Abstract
Recently, cereblon (CRBN) expression was found to be essential for the activity of thalidomide and lenalidomide. In the present study, we investigated whether the clinical efficacy of thalidomide in multiple myeloma is associated with CRBN expression in myeloma cells. Patients with newly diagnosed multiple myeloma were included in the HOVON-65/GMMG-HD4 trial, in which postintensification treatment in 1 arm consisted of daily thalidomide (50 mg) for 2 years. Gene-expression profiling, determined at the start of the trial, was available for 96 patients who started thalidomide maintenance. In this patient set, increase of CRBN gene expression was significantly associated with longerprogression-free survival (P = .005). In contrast, no association between CRBN expression and survival was observed in the arm with bortezomib maintenance. We conclude that CRBN expression may be associated with the clinical efficacy of thalidomide. This trial has been registered at the Nederlands Trial Register (www.trialregister.nl) as NTR213; at the European Union Drug Regulating Authorities Clinical Trials (EudraCT) as 2004-000944-26; and at the International Standard Randomized Controlled Trial Number (ISRCTN) as 64455289.
Key Points
Higher expression of the cereblon gene is associated with better outcome in newly diagnosed multiple myeloma patients treated with thalidomide maintenance.
Introduction
Introduction of thalidomide, bortezomib, and lenalidomide has greatly improved induction treatment for multiple myeloma (MM).1-4 Attention is now shifting toward improving consolidation and maintenance therapy.5 Thalidomide and lenalidomide represent immunomodulatory drugs (IMiDs) with variable efficacy during maintenance after high-dose therapy and in the nontransplantation setting.6-8 So far, there are no biomarkers for prediction of outcome after thalidomide and/or lenalidomide treatment. CRBN was recently identified as the target gene responsible for the teratogenic effects of thalidomide.9 CRBN levels were also shown to be critical for the antitumor activity of lenalidomide and thalidomide in both in vitro model systems and in lenalidomide-resistant patients.10 In the present study, we report that CRBN expression is associated with outcome of thalidomide maintenance in newly diagnosed MM patients.
Methods
Patients and procedures
In the HOVON-65/GMMG-HD4 trial, patients with newly diagnosed MM were randomly assigned to receive either VAD (vincristine, doxorubicin, and dexamethasone) induction, intensification with high-dose melphalan (HDM), and autologous stem cell transplantation (ASCT) followed by maintenance therapy with thalidomide or PAD (bortezomib, doxorubicin, and dexamethasone), HDM, and ASCT followed by maintenance with bortezomib. The maximum duration of maintenance therapy in both arms was 2 years.11 Patients randomized to VAD received maintenance with thalidomide 50 mg daily for 2 years starting 4 weeks after HDM. This study was approved by the ethics committees of the Erasmus University MC, the University of Heidelberg, and the participating sites. All patients gave written informed consent and the trial was conducted according to the European Clinical Trial Directive 2005 and the Declaration of Helsinki.
Response assessments and end points
Clinical characteristics were registered at diagnosis. Cytogenetic studies were performed as described previously.12 For this subanalysis, progression-free survival (PFS) and overall survival (OS) were measured from start of the maintenance treatment. For PFS, progression was used as the end point and for OS, death from any cause. Patients alive at the date of last contact were censored. Evaluation of response is described in detail in supplemental Table 4 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
GEP and statistical analysis
The gene-expression profiling (GEP) dataset GSE19784 was used, which was derived from patients included in the HOVON-65/GMMG-HD4 trial.11,13 CRBN expression was assessed using the intensity values of the probe sets 218142_s_at and 222533_at, combined using the method of Dai et al.14 Presence calls for CRBN expression were determined with the PANP algorithm using standard settings (see the PANP reference manual on the Bioconductor Web site, http://www.bioconductor.org/packages/release/bioc/html/panp.html).15 Details of the quantitative RT-PCR are given in supplemental Figure 3. Multivariate Cox regression analysis was performed to assess the value of CRBN as a prognostic factor in relation to the International Staging System (ISS) and high-risk cytogenetics, as described previously.11
Results and discussion
Patients and response
A total of 833 patients were enrolled in the HOVON65/GMMG-HD4 trial. Of the patients randomized to the VAD arm, 77 of 347 (22%) went off protocol after HDM because of allo-SCT (n = 21, 6%), persisting toxicity (n = 11, 3%), or other reasons (n = 45, 13%), whereas 270 (78%) patients started thalidomide maintenance treatment. Normal completion of thalidomide maintenance was achieved in 73 of 270 (27%) patients. Eleven of 270 thalidomide maintenance patients underwent allo-SCT and were not considered in this subanalysis. Of the remaining 259 patients, GEP and survival data were available for 96. Baseline characteristics between this subgroup (n = 96) and the remainder (n = 163) were comparable (supplemental Table 1). Present calls were found for both CRBN probe sets in 95 of 96 thalidomide maintenance cases, with one patient demonstrating a borderline present call (“M”) for one probe set and a present call for the other. A significant correlation was found between CRBN gene expression measured by microarray (National Center for Biotechnology Gene Expression Omnibus [NCBI-GEO] repository: GSE19784) and quantitative RT-PCR (Spearman rho: 0.67, P = .002, n = 18; supplemental Figure 3). The EMC clustering represents our gene expression based classification of MM.16 Of the clusters evaluated, the CTA cluster demonstrated a significantly higher CRBN expression compared with the other clusters (Bonferroni-Holm corrected P = .01, supplemental Figure 2).16
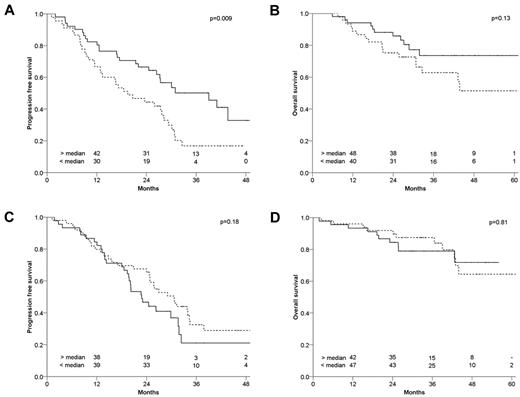
In univariate Cox regression analysis, CRBN expression was significantly associated with PFS (hazard ratio = 0.68; 95% confidence interval, 0.52-0.89; P = .005) and with OS (hazard ratio = 0.65; 95% confidence interval, 0.43-0.97; P = .04; Table 1). Kaplan-Meier analysis was used solely for visualization with CRBN expression split in 2 or 4 groups using median or quartile intensities: patients with CRBN expression above the median demonstrated longer PFS compared with patients with CRBN levels below the median (P = .009; Figure 1A-B quartile intensities and supplemental Figure 4). In addition, an optimal CRBN cutoff was calculated (supplemental Table 2). For this calculation, the PFS data that prohibit use of this cutoff in this dataset for any analyses related to PFS were used. In contrast, the median expression value was arbitrarily chosen and used for analysis in relation to response upgrade. Multivariate Cox regression analysis was performed on 81 patients for whom the following covariates were available: ISS, continuous CRBN levels, and high-risk FISH [del(17p) and/or 1q gain and/or t(4;14)]. Higher CRBN levels remained significantly related to longer PFS, but not OS, with a hazard ratio of 0.66 (P = .03) and 0.75 (P = .3), respectively (Table 1). No significant correlation was found between any of these covariates and CRBN, but lower CRBN expression was found in ISSIII compared with either ISSI or ISSII (Bonferroni corrected P = .10 by Kruskal Wallis test). The CRBN gene is positioned on chromosome 3. Chromosome 3 trisomies are frequently found in patients with hyperdiploidy and, indeed, CRBN levels were significantly higher in hyperdiploid patients compared with nonhyperdiploid patients (P = .005). However, in a multivariate Cox regression analysis, CRBN levels, but not hyperdiploidy, were found to be related to PFS (P = .006 and P = .8, respectively; data not shown).
Cox regression analyses
| Covariate . | HR . | 95% CI . | P . |
|---|---|---|---|
| Univariate Cox regression analysis of CRBN expression in relation to PFS | |||
| CRBN expression | 0.68 | 0.52-0.89 | .005 |
| Univariate Cox regression analysis of CRBN expression in relation to OS | |||
| CRBN expression | 0.65 | 0.43-0.97 | .04 |
| Multivariate Cox regression analysis of CRBN expression in relation to PFS | |||
| CRBN expression | 0.66 | 0.45-0.96 | .03 |
| ISS2 | 2.35 | 1.15-4.82 | .02 |
| ISS3 | 2.55 | 1.21-5.36 | .01 |
| High-risk FISH | 2.82 | 1.59-5.00 | .0004 |
| Multivariate Cox regression analysis of CRBN expression in relation to OS | |||
| CRBN expression | 0.75 | 0.43-1.32 | .32 |
| ISS2 | 4.66 | 1.38-15.81 | .01 |
| ISS3 | 5.49 | 1.67-18.08 | .005 |
| High-risk FISH | 3.65 | 1.53-8.70 | .003 |
| Covariate . | HR . | 95% CI . | P . |
|---|---|---|---|
| Univariate Cox regression analysis of CRBN expression in relation to PFS | |||
| CRBN expression | 0.68 | 0.52-0.89 | .005 |
| Univariate Cox regression analysis of CRBN expression in relation to OS | |||
| CRBN expression | 0.65 | 0.43-0.97 | .04 |
| Multivariate Cox regression analysis of CRBN expression in relation to PFS | |||
| CRBN expression | 0.66 | 0.45-0.96 | .03 |
| ISS2 | 2.35 | 1.15-4.82 | .02 |
| ISS3 | 2.55 | 1.21-5.36 | .01 |
| High-risk FISH | 2.82 | 1.59-5.00 | .0004 |
| Multivariate Cox regression analysis of CRBN expression in relation to OS | |||
| CRBN expression | 0.75 | 0.43-1.32 | .32 |
| ISS2 | 4.66 | 1.38-15.81 | .01 |
| ISS3 | 5.49 | 1.67-18.08 | .005 |
| High-risk FISH | 3.65 | 1.53-8.70 | .003 |
High-risk cytogenetics is defined as having del(17p) and/or 1q gain and/or t(4;14).
HR indicates hazard ratio; and CI, confidence interval.
CRBN expression in HOVON-65/GMMG-HD4. Shown is CRBN expression in relation to PFS and OS Kaplan-Meier curves of CRBN expression in relation to survival in thalidomide-treated patients (A-B) and in relation to bortezomib-treated patients (C-D). PFS is shown at left; OS on the right. Log-rank P values are shown in the right corner of each panel. Broken lines indicate CRBN expression levels below the median and solid lines indicate expression levels above the median. Remaining patients at risk are shown above the x-axis (PFS at 1, 2, 3, and 4 years and OS at 1, 2, 3, 4, and 5 years). The median CRBN expression was determined on the combined data of both thalidomide- and bortezomib-treated patients: 45 of 96 patients were below the median in the thalidomide set, whereas 50 of 95 were below the median in the bortezomib set.
CRBN expression in HOVON-65/GMMG-HD4. Shown is CRBN expression in relation to PFS and OS Kaplan-Meier curves of CRBN expression in relation to survival in thalidomide-treated patients (A-B) and in relation to bortezomib-treated patients (C-D). PFS is shown at left; OS on the right. Log-rank P values are shown in the right corner of each panel. Broken lines indicate CRBN expression levels below the median and solid lines indicate expression levels above the median. Remaining patients at risk are shown above the x-axis (PFS at 1, 2, 3, and 4 years and OS at 1, 2, 3, 4, and 5 years). The median CRBN expression was determined on the combined data of both thalidomide- and bortezomib-treated patients: 45 of 96 patients were below the median in the thalidomide set, whereas 50 of 95 were below the median in the bortezomib set.
CRBN expression was not associated with an upgrade of response, considered to be improvement of response during thalidomide maintenance (P = .3, supplemental Table 4). To determine whether CRBN expression was specifically relevant for the outcome of thalidomide treatment, we also examined the relationship between CRBN expression and survival in patients treated with bortezomib maintenance. No association was observed between CRBN expression and PFS/OS after bortezomib maintenance (Figure 1C-D). For validation of these results, the MRC-IX study was evaluated.17 Only 30 patients with gene expression were available who received thalidomide during maintenance but not during induction. This subset was too small to allow solid analysis of the relationship between CRBN expression and outcome after thalidomide maintenance. Finally, CRBN forms an E3 ubiquitin ligase complex with the proteins DDB1 and CUL4A.9 This complex has been suggested to be involved in the regulation of β-catenin activity, which in turn affects downstream targets such as CCND1 and C-MYC. CRBN was also found to bind to AMPKα1 (PRKAA1) and to the large conductance Ca2+-activated potassium channel KCNMA1.18 In a multivariate model with CRBN levels, only CCND1 and CRBN were found to be independently related to longer PFS (supplemental Table 3). A relationship with PFS was not found for either CCND1 or CRBN in the patients treated with bortezomib in the maintenance phase.
In conclusion, in the present study, we observed that higher expression of CRBN was associated with increased PFS during maintenance treatment with thalidomide, but not in patients with bortezomib maintenance. This corresponds well to the report of reduced CRBN expression in > 85% of MM patients who were lenalidomide resistant.10 Our observations warrant analysis of the predictive effect of CRBN expression in newly diagnosed and relapsed/refractory patients treated with IMiDs as part of induction and consolidation treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Yvonne de Knegt for technical support.
This trial was supported by the Dutch Cancer Foundation, the German Federal Ministry of Education and Research, and an unrestricted grant from Janssen-Cilag-Ortho Biotech. The GMMG study group received further grants to support this trial by Novartis, AMGEN, Chugai, MSCNET, and Roche.
Authorship
Contribution: A.B. and M.v.D. analyzed and interpreted the data and wrote the manuscript; R.K. collected, analyzed, and interpreted the data; B.v.d.H. performed the statistical analysis and collected the data; L.e.J., U.B., and S.Z. collected the data and managed the trial; A.B. collected the cytogenetic data; D.H. managed the trial and contributed essential materials; H.M.L. managed the trial; H.G. supervised the trial and contributed essential materials; and P.S. designed the research, supervised the trial, and wrote the manuscript.
Conflict-of-interest disclosure: S.Z. serves on the advisory board of Celgene. H.M.L. serves on advisory boards for Celgene and Genmab. H.G. serves on the advisory board for Johnson & Johnson. P.S. serves on advisory boards for Skyline Diagnostics, Janssen, and Celgene. The remaining authors declare no competing financial interests.
Correspondence: Prof P. Sonneveld, MD, PhD, Erasmus MC, Department of Hematology, Room L 407, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: p.sonneveld@erasmusmc.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal