Key Points
BCR subsets 2 and 8 show specific genetic profiles influencing CLL course.
Abstract
Genetic lesions and B-cell receptor (BCR) signaling are both oncogenic drivers in chronic lymphocytic leukemia (CLL). However, scant data are available on preferential associations between specific genetic alterations and stereotyped BCR subsets. By analyzing 1419 cases, 2 CLL subsets (2 and 8) harboring stereotyped BCR are enriched in specific molecular alterations influencing disease course. SF3B1 mutations are the genetic hallmark of IGHV3-21-CLL belonging to subset 2 (52%) but are evenly represented in nonstereotyped IGHV3-21-CLL. Trisomy 12 (87%) and NOTCH1 mutations (62%) characterize IGHV4-39-CLL belonging to subset 8 but occur with the expected frequency in IGHV4-39-CLL with heterogeneous BCR. Clinically, co-occurrence of SF3B1 mutations and subset 2 BCR configuration prompts disease progression in IGHV3-21-CLL, whereas cooperation between NOTCH1 mutations, +12, and subset 8 BCR configuration invariably primes CLL transformation into Richter syndrome. These findings provide a proof of concept that specific stereotyped BCR may promote or select molecular lesions influencing outcome.
Introduction
Driver genetic lesions in chronic lymphocytic leukemia (CLL) include recurrent chromosomal aberrations and point mutations affecting the TP53, NOTCH1, SF3B1, BIRC3, and MYD88 genes.1-7 Genetic abnormalities arise at the time of neoplastic transformation and during clonal evolution likely following various microenvironmental signals, including those provided by the B-cell receptor (BCR).8
CLL often carries surface BCR with similar stereotypic patterns resulting from IGHV-IGHD-IGHJ rearrangements already present in the natural B-cell repertoire of normal individuals prior to transformation, thus suggesting that recognition of common epitopes or classes of structurally similar epitopes is likely involved in leukemic clone selection.9-12 So far, more than 200 different subsets have been defined with stereotyped BCR, with 8 subsets (1-8) accounting for ∼30% of all stereotyped CLL.10
Despite the established role of genetics and BCR features in the pathobiology of CLL, little is known regarding the association between specific genetic aberrations and distinct CLL subsets with stereotyped BCR.13,14 By investigating a large cohort of more than 1400 CLL cases, here we report that 2 CLL immunogenetic subgroups with proven prognostic/predictive relevance (stereotyped BCR subsets 2 and 8)15-17 show a subset-specific genetic profile influencing disease course. Our results point to the potential role of specific stereotyped BCR patterns in promoting the occurrence or selection of genetic lesions that impact on clinical outcome of CLL.
Study design
Samples
The study was based on 1419 newly diagnosed CLL cases (supplemental Figure 1 and Table 1; see the Blood Web site). According to sample size analysis, the number of cases was sufficient to allow identification of all possible genetic lesions co-occurring with each of the most represented BCR subsets (ie, those including >5 cases) (power, 80%; α-error, 0.5). The study was approved by the institutional ethical committee (Protocol Code 59/CE; Study Number CE 8/11). Patients provided informed consent in accordance with local Institutional Review Board requirements and the Declaration of Helsinki.
Molecular studies
Molecular studies were performed on biological samples collected at the time of CLL diagnosis. IGHV-IGHD-IGHJ and IGLV-J rearrangements, and mutations of the TP53, NOTCH1, SF3B1, MYD88, and BIRC3 genes were analyzed by Sanger sequencing.5,7,17 Probes used for fluorescence in situ hybridization were the following: (1) LSID13S319, CEP12, LSIp53, and LSIATM (Abbott); and (2) RP11-177O8 (BIRC3) BAC clone.7 Further details of the molecular studies are available in the supplemental Appendix.
Statistical analysis
Immunoglobulin heavy variable gene complementarity determining region 3 sequences from our CLL data set (supplemental Tables 2 and 3) and from public CLL databases were aligned, and stereotyped rearrangements were clustered according to a previously reported bioinformatic algorithm.10
The binomial distribution of the frequency of the genetic lesions within each of the 8 most represented BCR subsets was calculated according to the π of the genetic lesions in IGHV-mutated or IGHV-unmutated CLL, depending on the IGHV mutation status that preferentially or exclusively associated with a given BCR subset. Deviations of the observed binomial distribution from the theoretically expected binomial distribution were assessed by the nonparametric binomial test. All statistical tests were 2-sided, and P values were corrected for multiple comparisons using the Bonferroni method. Statistical significance was defined as P < .05. The analysis was performed with SPSS software v20.0 (Chicago, IL). Further details of the statistical analysis are available in the supplemental Appendix.
Results and discussion
To uncover the interactions between molecular lesions of cancer genes and BCR-driven mechanisms in CLL, we performed a cross-sectional investigation of the associations between the most recurrent mutations (TP53, NOTCH1, SF3B1, BIRC3, and MYD88), chromosomal abnormalities (del17p, del11q, del13q, +12, and BIRC3 deletion), and BCR features in 1419 newly presented CLL cases. The molecular and immunogenetic features of the study cohort were consistent with those expected in newly diagnosed CLL (supplemental Figures 2A and 3). The overall frequency of BCR stereotypy (21.4%) was in the range of that reported in CLL series of similar size.18,19
The genetics of CLL differed according to IGHV mutation status, as documented by the significantly higher prevalence of favorable lesions in IGHV-mutated CLL and of unfavorable lesions in IGHV-unmutated cases (supplemental Figure 2B). On the contrary, after compensating for IGHV mutation status, BCR stereotypy did not affect the distribution of genetic lesions (supplemental Figure 4). Although IGHV-unmutated CLL showed a broad association with all unfavorable genetic lesions, expression of zeta-chain-associated protein kinase 70 and CD38, 2 molecules that may cooperate with the BCR in CLL,1 appeared to cluster with specific genetic subgroups (supplemental Figure 5). Overall, these data are consistent with the notion that (1) IGHV mutation status, but not BCR stereotypy as a whole, distinguishes different clinical and biological subgroups of CLL; (2) not only BCR stereotypy, but also molecular genetics is highly dependent on IGHV mutational status in CLL; and (3) different components of the CLL signaling machinery may favor distinct genetic events in this leukemia.
Consistent with previous reports, BCR subsets 1 to 8 were the most represented in this CLL cohort, overall accounting for ∼30% of all stereotyped cases.10 Two major subsets, namely, subsets 2 (IGHV3-21) and 8 (IGHV4-39), recurrently showed a distinctive pattern of molecular alterations (Figure 1A-B). In contrast, in all the other major subsets the prevalence of genetic lesions did not differ significantly from what was expected based on chance alone (Figure 1A-B).
Distribution of CLL driver genetic lesions across major stereotyped BCR subsets. (A) Circos plot representing the relative frequency and pairwise co-occurrence of CLL driver genetic lesions and usage of stereotyped BCR belonging to majors subsets (1, 2, 3, 4, 5, 6, 7B, and 8) in newly diagnosed CLL. The length of the arc corresponds to the frequency of the genetic lesions. The width of the ribbon corresponds to the percentage of patients in which the 2 connected variables co-occur. (B) Prevalence of CLL driver genetic lesions within the major BCR subsets (1, 2, 3, 4, 5, 6, 7B, and 8) in newly diagnosed CLL. p, P value from the nonparametric binomial test; p corrected, P value from the nonparametric binomial test corrected for multiple hypothesis testing.
Distribution of CLL driver genetic lesions across major stereotyped BCR subsets. (A) Circos plot representing the relative frequency and pairwise co-occurrence of CLL driver genetic lesions and usage of stereotyped BCR belonging to majors subsets (1, 2, 3, 4, 5, 6, 7B, and 8) in newly diagnosed CLL. The length of the arc corresponds to the frequency of the genetic lesions. The width of the ribbon corresponds to the percentage of patients in which the 2 connected variables co-occur. (B) Prevalence of CLL driver genetic lesions within the major BCR subsets (1, 2, 3, 4, 5, 6, 7B, and 8) in newly diagnosed CLL. p, P value from the nonparametric binomial test; p corrected, P value from the nonparametric binomial test corrected for multiple hypothesis testing.
Compared with other major BCR subsets, CLL subset 2 was significantly enriched in SF3B1 mutations, which occurred in 45.4% of cases (P = 1.58 × 10−4) (Figure 1A-B). The prevalence of SF3B1 mutations was higher when the analysis was restricted to subset 2 cases utilizing both the IGHV3-21 and IGHVL3-21 genes (52.6%). In contrast, IGHV3-21-CLL with heterogeneous BCR displayed a significantly lower prevalence of SF3B1 mutations (13.8%; P = .002) (supplemental Figure 6A), thus suggesting that the association between subset 2 and SF3B1 mutations is driven by the homology in the BCR rather than mere usage of the IGHV3-21 gene. Of note, subset 2 CLL consistently lacked TP53 abnormalities, which were otherwise evenly represented in all the other major subsets (Figure 1A-B). This observation is in agreement with the known anticorrelation between SF3B1 and TP53 lesions in CLL and points to SF3B1 mutations, rather than TP53 abnormalities, as the main driver of progressiveness in subset 2 CLL.5
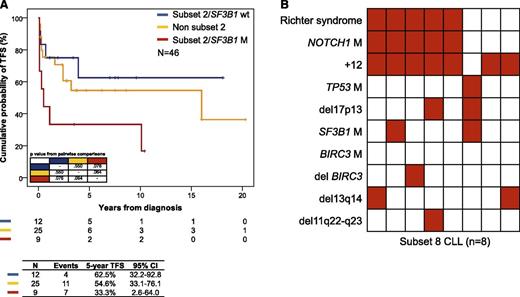
Consistently, subset 2 CLL harboring SF3B1 mutations showed a higher progression rate to a disease requiring treatment (5-year treatment-free survival [TFS], 33.3%) compared with subset 2 CLL harboring a wild-type SF3B1 (5-year TFS, 62.5%; P = .064) and to IGHV3-21-CLL with heterogeneous BCR (5-year TFS, 54.6%; P = .076) (Figure 2A). These data suggest that, among IGHV3-21-CLL, the association between SF3B1 mutations and subset 2 BCR configuration contributes to promote disease progression, rather than the sole stereotypic features of IGHV3-21–bearing BCR.
Clinical impact of the genotype in stereotyped BCR subset 2 and subset 8 CLL. (A) Kaplan-Meier estimates of TFS according to SF3B1 mutation status in CLL utilizing the IGHV3-21 gene. (B) Heat map showing the relationship between genetic lesions and Richter syndrome transformation in subset 8 CLL. Rows correspond to identical variables, and columns represent individual patients color-coded based on the molecular status (white, absence of the variable; red, presence of the variable).
Clinical impact of the genotype in stereotyped BCR subset 2 and subset 8 CLL. (A) Kaplan-Meier estimates of TFS according to SF3B1 mutation status in CLL utilizing the IGHV3-21 gene. (B) Heat map showing the relationship between genetic lesions and Richter syndrome transformation in subset 8 CLL. Rows correspond to identical variables, and columns represent individual patients color-coded based on the molecular status (white, absence of the variable; red, presence of the variable).
Subset 8 CLL was significantly enriched in +12, which occurred in 87.5% of cases (P = 9.6 × 10−4) (Figure 1A-B). Consistent with the relationship between +12 and NOTCH1 lesions,20,21 subset 8 CLL was also enriched in NOTCH1 mutations (62.5% of cases), although at a borderline significance probably due to sample size. The prevalence of +12 and NOTCH1 mutations was significantly higher in subset 8 CLL than in IGHV4-39-CLL with nonstereotyped BCR (+12, 27.9%, P = .003; NOTCH1 mutations, 8.1%, P = .006; supplemental Figure 6B), implying that these genetic-immunogenetic associations are influenced by the homology in the BCR rather than shared usage of the IGHV4-39 gene.
From a clinical standpoint, as already documented in a partially overlapping CLL cohort,17 the majority (5/8, 62.5%) of patients belonging to subset 8 had transformed to Richter syndrome. All transformed cases carried both NOTCH1 mutations and +12, whereas this genetic association was never observed among subset 8 patients that had not transformed to Richter syndrome (P = .017) (Figure 2B). These data indicate a cooperation between NOTCH1 mutations, +12, and subset 8 BCR configuration in priming Richter syndrome transformation.22,23
Taken together, our observations provide a proof of concept that specific BCR configurations may contribute to clonal selection of specific genetic lesions in CLL. Although our analysis was focused on the most represented BCR subsets, the interaction between BCR configuration and specific genetic lesions might be a more general phenomenon in CLL. In order to extend this observation also to minor BCR subsets, investigation of large case series of thousands of CLL patients is warranted. In addition, our data prompt studies aimed at assessing the role of BCR signaling in clonal evolution and selection of specific genetic abnormalities in lymphoid tumors, nonneoplastic conditions,24 and normal B-cell subsets25 bearing stereotyped BCR.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) Foundation Milan, Italy (Special Program Molecular Clinical Oncology 5 x 1000, 10007; My First AIRC Grant 13470 and 10327; and Investigator Grant 13227); Futuro in Ricerca and Programma di Ricerca di Rilevante Interesse Nazionale Ministero dell'Istruzione, dell'Università e della Ricerca, Rome, Italy; Progetto Giovani Ricercatori and Ricerca Sanitaria Finalizzata, Ministero della Salute, Rome, Italy; Compagnia di San Paolo, Turin, Italy (PMN_call_2012_0071); Fondazione Cariplo, Milan, Italy; Centro di Riferimento Oncologico, Aviano, Italy (2012-0689, “5x1000 Intramural Program”); and fellowships from Novara-AIL Onlus Foundation, Novara, Italy (S.M.).
Authorship
Contribution: D.R., G.G., and V.G. designed the study, interpreted data, and wrote the manuscript; D.R. and R.S. performed statistical analysis; V.S. and R.B. performed immunogenetic analysis; S.R., M.D.-B., A.B., M.D., C.C., and A.Z. performed molecular analysis; F.M.R. and S.M. performed fluorescence in situ hybridization analysis; F.F. contributed to carrying out the study and helped draft the manuscript for intellectual content; and J.N., P.B., A.G., F.Z., G.P., L.L., D.G.E., F.D.-R., R.M., and G.D.P. provided well-characterized biological samples and clinical data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Davide Rossi, Division of Hematology, Department of Translational Medicine, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail: rossidav@med.unipmn.it.
References
Author notes
G.G. and V.G. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal