21q22.3) | The encoded protein is the small subunit of U2 auxiliary factor, which is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. The small subunit plays a critical role in both constitutive and enhancer-dependent RNA splicing by directly mediating interactions between the large subunit and proteins bound to the enhancers. | Myeloid neoplasms |
| U2AF2 (U2 small nuclear RNA auxiliary factor 2; 19q13.42) | The encoded protein is the large subunit of U2 auxiliary factor (U2AF), which is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. The U2AF large subunit contains a sequence-specific RNA-binding region with 3 RNA recognition motifs and an arginine/serine-rich domain necessary for splicing. | Myeloid neoplasms |
| ZRSR2 (zinc finger CCCH type, RNA-binding motif and serine/arginine rich 2; Xp22.1) | The encoded protein is an essential splicing factor that associates with the U2 auxiliary factor heterodimer. This latter is required for the recognition of a functional 3′ splice site in pre-mRNA splicing. | Myeloid neoplasms |
| PRPF40B (PRP40 pre-mRNA processing factor 40 homolog B, S cerevisiae) | The encoded protein may serve in early spliceosome assembly. | Myeloid neoplasms |
| Gene (symbol, full official name, and chromosomal location) . | Encoded protein . | Human malignancies in which somatic mutations have been detected . |
|---|---|---|
| SRSF2 (serine/arginine-rich splicing factor 2; 17q25.1) | The encoded protein is a member of the serine/arginine (SR)–rich family of pre-mRNA splicing factors. Each of these factors contains an RNA recognition motif (RRM) for binding RNA and an RS domain for binding other proteins. SRSF2 enhances the splicing process both at 5′ and 3′ and is also involved in mRNA export from the nucleus and in translation. | Myeloid neoplasms with a particular enrichment in chronic myelomonocytic leukemia |
| SF1 (splicing factor 1; 11q13) | The encoded protein specifically recognizes the intron branch point sequence and is required for the early stages of spliceosome assembly. | Myeloid neoplasms |
| SF3B1 (splicing factor 3b, subunit 1, 155kDa; 2q33.1) | The encoded protein is subunit 1 of the splicing factor 3b protein complex. Splicing factor 3b, together with splicing factor 3a and a 12S RNA unit, forms the U2 snRNP. The splicing factor 3b/3a complex binds pre-mRNA upstream of the intron's branch site in a sequence-independent manner and may anchor the U2 snRNP to the pre-mRNA. | Myeloid neoplasms with a specific enrichment in conditions characterized by ring sideroblasts Chronic lymphocytic leukemia Various tumors |
| SF3A1 (splicing factor 3a, subunit 1, 120kDa; 22q12.2) | The encoded protein is subunit 1 of the splicing factor 3a protein complex. | Myeloid neoplasms |
| U2AF1 (U2 small nuclear RNA auxiliary factor 1; 21q22.3) | The encoded protein is the small subunit of U2 auxiliary factor, which is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. The small subunit plays a critical role in both constitutive and enhancer-dependent RNA splicing by directly mediating interactions between the large subunit and proteins bound to the enhancers. | Myeloid neoplasms |
| U2AF2 (U2 small nuclear RNA auxiliary factor 2; 19q13.42) | The encoded protein is the large subunit of U2 auxiliary factor (U2AF), which is a non-snRNP protein required for the binding of U2 snRNP to the pre-mRNA branch site. The U2AF large subunit contains a sequence-specific RNA-binding region with 3 RNA recognition motifs and an arginine/serine-rich domain necessary for splicing. | Myeloid neoplasms |
| ZRSR2 (zinc finger CCCH type, RNA-binding motif and serine/arginine rich 2; Xp22.1) | The encoded protein is an essential splicing factor that associates with the U2 auxiliary factor heterodimer. This latter is required for the recognition of a functional 3′ splice site in pre-mRNA splicing. | Myeloid neoplasms |
| PRPF40B (PRP40 pre-mRNA processing factor 40 homolog B, S cerevisiae) | The encoded protein may serve in early spliceosome assembly. | Myeloid neoplasms |
Information on genes and their encoded proteins is from http://www.ncbi.nlm.nih.gov/gene.
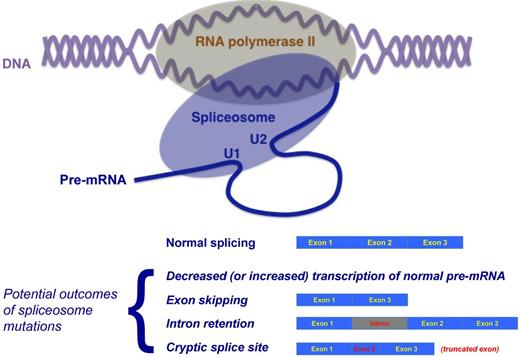
Although transcription of nucleosome-free DNA into pre-mRNA by RNA polymerase II and pre-mRNA splicing are traditionally considered as separate and sequential processes, increasing evidence suggests that transcription and RNA processing are strictly coupled in vivo and that several splicing factors also play a role in the regulation of transcription.8 For an in-depth analysis of this topic, the reader is referred to a recent review by Han et al.9 The concept of cotranscriptional splicing is represented schematically in Figure 2.
Schematic representation of the concept of cotranscriptional RNA splicing and potential outcomes of mutations of genes encoding proteins of the spliceosome. Top: Although transcription of nucleosome-free DNA into pre-mRNA by RNA polymerase II and pre-mRNA splicing are traditionally considered as separate and sequential processes, increasing evidence suggests that transcription and RNA processing are strictly coupled in vivo and that several splicing factors also play a role in the regulation of transcription. According to Han et al,9 cotranscriptional splicing represent a general rule with exceptions. Bottom: Potential outcomes of mutations of genes encoding proteins of the spliceosome. These outcomes include reduced (or also increased) transcription, exon skipping, intron retention, and cryptic exonic (or intronic) splice site activation with truncated (or elongated) exon. Adapted from Ebert and Bernard.4
Schematic representation of the concept of cotranscriptional RNA splicing and potential outcomes of mutations of genes encoding proteins of the spliceosome. Top: Although transcription of nucleosome-free DNA into pre-mRNA by RNA polymerase II and pre-mRNA splicing are traditionally considered as separate and sequential processes, increasing evidence suggests that transcription and RNA processing are strictly coupled in vivo and that several splicing factors also play a role in the regulation of transcription. According to Han et al,9 cotranscriptional splicing represent a general rule with exceptions. Bottom: Potential outcomes of mutations of genes encoding proteins of the spliceosome. These outcomes include reduced (or also increased) transcription, exon skipping, intron retention, and cryptic exonic (or intronic) splice site activation with truncated (or elongated) exon. Adapted from Ebert and Bernard.4
Abnormalities of mRNA splicing and human disease
Abnormal mRNA splicing may result from mutations of splice-site sequences, mutations in splicing regulatory sequences, and mutations in genes of the splicing machinery itself (ie, spliceosome mutations).
Mutations of the splice-site sequences or splicing regulatory sequences are responsible for a large fraction of human inherited diseases.6,10 For example, β-thalassemia may derive from mutations in splice-site sequences of the HBB gene that cause abnormal splicing of HBB and defective synthesis of its protein, β-globin.11 Conversely, inherited disorders caused by spliceosome mutations (ie, mutations of genes encoding spliceosomal proteins or mutations in spliceosomal RNA) are rare, at least based on our current understanding. For detailed information, the reader is referred to a recent comprehensive review article by Padgett.6
Somatic mutations of spliceosome genes were unknown until recently. From a theoretical point of view, they disrupt several processes downstream, because an altered spliceosome may cause abnormal transcription and altered splicing outcomes of thousands of genes. These outcomes include reduced (or also increased) transcription, exon skipping, intron retention, and cryptic splice site activation with truncated (or elongated) exon (Figure 2). It is now becoming apparent that through these mechanisms, somatic mutations of the RNA splicing machinery can play a role in the pathogenesis of human cancers, in particular in the pathophysiology of hematologic malignancies.
SF3B1 mutations in myeloid neoplasms
The first identification of a somatic mutation of SF3B1 was done at the end of 2010 by investigators of the Chronic Myeloid Disorders Working Group (CMD-WG) of the International Cancer Genome Consortium. When this collaboration was established in the first months of 2010, it was decided to perform whole-exome sequencing studies in patients with MDS, and more specifically in those with myelodysplasia with ring sideroblasts.
MDS are clonal disorders of hematopoietic stem cells characterized by peripheral cytopenia and propensity to evolve into acute myeloid leukemia (AML).12,13 These disorders show impressive clinical heterogeneity, ranging from conditions with a near-normal standardized mortality ratio to entities that are very close to AML.14-16 Refractory anemia with ring sideroblasts (RARS) is a phenotypically well-defined subtype of MDS characterized by erythroblasts with iron-loaded mitochondria, which are visualized by Prussian blue staining (Perls reaction) as a perinuclear ring of blue granules. Ring sideroblasts are also the morphologic hallmark of congenital sideroblastic anemias, inherited disorders because of germline mutations of genes such ALAS2 (X-linked sideroblastic anemia, OMIM #300751), ABCB7 (sideroblastic anemia and spinocerebellar ataxia, OMIM #301310), and SLC25A38 (autosomal recessive pyridoxine-refractory sideroblastic anemia, OMIM #205950).17-20 In patients with RARS, the iron deposited in perinuclear mitochondria of ring sideroblasts is present in the form of mitochondrial ferritin, which is encoded by the FTMT gene and is undetectable in normal erythropoiesis.21,22 In addition, both CD34-positive cells and immature RBCs from RARS patients are characterized by reduced expression of ABCB7,23,24 a gene that encodes an ATP-binding cassette (ABC) transporter involved in the regulation of mitochondrial iron homeostasis.25
The CMD-WG investigators reasoned that the peculiar phenotype of RARS could make easier the identification of recurrently mutated genes and used massively parallel sequencing technology to this purpose. Using this approach, Papaemmanuil et al identified 62 point mutations with a predominance of transitions across 8 RARS patients.2 Very interestingly, 6 of the 8 subjects studied had somatic mutations in SF3B1.
To characterize the spectrum and frequency of SF3B1 mutations, the CMD-WG investigators undertook targeted resequencing of the gene in hematologic malignancies and solid tumors.14,26 Somatic mutations of SF3B1 were found in approximately 30% of patients with MDS and in 20% of patients with myelodysplastic/myeloproliferative neoplasm (MDS/MPN). SF3B1 was also mutated in smaller proportions of patients with other tumor types. All mutations appeared to be heterozygous substitutions clustered in exons 12-16 of the gene. The K700E mutation accounted for more than 50% of the variants observed, and additional codons (666, 662, 622, and 625) were found to be hot spots for mutation. Results of quantitative evaluation of SF3B1 mutant allele burden were consistent with the presence of a dominant clone with heterozygous SF3B1 mutation in most of the cases and with the existence of a minority mutant clone in a smaller fraction of patients.26
Additional investigations performed by Papaemmanuil et al demonstrated that SF3B1 mutations were less deleterious than expected by chance, implying that the mutated protein retains structural integrity with altered function.2 Furthermore, gene-expression profiling revealed that SF3B1 mutations were associated with down-regulation of key gene networks, including core mitochondrial pathways.2
In the same period that the CMD-WG investigators were conducting whole-exome studies in myelodysplasia with ring sideroblasts, Yoshida et al performed whole-exome sequencing of paired tumor/control DNA from patients with various subtypes of MDS.1 Again, this study unexpectedly revealed novel pathway mutations involving multiple components of the RNA splicing machinery, including U2AF1, ZRSR2, SRSF2, and SF3B1. Targeted analysis in a larger patient population showed that RNA splicing mutations were frequent in patients with MDS and MDS/MPN. More specifically, somatic mutations of SF3B1 were found in 75% of patient with ring sideroblasts, whereas mutations of U2AF1, ZRSR2, and SRSF2 were more common in myeloid disorders without ring sideroblasts. In particular, approximately one-third of patients with chronic myelomonocytic leukemia had somatic mutations of SRSF2.
Relationship between SF3B1 mutations and ring sideroblasts in myeloid neoplasms
In patients with MDS or MDS/MPN, we found a close relationship between ring sideroblasts and SF3B1 mutations.26 In fact, the SF3B1 mutation status had a positive predictive value for disease phenotype with ring sideroblasts of 97.7% (95% confidence interval, 93.5%-99.5%), whereas the absence of ring sideroblasts had a negative predictive value for SF3B1 mutation of 97.8% (95% confidence interval, 93.8%-99.5%).
The close relationship between SF3B1 mutations and ring sideroblasts in myeloid neoplasms has been confirmed in all subsequent studies.27-32 This is consistent with a causal relationship and makes SF3B1 the first gene to be strongly associated with a specific morphologic feature in myeloid neoplasms.
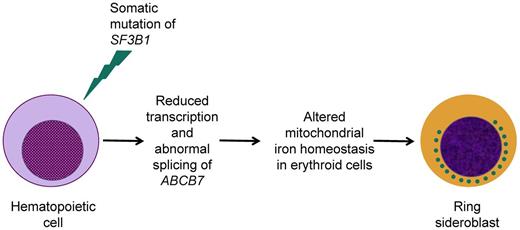
The molecular mechanisms underlying iron accumulation in the mitochondria of immature RBCs from patients with RARS are currently being investigated. One potential mechanism of ring sideroblast formation might be the reduced expression of the gene ABCB7, which is essential for hematopoiesis33 and mitochondrial iron homeostasis25 and is mutated in the congenital sideroblastic anemia associated with spinocerebellar ataxia.18 Overall, ABCB7 protein deficiency results in mitochondrial iron overload, reduced heme synthesis in immature RBCs, and ineffective erythropoiesis.25 In hematopoietic progenitors and precursors from RARS patients, low levels of ABCB7 expression are typically associated with high levels of FTMT expression.21,34,35 Overexpression of mitochondrial ferritin in RARS erythroblasts occurs at a very early stage of erythroid differentiation,34 suggesting that it might be a cause rather than a consequence of mitochondrial iron overload. A recent study by Nikpour et al has tested the hypothesis that a reduced expression of ABCB7 plays a role in the pathophysiology of RARS.36 The investigators showed that down-regulation of ABCB7 led to markedly reduced erythroid growth in vitro with accumulation of mitochondrial ferritin in immature RBCs. In addition, normalization of ABCB7 expression in RARS erythroblasts rescued aspects of the RARS phenotype. Finally, an altered ABCB7 exon usage was found in primary cells from patients with SF3B1-mutated RARS. These findings suggest that reduced transcription and abnormal splicing of ABCB7 in patients carrying a somatic mutation of SF3B1 may be at least one of the mechanisms that lead to ring sideroblast formation and RARS phenotype, as represented schematically in Figure 3.
Schematic representation of our current understanding of the relationship between the occurrence of a somatic SF3B1 mutation and the formation of ring sideroblasts in patients with RARS. Previous studies found reduced expression of ABCB7 in CD34-positive cells and cultured erythroblasts from RARS patients,23,24 whereas a recent study has shown altered ABCB7 exon usage in primary RARS cells.36 The interconnection between DNA transcription and RNA splicing, illustrated in Figure 2, is consistent with the conclusion that a somatic SF3B1 mutation may result in reduced transcription and abnormal splicing of ABCB7.
Schematic representation of our current understanding of the relationship between the occurrence of a somatic SF3B1 mutation and the formation of ring sideroblasts in patients with RARS. Previous studies found reduced expression of ABCB7 in CD34-positive cells and cultured erythroblasts from RARS patients,23,24 whereas a recent study has shown altered ABCB7 exon usage in primary RARS cells.36 The interconnection between DNA transcription and RNA splicing, illustrated in Figure 2, is consistent with the conclusion that a somatic SF3B1 mutation may result in reduced transcription and abnormal splicing of ABCB7.
In a study aimed at defining the relationship between dysfunction in SF3B1 and formation of ring sideroblasts, Visconte et al37 found that meayamycin, a pharmacologic inhibitor of the splicing factor 3b complex,38 induced the formation of ring sideroblasts in healthy BM cells in vitro. They were also able to detect ring sideroblasts by Perls staining in Sf3b1 heterozygous-knockout mice and therefore concluded that SF3B1 haploinsufficiency leads to ring sideroblast formation.
Although the majority of SF3B1-mutated RARS patients have a fully dominant clone that is heterozygous for the mutation, their percentage of ring sideroblasts ranges considerably from less than 20% to more than 90%. Although we found a significant relationship between this percentage and SF3B1 mutant allele burden,26 this does not fully explain the above variability, suggesting that additional factors play a role in the RARS phenotypic expression.
In all of the studies published so far, approximately 10%-20% of RARS patients have no evidence of mutations in SF3B1. This may partly reflect inaccuracies in the diagnosis of RARS related to difficulties in the identification of ring sideroblasts.39 However, the possibility exists that other mutant genes of RNA splicing machinery may lead to ring sideroblast formation.1
Prognostic relevance of SF3B1 mutations in MDS
The close relationship between ring sideroblasts and SF3B1 mutations in MDS would in principle imply that the mutant gene be associated with a mild clinical course. In fact, RARS is mainly characterized by erythroid dysplasia and ineffective erythropoiesis, and in most patients its clinical course is stable for years with a low risk of leukemic evolution.40-42 In our study on prognostic factors in MDS,15 the life expectancy of RARS patients 70 years or older was not significantly different from that of the general reference population.
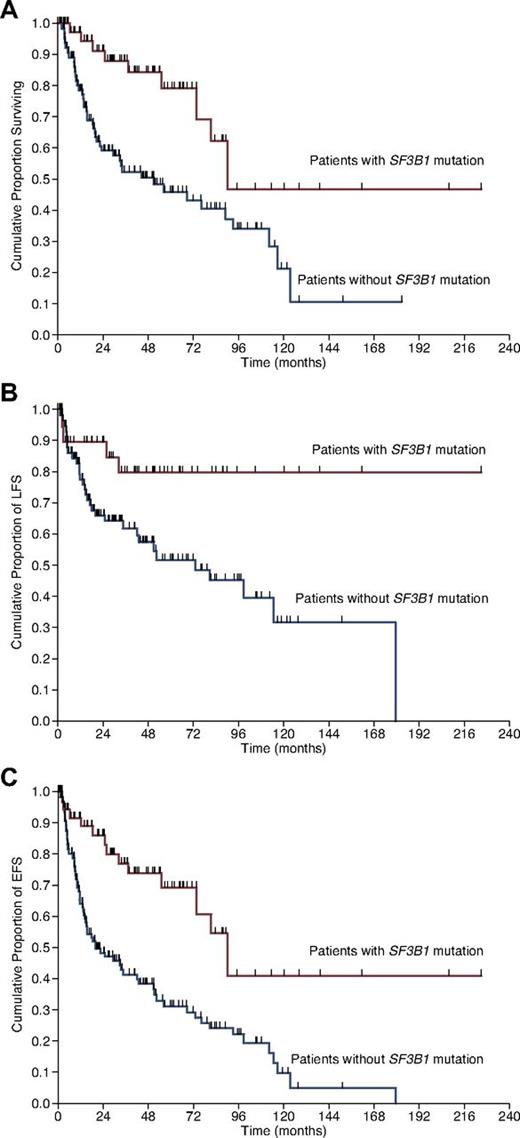
The studies published so far on the prognostic relevance of SF3B1 mutations in MDS have provided conflicting results, as detailed in Table 2. Before examining this issue, we want to emphasize that, for a proper evaluation of the prognostic relevance of any somatic mutation in MDS patients, ad hoc analyses should account for left censoring of the observations at the time of mutation assessment and for right censoring of the observations at the time of any potentially disease-modifying treatment. The striking enrichment of SF3B1 mutation in patients with RARS and its low frequency in patients with excess of blasts would a priori imply that in a population of MDS patients, those carrying a mutant SF3B1 had a better clinical outcome than those with a wild-type gene. This observation has indeed been made by us (Figure 4)26 and others.28 In contrast, 2 studies did not find any significant effect of the mutation on clinical outcome29,30 ; apparently, these studies did not account for right censoring of the observations at the time of any potentially disease-modifying treatment. In another study including only MDS patients with ring sideroblasts, the presence of SF3B1 mutations was associated with better overall and leukemia-free survival in univariate analysis.27 However, this prognostic value was lost in multivariable analysis including World Health Organization (WHO) morphologic category.27 This was the case also in our study,26 mainly because of the strict collinearity between ring sideroblasts and SF3B1 mutations. Lastly, in a study on MDS patients with lower-risk disease, mutations of SF3B1 showed only a trend toward longer survival.31
Recent studies on the prognostic relevance of SF3B1 mutations in patients with MDS
| Study . | No. of patients studied for clinical outcome . | Survival analysis . | Prognostic relevance of SF3B1 mutation . |
|---|---|---|---|
| Malcovati et al26 | 323 MDS patients with various WHO subtypes | Survival analyses were performed with the Kaplan-Meier method. Comparison between survival curves was carried out by the Wilcoxon test and multivariate survival analyses were performed by Cox proportional hazards regression. These latter analyses accounted for left censoring of the observations at the time of mutation assessment, and for right censoring of the observations at the time of potentially disease-modifying treatment. | Patients carrying an SF3B1 mutation showed a significantly better overall survival compared with those without mutation. The significant value of SF3B1 mutation was retained in a multivariate analysis including age, sex, hemoglobin level, absolute neutrophil count, platelet count, cytogenetic risk, BM blasts, and ring sideroblasts. Patients with the SF3B1 mutation showed a significantly better leukemia-free survival than those without mutation and this was confirmed in multivariate analysis. The SF3B1 mutation status remained an independent predictor of better survival even after adjustment for IPSS risk, whereas its significance was borderline after adjustment for WPSS risk. |
| Patnaik et al27 | 107 MDS patients, all with ring sideroblasts: 48 RARS, 43 with RCMD-RS, and 11 with excess of blasts and 15% or more ring sideroblasts | Survival analyses were performed with the Kaplan-Meier method and multivariate survival analyses were performed by Cox proportional hazards regression. | In univariate analysis, the presence of SF3B1 mutations was significantly associated with better overall and leukemia-free survival. In multivariate analysis, only age, transfusion dependence, and IPSS risk retained independent prognostic relevance. |
| Makishima et al28 | 88 MDS and 66 MDS/MPN patients | Survival analyses were performed with the Kaplan-Meier method. | The presence of SF3B1 mutations was associated with longer survival in univariate analysis. |
| Damm et al29 | 221 patients with MDS or MDS/MPN | Overall survival end points, measured from the date of diagnosis, were death (failure) and alive at last follow-up (censored). | There was no survival difference between patients with and those without SF3B1 mutation. |
| Thol et al30 | 193 MDS patients | Overall survival end points, measured from the date of first sample collection, were death (failure) and alive at last follow-up (censored). | SF3B1 mutations did not represent an independent prognostic factor. |
| Bejar et al31 | 288 patients with lower-risk MDS | Overall survival was calculated from the time of sample collection to the time of death from any cause. | SF3B1 mutations showed a non significant trend toward longer survival. |
| Study . | No. of patients studied for clinical outcome . | Survival analysis . | Prognostic relevance of SF3B1 mutation . |
|---|---|---|---|
| Malcovati et al26 | 323 MDS patients with various WHO subtypes | Survival analyses were performed with the Kaplan-Meier method. Comparison between survival curves was carried out by the Wilcoxon test and multivariate survival analyses were performed by Cox proportional hazards regression. These latter analyses accounted for left censoring of the observations at the time of mutation assessment, and for right censoring of the observations at the time of potentially disease-modifying treatment. | Patients carrying an SF3B1 mutation showed a significantly better overall survival compared with those without mutation. The significant value of SF3B1 mutation was retained in a multivariate analysis including age, sex, hemoglobin level, absolute neutrophil count, platelet count, cytogenetic risk, BM blasts, and ring sideroblasts. Patients with the SF3B1 mutation showed a significantly better leukemia-free survival than those without mutation and this was confirmed in multivariate analysis. The SF3B1 mutation status remained an independent predictor of better survival even after adjustment for IPSS risk, whereas its significance was borderline after adjustment for WPSS risk. |
| Patnaik et al27 | 107 MDS patients, all with ring sideroblasts: 48 RARS, 43 with RCMD-RS, and 11 with excess of blasts and 15% or more ring sideroblasts | Survival analyses were performed with the Kaplan-Meier method and multivariate survival analyses were performed by Cox proportional hazards regression. | In univariate analysis, the presence of SF3B1 mutations was significantly associated with better overall and leukemia-free survival. In multivariate analysis, only age, transfusion dependence, and IPSS risk retained independent prognostic relevance. |
| Makishima et al28 | 88 MDS and 66 MDS/MPN patients | Survival analyses were performed with the Kaplan-Meier method. | The presence of SF3B1 mutations was associated with longer survival in univariate analysis. |
| Damm et al29 | 221 patients with MDS or MDS/MPN | Overall survival end points, measured from the date of diagnosis, were death (failure) and alive at last follow-up (censored). | There was no survival difference between patients with and those without SF3B1 mutation. |
| Thol et al30 | 193 MDS patients | Overall survival end points, measured from the date of first sample collection, were death (failure) and alive at last follow-up (censored). | SF3B1 mutations did not represent an independent prognostic factor. |
| Bejar et al31 | 288 patients with lower-risk MDS | Overall survival was calculated from the time of sample collection to the time of death from any cause. | SF3B1 mutations showed a non significant trend toward longer survival. |
RCMD-RS indicates refractory cytopenia with multilineage dysplasia and ring sideroblasts.
Kaplan-Meier analysis of survival in MDS patients stratified according to SF3B1 mutation status. (A) Overall survival, defined here as the time (in months) between the date of diagnosis and the date of death (for patients who deceased) or last follow-up (for censored patients). (B) Leukemia-free survival (LFS), defined as the time between the date of diagnosis and the date of leukemic transformation (for patients who progressed to AML) or last follow-up. (C) Event-free survival (EFS), defined as the time between the date of diagnosis and the date of first event (death or leukemic transformation) or last follow-up. Vertical tick marks indicate right-censored patients. Reproduced from Malcovati et al.26
Kaplan-Meier analysis of survival in MDS patients stratified according to SF3B1 mutation status. (A) Overall survival, defined here as the time (in months) between the date of diagnosis and the date of death (for patients who deceased) or last follow-up (for censored patients). (B) Leukemia-free survival (LFS), defined as the time between the date of diagnosis and the date of leukemic transformation (for patients who progressed to AML) or last follow-up. (C) Event-free survival (EFS), defined as the time between the date of diagnosis and the date of first event (death or leukemic transformation) or last follow-up. Vertical tick marks indicate right-censored patients. Reproduced from Malcovati et al.26
In our study,26 SF3B1 mutation remained an independent predictor of better survival even after adjustment for the International Prognostic Scoring System (IPSS) risk, whereas its significance was borderline after adjustment for the WHO classification–based Prognostic Scoring System (WPSS) risk. In a multivariate analysis including WHO categories, when stratifying MDS patients according to unilineage dysplasia, multilineage dysplasia, and excess of blasts, SF3B1 mutation remained an independent predictor of better clinical outcome (M.C., L.M., unpublished observations, October 2012). This effect was lost when stratification included also a “ring sideroblast” category, again because of the strict collinearity between SF3B1 mutation and ring sideroblasts.
Clearly, the positive prognostic value of SF3B1 mutation may be partly or completely lost after acquisition of other somatic mutations, in particular those associated with disease progression. This might be the case, for example, if a patient with SF3B1-mutated RARS acquires an ASXL1 mutation and progresses to a condition characterized by multilineage dysplasia or excess of blasts in addition to ring sideroblasts. Alternatively, an SF3B1 mutation may mitigate the negative impact of other concomitant molecular lesions. A recent study has shown that DNMT3A and SF3B1 mutations commonly cooccur, suggesting a possible biologic cooperativity between the 2 molecular lesions.31 In that study, SF3B1 mutations had a trend toward longer survival and patients with concomitant DNMT3A and SF3B1mutations had a longer median survival than patients with DNMT3A mutations alone. This observation underlines the importance of simultaneously evaluating the mutation status of several genes of potential clinical relevance in the molecular prognostication of any myeloid neoplasm, a concept clearly illustrated also by a recent study in AML patients.43
SF3B1 mutations in refractory anemia with ring sideroblasts associated with marked thrombocytosis
RARS associated with marked thrombocytosis (RARS-T) is a MDS/MPN characterized by refractory anemia associated with erythroid dysplasia and ringed sideroblasts, platelet count 450 × 109/L or greater, and the presence of large atypical megakaryocytes similar to those observed in BCR/ABL1-negative MPN.44
The majority of patients with RARS-T carry a somatic mutation of SF3B1,26,28 and approximately 50% of them have a somatic mutation of JAK2 or MPL.44-46 A recent analysis of 47 patients with RARS-T strictly matching the 2008 WHO criteria showed that 41 of 47 (87%) subjects carried an SF3B1 mutation, 36 (77%) had JAK2 (V617F), and 30 (64%) had concomitant mutations in SF3B1 and JAK2.32 This reinforces the opinion that RARS-T is a true MDS/MPN at both the clinical and the molecular levels,44 and should no longer be considered a provisional entity in the WHO classification. A recent European collaborative study has shown that patients with RARS-T have better overall survival than those with RARS and shorter overall survival than those with essential thrombocythemia,46 underlying the clinical relevance of interactions between different mutant genes.
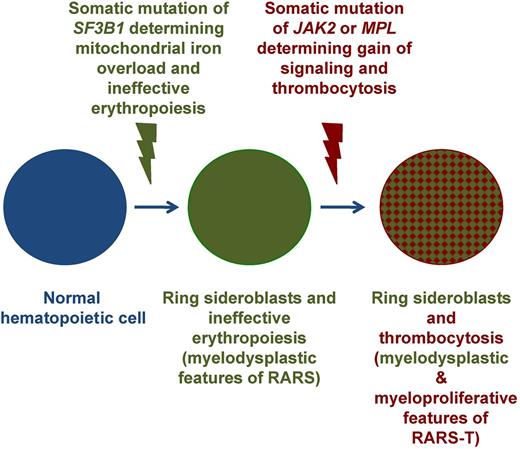
We demonstrated previously that RARS-T may develop from RARS through the acquisition of somatic mutations of JAK2, MPL, or other, as-yet-unknown genes.45 In keeping with previous observations, the assessment of the mutant allele burden in RARS-T patients carrying concomitant mutations of SF3B1 and JAK2 showed higher values for SF3B1 mutants compared with JAK2 mutants.26 These considerations suggest that, in a portion of patients, RARS-T may develop through sequential acquisition of SF3B1 and JAK2 mutations, as represented schematically in Figure 5.44 It has been suggested that RARS-T might also develop from a JAK2 (V617F)–mutated MPN through the acquisition of an SF3B1 mutation.32 This is theoretically possible, but less likely than the transition reported in Figure 5, because it would imply that a JAK2 (V617F)–mutated/SF3B1-mutated subclone has a competitive advantage over a JAK2 (V617F)–mutated dominant clone. It should be noted that SF3B1 mutations have not been detected so far in patients with polycythemia vera,2 whereas they have been identified in only 3% of those with essential thrombocythemia2 and in 7% of those with primary myelofibrosis.47
Schematic representation of the multistep molecular pathogenesis of RARS-T through sequential acquisition of SF3B1 and JAK2 or MPL mutations. The occurrence of a somatic mutation of SF3B1 causes mitochondrial iron overload, ineffective erythropoiesis, and anemia, typical myelodysplastic features of RARS. The subsequent occurrence of a somatic mutation of JAK2 or MPL involves the emergence of a subclone that carries 2 mutant genes (SF3B1 and JAK2 or MPL) and determines gain of thrombopoietin signaling, proliferation of large atypical megakaryocytes, and thrombocytosis, typical myeloproliferative feature of RARS-T. Adapted from Cazzola et al.44
Schematic representation of the multistep molecular pathogenesis of RARS-T through sequential acquisition of SF3B1 and JAK2 or MPL mutations. The occurrence of a somatic mutation of SF3B1 causes mitochondrial iron overload, ineffective erythropoiesis, and anemia, typical myelodysplastic features of RARS. The subsequent occurrence of a somatic mutation of JAK2 or MPL involves the emergence of a subclone that carries 2 mutant genes (SF3B1 and JAK2 or MPL) and determines gain of thrombopoietin signaling, proliferation of large atypical megakaryocytes, and thrombocytosis, typical myeloproliferative feature of RARS-T. Adapted from Cazzola et al.44
SF3B1 mutations in CLL
Our understanding of the genetics of chronic lymphocytic leukemia (CLL) has advanced significantly in recent years and the biology of CLL is currently used to choose treatment.48 However, the most striking advances regarding mutant genes have been made possible in the last 2 years by the use of next-generation sequencing technology.
In a comprehensive analysis of CLL combining whole-genome sequencing in 4 cases with targeted resequencing in 363 patients, Puente et al identified 4 genes that were recurrently mutated in CLL patients (NOTCH1, XPO1, MYD88, and KLHL6).49 Wang et al used a similar strategy and analyzed DNA samples from 91 CLL patients (88 exomes and 3 genomes).3 They identified 9 genes that were recurrently mutated: 4 (TP53, ATM, MYD88, and NOTCH1) were already known for their established role in CLL, whereas the remaining 5 genes (SF3B1, FBXW7, ZMYM3, DDX3X, and MAPK1) had not been previously associated with CLL.
In the study by Wang et al, somatic mutations of SF3B1 were found in approximately 15% of patients, making this the second most frequently mutated gene in CLL, and were highly enriched in patients with del(11q).3 Considering that the ATM gene maps on chromosome 11q22-q23, and that 2 patients had point mutations in both ATM and SF3B1 but did not have del(11q), the above enrichment suggests that SF3B1 mutations may be synergistic with loss of ATM. Using an ad hoc assay, Wang et al found alterations in pre-mRNA splicing in tumor samples from CLL patients carrying a somatic mutation of SF3B1.3 With respect to clinical implications, they concluded that SF3B1 mutations are associated with a poor prognosis in CLL, because a Cox multivariate regression model revealed that an SF3B1 mutation was predictive of an earlier need for treatment independently of other established predictive factors. This analysis had limitations, mainly because of lack of information on the timing of acquisition of the SF3B1 mutation.50
While investigating the coding genome of fludarabine-refractory CLL, Rossi et al found recurrent mutations of SF3B1, and therefore did targeted resequencing of the gene in 3 different patient cohorts.51 The incidence of SF3B1 mutation was 17% in fludarabine-refractory patients, 5% in newly diagnosed patients, and 6% in patients with Richter syndrome. Serial samples of mutated patients were available in 3 cases and the SF3B1 mutation was found to be acquired during follow-up in 2 of these. SF3B1 mutations were absent among mature B-cell neoplasms other than CLL, and in a separate study,52 a mutant SF3B1 was detected in only one of 63 subjects with clinical monoclonal B-cell lymphocytosis, a condition that might represent the initial phases of CLL.53
Quesada et al studied 279 untreated patients with CLL and found SF3B1 mutations in 27 (10%).54 As in previous studies, the most frequent mutation was K700E, occurring in almost half of the cases, followed by G742D, a mutation rarely found in myeloid neoplasms. No SF3B1 mutation was detected in 156 patients with non-Hodgkin lymphoma.54 CLL patients with mutations in SF3B1 had more aggressive disease and shorter overall survival than patients without these mutations. The fact that these patients were untreated suggests that SF3B1 mutations may occur during disease development without necessarily being a result of chemotherapy-induced selective pressure.55
Several groups are currently evaluating the clinical significance of SF3B1 mutations in CLL patients. Oscier et al have recently assessed the effect of NOTCH1 and SF3B1 mutations in a cohort of 494 newly diagnosed CLL patients enrolled in the randomized phase 3 CLL4 trial comparing chlorambucil with fludarabine and cyclophosphamide or fludarabine alone.56 NOTCH1 and SF3B1 mutations were found in 10% and 17% of untreated CLL patients and were independently associated with shorter survival in multivariate analysis. SF3B1 mutations were enriched in CD38-positive patients. In that study, TP53 abnormalities remained the most important negative prognostic factor, as was found previously in the German CLL Study Group CLL4 trial.57
The available evidence indicates that SF3B1 mutations are specific for CLL among mature B-cell neoplasms, have a lower incidence in early stages of CLL, occur more commonly in advanced disease, and tend to be associated with poor prognosis. Ongoing analyses from the German CLL Study Group will likely provide additional information.
SF3B1 mutation, clonal dominance, and disease progression in hematologic malignancies
Most patients with RARS carrying an SF3B1 mutation have a mutant allele burden between 40% and 50% in both circulating granulocytes and BM CD34-positive cells,26 indicating that the mutation occurs in a hematopoietic progenitor, determines expansion and dominance of a heterozygous clone, and is then transmitted to its myeloid progeny.
The mechanisms by which a mutant SF3B1 protein determines clonal proliferation of a CD34-positive cell have not yet been elucidated. As shown in Figure 2, a spliceosome mutation may cause abnormal transcription and various types of abnormal splicing or alternative splicing events. The fundamental problem is that the spliceosome controls the expression of thousands of genes and it is therefore difficult to identify those that are relevant for clonal expansion of a hematopoietic cell. The current debate as to whether an SF3B1 mutation is a gain-of-function or a loss-of-function mutation is partly meaningless. Because the mutant SF3B1 protein alters transcription and splicing of several genes, deciphering gains and losses is extremely difficult. In addition, even assuming that an SF3B1 mutation can cause clonal expansion and dominance of hematopoietic progenitors, it remains to be explained how the same mutation results in defective maturation of immature RBCs leading to ineffective erythropoiesis. One interpretation is that the abnormal mitochondrial iron homeostasis (Figure 3) results in excessive apoptosis of erythroid cells.
The effects of SF3B1 mutations may also be unrelated to the splicing activity of the protein. In fact, Sf3b1 heterozygous knockout mice have skeletal abnormalities associated with ectopic Hox gene expression.58 This is independent of the alteration of general gene expression and derives from an abnormal interaction with the polycomb group proteins that are responsible for the stable repression of Hox genes.
With respect to CLL, the available evidence indicates that SF3B1 mutations are very unlikely to be founding mutations in this lymphoid neoplasm. More likely, they occur during the clonal progression of the disease and determine the emergence of more aggressive subclones.
Ebert and Bernard4 have proposed the provocative hypothesis that SF3B1 mutations might occur initially in multipotent hematopoietic stem cells and that additional somatic mutations may determine the disease phenotype. In the case of RARS, the SF3B1 mutation must occur in a hematopoietic cell capable of myeloid differentiation26 and can cause a myelodysplastic phenotype without the need of additional mutations.2 With respect to CLL, the SF3B1 mutation is unlikely to be an initiating event. Nonetheless, the notion that myeloid and lymphoid neoplasms can share mutant genes is potentially interesting, as shown by a study on TET2 mutations in malignant lymphomas.59 This is at least consistent that the concept that the same mutation can cause 2 completely different diseases depending on the cell type in which it occurs.
Potential implications of SF3B1 mutations for molecular diagnosis and prognosis of myeloid and lymphoid neoplasms
Molecular tools are increasingly being used in the diagnosis and prognostic definition of both myeloid and lymphoid neoplasms. The implications of SF3B1 mutations based on the findings of studies published so far are summarized in Table 3.
Potential implications of somatic mutations of SF3B1 for molecular diagnosis and prognostication of myeloid and lymphoid neoplasms
| Disorder . | SF3B1 mutation . | |
|---|---|---|
| Diagnostic significance . | Prognostic relevance . | |
| MDS | The 2008 WHO diagnostic criteria for MDS13 are essentially based on morphology (erythroid dysplasia, ring sideroblasts, multilineage dysplasia, and excess of blasts), and include cytogenetics with respect to MDS with del(5q). | Prospective studies are required to clarify the prognostic significance of SF3B1 mutations in MDS and to resolve the current controversy on this issue. |
| In perspective, searching for an SF3B1 mutation may become less expensive and more accurate than enumerating ring sideroblasts. In this context, the current RARS might become “MDS associated with SF3B1 mutation.” Combinations of SF3B1 mutation and additional mutant genes might define conditions that are associated with less favorable clinical outcome. | To date, SF3B1 appears to be the only gene for which somatic mutations are associated with a good prognosis in MDS. | |
| MDS/MPN | The 2008 WHO diagnostic criteria for RARS-T13 are as follows:
Future diagnostic criteria for RARS-T might include a combination of anemia and thrombocytosis as clinical parameters and a combination of SF3B1 and JAK2 or MPL mutations as molecular parameters. | Patients with RARS-T have shorter survival than those with essential thrombocythemia.46 Analysis of SF3B1 mutation status may be useful to distinguish between the two myeloid disorders. |
| CLL | Unlikely to be useful for diagnosis. | SF3B1 mutations occur more commonly in advanced disease and tend to be associated with poor prognosis. Therefore, analysis of SF3B1 mutation status might be included in a targeted gene-sequencing approach to molecular prognostication of CLL. The available evidence indicates that somatic mutations of TP53, NOTCH1, and SF3B1 have independent prognostic significance. |
| Disorder . | SF3B1 mutation . | |
|---|---|---|
| Diagnostic significance . | Prognostic relevance . | |
| MDS | The 2008 WHO diagnostic criteria for MDS13 are essentially based on morphology (erythroid dysplasia, ring sideroblasts, multilineage dysplasia, and excess of blasts), and include cytogenetics with respect to MDS with del(5q). | Prospective studies are required to clarify the prognostic significance of SF3B1 mutations in MDS and to resolve the current controversy on this issue. |
| In perspective, searching for an SF3B1 mutation may become less expensive and more accurate than enumerating ring sideroblasts. In this context, the current RARS might become “MDS associated with SF3B1 mutation.” Combinations of SF3B1 mutation and additional mutant genes might define conditions that are associated with less favorable clinical outcome. | To date, SF3B1 appears to be the only gene for which somatic mutations are associated with a good prognosis in MDS. | |
| MDS/MPN | The 2008 WHO diagnostic criteria for RARS-T13 are as follows:
Future diagnostic criteria for RARS-T might include a combination of anemia and thrombocytosis as clinical parameters and a combination of SF3B1 and JAK2 or MPL mutations as molecular parameters. | Patients with RARS-T have shorter survival than those with essential thrombocythemia.46 Analysis of SF3B1 mutation status may be useful to distinguish between the two myeloid disorders. |
| CLL | Unlikely to be useful for diagnosis. | SF3B1 mutations occur more commonly in advanced disease and tend to be associated with poor prognosis. Therefore, analysis of SF3B1 mutation status might be included in a targeted gene-sequencing approach to molecular prognostication of CLL. The available evidence indicates that somatic mutations of TP53, NOTCH1, and SF3B1 have independent prognostic significance. |
The 2008 WHO diagnostic criteria for MDS are essentially based on morphology (erythroid dysplasia, ring sideroblasts, multilineage dysplasia, and excess of blasts), and include cytogenetics with respect to MDS with del(5q). At present, the most important prognostic factors in MDS are WHO category, cytogenetic abnormality, and transfusion requirement or severity of anemia, all of which are included in the WPSS.16 The degree of neutropenia and thrombocytopenia have additional prognostic significance, as shown by the recent study on the revised IPSS (IPSS-R) for MDS.60 Nevertheless, there is a need for more reliable diagnostic and prognostic tools.
The elegant study by Bejar et al has illustrated the clinical effect of point mutations in MDS, in particular the potential of molecular data for facilitating diagnosis, improving prognostication, and allowing the evaluation of disease progression.61 In the next years, targeted gene sequencing is likely to become a frontline diagnostic and prognostic test for MDS, and mutation analysis of SF3B1 should be part of this approach, as suggested in Table 3.
Within MDS/MPN, future diagnostic criteria of RARS-T might include a combination of anemia and thrombocytosis as clinical parameters and a combination of SF3B1 and JAK2 or MPL mutations as molecular parameters.
Finally, several studies are currently being conducted to define the prognostic relevance of SF3B1 mutations in CLL.
Spliceosome as a target for therapy
Several compounds can modulate mRNA splicing, and a recent review article by Webb et al examines this topic comprehensively.62 Natural products include spliceostatin A,63 pladienolide,64 and meayamycin,38 and small synthetic molecule modulators of splicing factor 3b include sudemycins.65
Spliceosome modulators have antitumor activity in cellular and animal models, and the available evidence indicates that interaction with splicing factor 3b is crucial for their cytotoxic activity.62,64 These compounds likely alter the interaction between splicing factor 3b and pre-mRNA66 and induce abnormal transcription and alternative splicing in mammalian cells.
In the case of hematologic malignancies associated with SF3B1 mutations, the mutant splicing factor 3b causes abnormal transcription and alternative splicing per se. The effect of the above compounds on an abnormal splicing factor is currently unpredictable and preclinical studies are therefore needed to explore the effect of molecule modulators of splicing factor 3b in SF3B1-mutated myeloid or lymphoid cells. The working hypothesis is that cells harboring a spliceosome mutation may be more sensitive to spliceosome modulators.
Acknowledgments
The authors thank the many colleagues of the Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium for the wonderful collaborative experience.
The studies on the biologic and clinical significance of SF3B1 mutations in myeloid neoplasms conducted at the Fondazione Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo and University of Pavia (Pavia, Italy) were supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (Milan, Italy) Special Program Molecular Clinical Oncology 5 × 1000 to the Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative (project number number 1005) and by a grant from the Fondazione Cariplo (Milan, Italy) to M.C.
Authorship
Contribution: M.C. conceived the article, analyzed the literature, and wrote the manuscript; and M.R. and L.M. analyzed the literature and contributed to manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mario Cazzola, MD, Department of Hematology Oncology, Fondazione IRCCS Policlinico San Matteo, 27100 Pavia, Italy; e-mail: mario.cazzola@unipv.it.