Key Points
The frequency of CD161++ MAIT cells is dramatically decreased in the blood of HIV-infected patients, and they are nonrecoverable with HAART.
Gut sequestration and apoptosis in response to bacterial signals may, amongst others, be mechanisms that contribute to this.
Abstract
HIV infection is associated with immune dysfunction, perturbation of immune-cell subsets and opportunistic infections. CD161++CD8+ T cells are a tissue-infiltrating population that produce IL17A, IL22, IFNγ, and TNFα, cytokines important in mucosal immunity. In adults they dominantly express the semi-invariant TCR Vα7.2, the canonical feature of mucosal associated invariant T (MAIT) cells and have been recently implicated in host defense against pathogens. We analyzed the frequency and function of CD161++/MAIT cells in peripheral blood and tissue from patients with early stage or chronic-stage HIV infection. We show that the CD161++/MAIT cell population is significantly decreased in early HIV infection and fails to recover despite otherwise successful treatment. We provide evidence that CD161++/MAIT cells are not preferentially infected but may be depleted through diverse mechanisms including accumulation in tissues and activation-induced cell death. This loss may impact mucosal defense and could be important in susceptibility to specific opportunistic infections in HIV.
Introduction
The natural course of human immunodeficiency virus type 1 (HIV-1) infection is associated with progressive immune dysfunction, perturbation of immune-cell subsets and increased opportunistic infections. In early disease, there is a dramatic loss of CD4+ T cells from the gastrointestinal tract resulting in impaired mucosal immunity, reduced peripheral CD4+ T-cell count, and increased systemic T-cell activation.1-4 These factors contribute to an increased susceptibility to infection with specific organisms such as Mycobacterium tuberculosis and Candida albicans.5-7 In addition, more recent evidence suggests an important role for the loss of CD8+ T cells in susceptibility to bacterial pneumonia and all-cause mortality in HIV infection.8
MAIT cells are a distinct subset of tissue-infiltrating lymphocytes with antibacterial functions that account for up to one-third of the CD8+ T-cell population in the blood of healthy individuals.9-11 MAIT cells are identified by expression of a semi-invariant T-cell receptor (TCR), iVα7.2,10,12,13 which recognizes ligands presented by MHC class I related (MR1) protein.14 MR1 presentation occurs on dendritic cells, monocytes, and lung epithelial cells in response to bacterial pathogens.9,10,12 MAIT cells are activated in vitro in an MR1-dependent fashion by a range of bacterial and fungal pathogens, including Escherichia coli, M tuberculosis, and C albicans,9,10 and in mouse models have been shown to provide protection against bacterial infection.10,15 In addition, MAIT cells have been shown to be lost from the blood and present in the lungs of patients with active tuberculosis, suggesting they may play an important role in host immunity to M tuberculosis.9,10
Specific subsets of CD4+ and CD8+ T cells, termed Th17 and Tc17, are defined by their ability to produce IL17A and are important in the regulation of mucosal integrity and antibacterial immunity.16-20 Early in HIV infection, Th17 cells are lost from the gastrointestinal tract, but may be restored through long-term highly active antiretroviral therapy (HAART) concurrent with a reduction in immune activation levels.21 The loss of this IL17A and IL22-producing cell population during untreated HIV infection has been shown to impact on mucosal immunity and pathogen dissemination from the gut, suggesting a protective role for such cells in HIV infection.22
Recent work from our group and others has revealed an important intersection in humans between MAIT cells, Tc17 cell populations, and expression of the C-type lectin CD161.11,12 CD8+ T cells expressing high levels of CD161 exhibit a distinct phenotype associated with RORγt expression, accompanied by IL17/22 production.11 The CD161++CD8+ T-cell population is detectable in cord blood, where it exhibits a similar transcriptional profile to that of adult cells; expansion and narrowing of TCR usage toward a Vα7.2+ MAIT phenotype occurs with age.12,13 Thus in adults the CD161++CD8+ T-cell population is dominated by cells bearing the canonical Vα7.2 TCR (MAIT; 80%-90%).12,13 In this study, for simplicity, we have described this CD161++CD8+ T-cell population as CD161++/MAIT cells, although minority MAIT cell populations exist which do not express CD8α.10,23
The in vivo significance of CD161++/MAIT cells is not yet known but the potential impact on AIDS pathogenesis is substantial. Their prominent homing capacity for mucosal tissue suggests they may play a role in mucosal defense against pathogens. Furthermore, CD161++/MAIT cells may play a role in intestinal defense, where loss in HIV may be relevant to bacterial translocation and immune activation.
We hypothesized that the CD161++/MAIT population would be disturbed in HIV infection, linked to enhanced susceptibility to specific mucosal infections. We provide evidence for a major, early, and sustained impact of HIV on this unique and prominent T-cell subset.
Methods
Cohorts
Healthy donors (n = 23) were non–HIV-tested laboratory volunteers and leukocyte cones supplied by the local health authority. Patients chronically infected with HIV were from the King's College London Infectious Diseases Biobank (n = 13) or the Thames Valley cohort (n = 10) and were sampled between 1 and 15 years after diagnosis. Patients with early HIV infection were randomly chosen baseline samples from the SPARTAC cohort (n = 35). A separate cohort of treatment-naive patients from the Swiss HIV Cohort Study (n = 29) were sampled before, 1 and 2 years into HAART. Induction of apoptosis was assessed in samples from the Swiss-Spanish Intermittent Treatment Trial (SSITT; n = 10). All HIV+ patients were HCV and HBV-negative at the time of sampling.
Colon biopsies (30cm from the anal margin) were collected from HIV+ patients at the Chelsea and Westminster Hospital (n = 12) who were undergoing colonoscopy as part of their routine clinical care. Indications for colonoscopy were diarrhea (n = 5), rectal bleeding (n = 3), abdominal pain (n = 2), iron deficiency anemia (n = 1), and surveillance of polyps (n = 1). The colon of all patients had a normal endoscopic appearance. Four patients were taking HAART at the time of biopsy and 7 had nonspecific colitis/inflammatory infiltrate (referred to in the text as nonspecific colitis), which is common in HIV infection.24-26 Normal noninflamed surgically resected tissue from the rectosigmoid or descending colon of age-matched controls was obtained from the Oxford Pathology/GI Biobank.
Gut specimens for isolation of lamina propria mononuclear cells (LPMCs) were taken from patients recruited from the Gastroenterology Unit and Colorectal Surgery Department at the John Radcliffe Hospital, Oxford. All tissue samples were taken from macroscopically healthy and noninflamed colon of colorectal cancer or IBD patients undergoing surgery.
The clinical data of the HIV-infected patients used in this study are provided in supplemental Tables 1 through 3 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry
Whole blood was either stained directly and the erythrocytes lysed with BD FACS lysing solution (BD Bioscience) before analysis or peripheral blood mononuclear cells (PBMCs) were isolated using Lymphoprep (AxisShield). LPMCs were isolated as previously described.27 For intracellular staining, PBMCs were then stimulated with PMA (250 ng/mL) and ionomycin (500 ng/mL) for 6 hours or left unstimulated. Brefeldin A (Sigma-Aldrich) was added at 1 μg/mL 5 hours before the end of stimulation.
All antibodies were from BD Bioscience unless otherwise indicated. Dead cell were excluded with Near-IR Dead Cell Stain (Invitrogen). Antibodies used were: CD3 Pacific Orange (UCHT1, Invitrogen) or eFluor605 (OKT3, eBioscience), CD4 eFluor650 (eBioscience), Alexafluor700 (RPA-T4), QDot605 (S3.5, Invitrogen) or PECy-7 (L200), CD8 PerCP, PECy-7 (SK1) or V450 (RPA-T8), CD45 Alexafluor700 (HI30, Biolegend), CD56 PECy-7 (B159), CD69 FITC (FN50, eBioscience), CD161 PE, APC (191B8, Miltenyi Biotech) or PECy-7 (HP3G10, eBioscience), TCR Vα7.2 FITC, PE or APC (3C10, BioLegend), IFNγ FITC (4S.B3), IL17A PE (eBio64CAP17, eBioscience), IL22 PerCP-eFluor710 (22URT1, eBioscience), CCR5 PE (2D7/CCR5), CXCR4 PECy-7 (12G5), and CCR6 PerCPCy-5.5 or PECy7 (11A9), activated capsase-3 PE (C92-605), CD95 PECy7 (DX2, Biolegend), TNFRI PE (16 803, R&D Systems), TNFRII FITC (22 235, R&D Systems), CD261 Alexafluor488 (DR-4-02, Serotec), CD262 PE (DJR2-4 [7-8], Biolegend), Bcl-2 FITC (Bcl2/100), and anti–KC57-RD1 PE (FH190-1-1; Beckman Coulter). For proliferation assays, PBMCs were stained with CellTrace Violet (Invitrogen) as per the manufacturer's instructions.
Data were collected on an LSRII flow cytometer (BD Biosciences) or a MACSQuant (Miltenyi Biotec) and analyzed using FlowJo Version 9.3.1 (TreeStar).
Immunohistochemistry
Immunohistochemistry was performed on 5-μm thick sections of formalin-fixed, paraffin-embedded tissues. Heat-induced antigen retrieval was performed using a pressure cooker (The Retriever, Electron Microscopy Sciences) and R-Buffer A (lipolysaccharide) or B (MDR-1, CD3, CD8; Electron Microscopy Sciences). Endogenous peroxidase activity was quenched with 3% hydrogen peroxide and 0.13% sodium azide (both Sigma-Aldrich), and sections blocked with 0.5% blocking reagent (Perkin Elmer). Primary antibodies included anti–MDR-1 (5A12.2, mouse IgG2b, Merck Millipore), anti-CD3 (F7.2.38, mouse IgG1, Dako) anti-CD8 (rabbit polyclonal, Abcam), anti-lipopolysaccharide (LPS) core (WN1 222-5, mouse IgG2A, Hycult Biotech), and isotype-matched controls. For immunofluorescent staining, samples were stained sequentially, initially for MDR-1 (detected with peroxidase-conjugated donkey anti–mouse IgG secondary (Jackson ImmunoResearch Laboratories), and then for CD3 and CD8 (detected sequentially with peroxidase-conjugated donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories), and peroxidase-conjugated goat anti–mouse IgG1 (Invitrogen) secondaries. Tyramide signal amplification, with TSA-plus Cy5, Cy3, and FITC reagents (PerkinElmer), was used to visualize staining of MDR-1, CD8, and CD3, respectively. Samples were reblocked with hydrogen peroxide and sodium azide between each stain. Controls for peroxidase blocking were included in all experiments. Slides were mounted with Prolong Gold with DAPI (Invitrogen) and imaged at room temperature on a FluoView FV1000 confocal microscope (Olympus) using a 40×/1.30 Oil UPlan FLN objective lens. Images were acquired using FV10-ASW software. For LPS, staining was detected with N-Histofine simple stain MAX PO (M; Nichirei) and ImmPACT DAB peroxidase substrate, samples counterstained with hematoxylin QS and mounted with VectaMount permanent mounting medium (all Vector Labs). Images were collected with a Nikon Coolscope Slide Scanner (Nikon). Further detail is provided in supplemental Methods.
Immunofluorescent images were analyzed with CellProfiler 2.0 and CellProfiler Analyst 2.0 software (www.cellprofiler.org; Broad Institute).28 Further detail is provided in supplemental Methods. LPS+ cells in the lamina propria were manually counted from randomly collected 20× magnification images; a median of 9 images was counted per slide (range 6-14). Surface area analyzed, excluding the intestinal lumen, was calculated in ImageJ Version 10.2 (National Institutes of Health).
In vitro stimulation of PBMCs
E coli (DH5α) was fixed in 1% paraformaldehyde for 5 minutes, and washed extensively. A negative control was prepared in identical fashion. PBMCs were cultured for 20 hours, 44 hours, or 6 days (as noted in the appropriate figure legends) with either paraformaldehyde-fixed E coli at the indicated bacteria per cell (BpC) ratio or the negative control. In some experiments, blocking antibodies against MR1 (Clone 26.5) or CD95 (ZB4, Millipore), appropriate isotype controls or recombinant human TNFRI/TNFRSF1A Fc chimera (R&D Systems) were used. Alternatively, PBMCs were activated with PMA (50 ng/mL) and ionomycin (250 ng/mL) for 20 hours or with anti-CD2/CD3/CD28 microbeads (T-cell Activation/Expansion Kit, Miltenyi Biotec) for 20 or 44 hours.
In vitro PBMC infection with HIV
PBMCs were activated with phytohemagglutinin (PHA, 1 μg/mL), recombinant human (rh)IL2 (25 ng/mL) and rhIL7 (20 ng/mL; all Sigma-Aldrich) for 72 hours. Activated cells were incubated with no virus, a CCR5-tropic HIV strain (JR-CSF) or a CXCR4-tropic strain (MN) at a multiplicity of infection (MOI) of 10 and were cultured in R10 for 9 days. RhIL2 (20 ng/mL) and rhIL7 (20 ng/mL) were further added to the cultures on days 0, 3, and 6, and cell samples were harvested and stained for p24 expression on days 6 and 9.
Statistical analysis
Statistics were performed using GraphPad Prism Version 5.0a. Normal distribution was determined using a Kolmogorov-Smirnov test. Differences between groups were analyzed using student t tests, Mann-Whitney tests, or ANOVA with Tukey or Dunnett multiple comparison tests or a test for linear trend for posthoc analysis or Kruskal-Wallis test with Dunn posthoc analysis where appropriate. Pearson correlation coefficients or Spearman correlation coefficients were calculated where appropriate. All tests were 2-tailed. All data are presented as means unless otherwise indicated. Statistical significance was accepted at P < .05.
Ethics
Samples were taken after written informed consent was given by the participants and/or their legal guardians in accordance with the Declaration of Helsinki and after approval from the relevant Ethical Review Boards at all sites.
Results
E coli activates CD161++CD8+ T cells in a PBMC environment
The majority (∼ 90%) of CD161++CD8+ T cells express TCR Vα7.2 and are thus defined as MAIT cells.13,29 In contrast, a smaller fraction of peripheral CD8+ T cells express lower levels (+) of CD161; this population is phenotypically, functionally, and transcriptionally distinct from the CD161++ population, bearing a diverse range of TCRs.11,13 We used the high expression of CD161 on CD8+ T cells (CD161++CD8+ T cells) to define the CD161++/MAIT cell population.
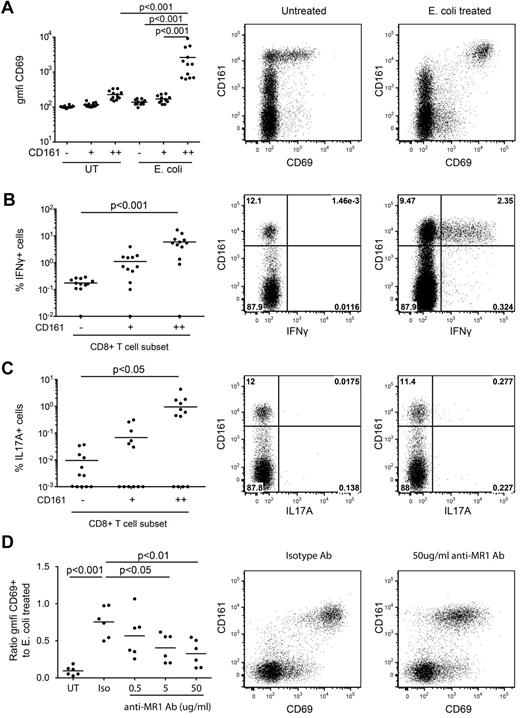
CD161++/MAIT cells isolated from healthy subjects can be activated by monocytes pre-exposed to a range of bacteria and yeast, triggering up-regulation of the activation marker CD69 and production of IFNγ in an MR1-dependent manner.10 Similar findings have been reported with MAIT cell lines.9 We confirmed this using unsorted PBMCs. PBMCs from healthy donors were exposed to paraformaldehyde-fixed E coli for 20 hours and cytokine production and CD69 expression assessed. Consistent with previous reports we observed dose-dependent activation of the CD161++CD8+ T-cell population (Figure 1A-C, supplemental Figure 1). E coli–induced activation was restricted to CD161++CD8+ T cells. Blocking of MR1 with specific antibody inhibited E coli–induced activation in a dose-dependent manner as measured by CD69 expression (Figure 1D) and IFNγ production (not shown), confirming that in our system, activation of CD161++CD8+ T cells by E coli exposure was MR1-dependent, consistent with previous studies.10 Similar results were obtained when gated on Vα7.2+CD161++CD8+ T cells (data not shown).
MR1-dependent activation of CD161++CD8+ T cells by E coli. Healthy isolated PBMCs were exposed to E coli overnight and analyzed for CD69 expression and IFNγ and IL17A production by CD8+ T cells as described in “Methods.” (A) There was a significant increase in the cell surface expression of CD69 on the CD161++ population in response to E coli exposure compared with the mock-infected control and to the CD161+ and CD161−E coli–exposed populations (n = 12). (B-C) A greater proportion of the E coli–exposed CD161++ population produced IFNγ or IL17A than the CD161− population (n = 12). Note data points lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.01% or 0.001% so as to appear on the log scale). (D) Increasing concentrations of anti-MR1 blocking antibody reduced the level of activation of the CD161++CD8+ T cells compared with the isotype antibody (n = 6). “Activation” on the y-axis is measured as a ratio of the geometric mean fluorescent intensity of CD69 on the antibody-treated E coli–exposed CD161++CD8+ T cells compared with that of the E coli–exposed CD161++CD8+ T cells. Plots are gated on CD8+ T cells.
MR1-dependent activation of CD161++CD8+ T cells by E coli. Healthy isolated PBMCs were exposed to E coli overnight and analyzed for CD69 expression and IFNγ and IL17A production by CD8+ T cells as described in “Methods.” (A) There was a significant increase in the cell surface expression of CD69 on the CD161++ population in response to E coli exposure compared with the mock-infected control and to the CD161+ and CD161−E coli–exposed populations (n = 12). (B-C) A greater proportion of the E coli–exposed CD161++ population produced IFNγ or IL17A than the CD161− population (n = 12). Note data points lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.01% or 0.001% so as to appear on the log scale). (D) Increasing concentrations of anti-MR1 blocking antibody reduced the level of activation of the CD161++CD8+ T cells compared with the isotype antibody (n = 6). “Activation” on the y-axis is measured as a ratio of the geometric mean fluorescent intensity of CD69 on the antibody-treated E coli–exposed CD161++CD8+ T cells compared with that of the E coli–exposed CD161++CD8+ T cells. Plots are gated on CD8+ T cells.
CD161++CD8+ T cells are lost from the blood in early HIV infection
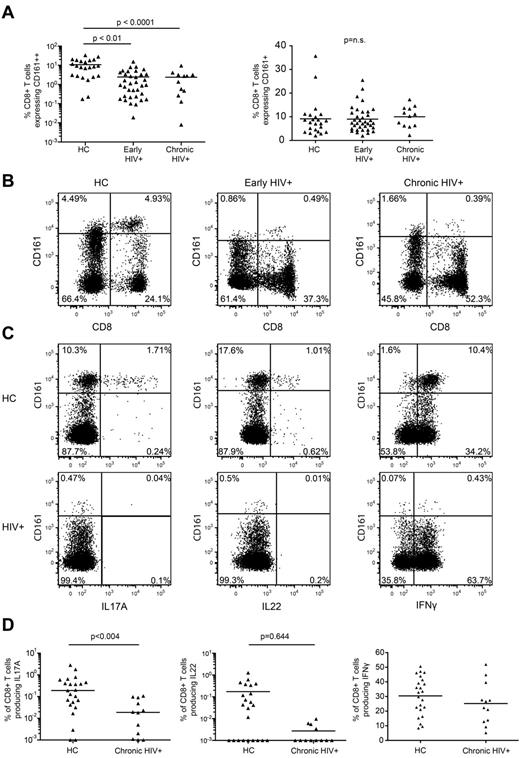
To assess the effect of HIV infection on CD161++/MAIT cells we first compared the frequency of CD161-expressing CD8+ T-cell populations in the peripheral blood of patients recently infected with HIV, patients chronically infected who were not currently undergoing treatment, and healthy controls (supplemental Table 1). The frequency of CD8+ T cells expressing CD161++ was markedly reduced in patients with early (2.44%; P < .0001) or chronic (2.38%; P < .01) HIV infection versus healthy controls (10.74%; Figure 2A-B). This was observed in both frozen PBMCs and freshly prepared whole blood (supplemental Figure 2A-B). The frequency of CD161++CD8+ T cells did not correlate with either viral load or CD4 count (supplemental Figure 2C-D). Some overlap between the frequency of CD161++CD8+ T cells in healthy controls and HIV-infected patients was noted, with normal or near normal frequencies in a minority of HIV-infected individuals and low frequencies in some healthy controls. No changes were noted in the non-MAIT CD161+(mid) CD8+ T-cell populations (Figure 2A).
Loss of CD161++/MAIT cells in HIV infection. Isolated PBMCs from healthy controls (n = 23) and patients with early (n = 35) or chronic stage HIV infection (n = 13) were stained for CD161 expression on CD8+ T cells. (A) There was a lower frequency of CD161++ cells in early and chronic HIV infection compared with the healthy control cohort, with no differences seen in the CD161+ populations. Results are displayed as a proportion of the CD8+ T cell population. (B) Representative FACS plots showing differences in the frequency of CD161++CD8+ T cells. Plots are gated on CD3+ cells. (C-D) PBMCs from the healthy control and chronic HIV infection cohorts were stimulated with PMA/ionomycin as described in “Methods.” The CD8+ T cells were analyzed for production of IFNγ, IL17A, and IL22. The gating strategy is shown in supplemental Figure 9a. (C) Representative FACS plots of PMA and ionomycin stimulated PBMCs in HIV+ patients and healthy subjects. Plots are gated on CD8+ T cells. (D) There were lower frequencies of IL17A producing cells in the HIV+ patients but no difference in the frequency of IFNγ or IL22 producing cells. Note data points lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.001% so as to appear on the log scale).
Loss of CD161++/MAIT cells in HIV infection. Isolated PBMCs from healthy controls (n = 23) and patients with early (n = 35) or chronic stage HIV infection (n = 13) were stained for CD161 expression on CD8+ T cells. (A) There was a lower frequency of CD161++ cells in early and chronic HIV infection compared with the healthy control cohort, with no differences seen in the CD161+ populations. Results are displayed as a proportion of the CD8+ T cell population. (B) Representative FACS plots showing differences in the frequency of CD161++CD8+ T cells. Plots are gated on CD3+ cells. (C-D) PBMCs from the healthy control and chronic HIV infection cohorts were stimulated with PMA/ionomycin as described in “Methods.” The CD8+ T cells were analyzed for production of IFNγ, IL17A, and IL22. The gating strategy is shown in supplemental Figure 9a. (C) Representative FACS plots of PMA and ionomycin stimulated PBMCs in HIV+ patients and healthy subjects. Plots are gated on CD8+ T cells. (D) There were lower frequencies of IL17A producing cells in the HIV+ patients but no difference in the frequency of IFNγ or IL22 producing cells. Note data points lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.001% so as to appear on the log scale).
Loss of Tc17 cells in HIV infection
Consistent with previous work,11 IL17A production within the CD8+ T-cell population was primarily restricted to the CD161++CD8+ T-cell population whereas IL22 and IFNγ production was observed in the CD161++, CD161+, and CD161− populations (Figure 2D). In agreement with the disappearance of the CD161++CD8+ T-cell population from the circulating blood of HIV+ patients, a lower frequency of Tc17 cells in chronically infected patients versus healthy controls was observed (0.034% versus 0.41%; P = .004). There was no difference in the frequency of IL22 or IFNγ-producing CD8+ T cells (Figure 2C-D).
CD161++CD8+ T cells express tissue homing markers
To determine the homing capacity of CD161++/MAIT cells, we examined expression of the tissue-homing marker CCR6, as well as the HIV entry coreceptor molecules CCR5 and CXCR4, on CD161++CD8+ T cells from healthy controls and patients with chronic HIV infection.
In healthy controls, the majority of CD161++CD8+ T cells (∼ 85%) expressed high levels of CCR6 and CCR5 with little CXCR4 expression evident (supplemental Figure 3A). compared with healthy controls, HIV+ patients had a lower frequency of circulating CCR6+CD8+ T cells (2.21% vesus 16.43%; P < .001) with no difference in the frequency of CCR5+ or CXCR4+CD8+ T cells. There was no difference in the frequencies of circulating CCR6+CD4+ T cells (supplemental Figure 3C).
Analysis of CD161++/MAIT cells in the colon in HIV infection
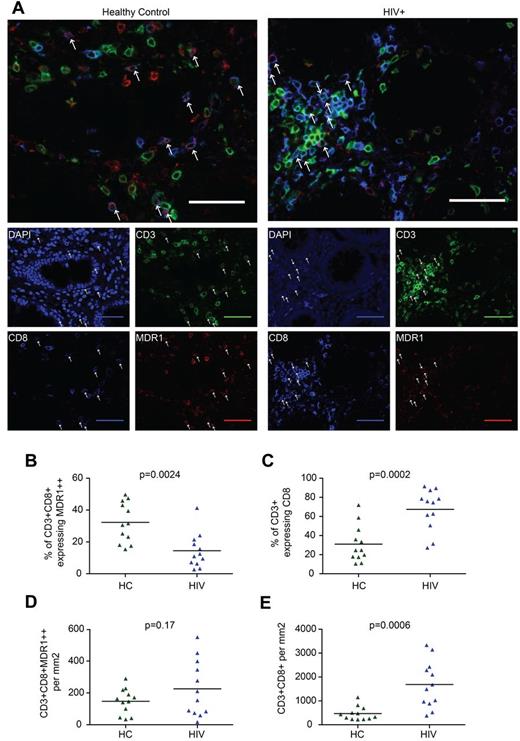
Given the expression of tissue homing receptors by CD161++/MAIT cells, their steady-state enrichment in the gut,10,14 their tendency to traffic to inflamed tissues,11 and the inflammatory response seen in the gut in HIV infection,1,2,26,30 we investigated the relationship between loss of CD161++/MAIT cells from the blood in HIV infection and trafficking to and retention in the gut. Colon biopsies from 12 HIV-infected patients were compared with tissue from age-matched controls (supplemental Table 3). Because of technical limitations in staining of formalin-fixed paraffin-embedded tissue with surface markers such as CD161, Vα7.2, IL18R, and CCR6, CD161++/MAIT cells were characterized by the coexpression of CD3, CD8, and high-level expression of the multi-drug efflux pump, MDR1 (Figure 3A).12,31 Although MDR1 expression is not uniquely linked to CD161++ status by flow cytometry, high levels of MDR1 are associated with CD161++CD8+ T cells in the intestine (supplemental Figure 4A-C). In addition, MDR1 expression has previously been linked to CD161++/MAIT cells at the levels of transcription, protein expression, and function.11-13,31 Although we cannot exclude contamination of the MDR1++CD8+CD3+ population with non-CD161++/MAIT cells, the frequency of CD3+CD8+MDR1++ cells in healthy controls (median 8.0% of CD3+ cells, range 2.0%-22.9%) was consistent with previous estimates of MAIT cell frequency in the intestine.12
CD161++/MAIT cell analysis in the colon in HIV. Sections of colon from HIV-infected patients (n = 12) or controls (n = 12) were stained for CD3, CD8, and MDR-1. CD161++/MAIT cells were defined as CD3+CD8+MDR1++. (A) Representative images from control (left) and HIV-infected tissue (right) are shown. Arrows indicate CD3+CD8+MDR1++ cells. Scale bars, 50 μm. (B) The proportion of CD3+CD8+ cells expressing high (++) levels of MDR1 was reduced in HIV infection. (C) The proportion of CD3+ cells expressing CD8 was increased in HIV infection. (D) No significant difference was seen in the number of CD3+CD8+MDR1++ cells per mm2. (E) The number of CD3+CD8+ cells per mm2 was increased in HIV infection.
CD161++/MAIT cell analysis in the colon in HIV. Sections of colon from HIV-infected patients (n = 12) or controls (n = 12) were stained for CD3, CD8, and MDR-1. CD161++/MAIT cells were defined as CD3+CD8+MDR1++. (A) Representative images from control (left) and HIV-infected tissue (right) are shown. Arrows indicate CD3+CD8+MDR1++ cells. Scale bars, 50 μm. (B) The proportion of CD3+CD8+ cells expressing high (++) levels of MDR1 was reduced in HIV infection. (C) The proportion of CD3+ cells expressing CD8 was increased in HIV infection. (D) No significant difference was seen in the number of CD3+CD8+MDR1++ cells per mm2. (E) The number of CD3+CD8+ cells per mm2 was increased in HIV infection.
We observed that the proportion of CD3+CD8+ cells expressing MDR1++ was reduced in HIV-infected patients versus controls (14.5% versus 32.3%, P = .0024; Figure 3B). However, the number of CD3+CD8+MDR1++ cells/mm2 was not significantly different between groups (226 versus 147, P = .17; Figure 3D). This was probably because of enrichment of CD3+CD8+ cells, both as a proportion of CD3+ cells (67.5% versus 31.0%, P = .0002; Figure 3C), and in absolute numbers (1687 versus 469/mm2, P = .0006; Figure 3E, supplemental Figure 5A-B). Furthermore, although not statistically significant, both the number of CD3+CD8+MDR1++ and CD3+CD8+ cells/mm2 were lower in patients on HAART and in patients without nonspecific colitis (supplemental Figure 5C-F). Although there are methodologic constraints in this approach, including variation in the exact site of biopsy or resection and the small area of colon assessed in both the HIV-infected (median = 0.99/mm2) and control samples (median = 1.54/mm2), our results were consistent with those of others which suggest an overall increase in CD8+ T-cell numbers in the gut during HIV infection.1,3,4,32 Overall, despite the clear loss of CD161++/MAIT cells from blood, they are still detectable in gut, as judged by analysis of CD8+CD3+MDR1++ cells.
CD161++/MAIT cells do not recover in the periphery during treatment
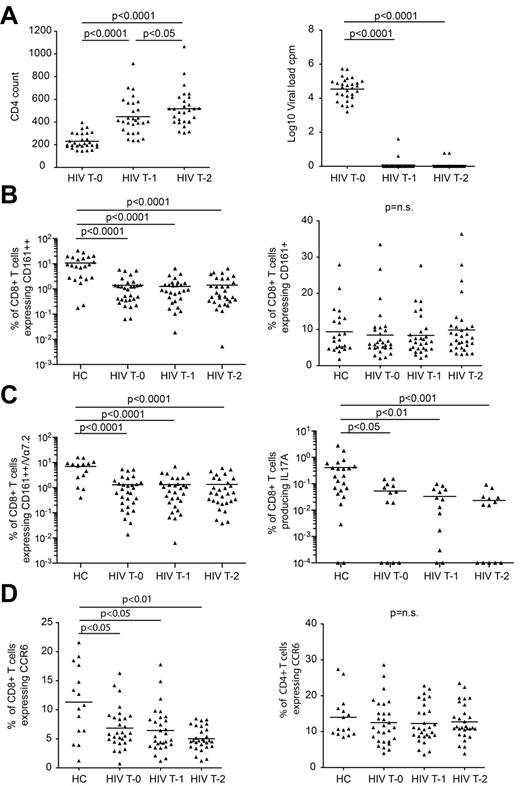
To assess whether MAIT cells are restored through HAART, we analyzed longitudinal PBMC samples from 29 treatment-naive patients enrolled in the Swiss HIV Cohort Study (supplemental Table 2). Samples were collected the week before starting treatment, and at 1 and 2 years after starting treatment. All patients showed complete suppression of viral load and recovery of CD4+ T cells with HAART (supplemental Table 2; Figure 4A).
CD161++/MAIT cells fail to recover with HAART. Patients with chronic-stage HIV infection were followed for 2 years of HAART, with samples taken before the start of treatment (T0), then at 1 year (T1) and 2 years (T2) into treatment (n = 29). PBMCs were stained for markers of interest including CD161, TCR Vα7.2 and CCR6, and a random selection were stimulated with PMA and ionomycin to analyze the production of IFNγ, IL17A and IL22 (n = 13). PBMCs from healthy controls were stained for surface markers of interest including CD161 (n = 23), TCR Vα7.2 (n = 15), and CCR6 (n = 15) or were stimulated with PMA and ionomycin to analyze the production of IFNγ, IL17A and IL22 (n = 23). (A) All HIV+ patients showed complete suppression of viral load and recovery of CD4+ T cells over the course of treatment. (B) There were no changes in the frequency of CD161++CD8+ T cells or CD161+CD8+ T cells during treatment. (C) CD161++Vα7.2+CD8+ T cells and IL17A-producing CD8+ T cells failed to recover over the course of treatment. Note data points in plots (B) and (C) lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.001% or 0.0001% so as to appear on the log scale). (D) There was a lower frequency of CCR6+CD8+ T cells in the HIV+ cohort at all treatment time points compared with healthy controls, and this population of cells failed to recover over the course of treatment. There was no difference in the frequency of CCR6+CD4+ T cells between healthy controls and the HIV+ cohort at any of the treatment time points, and no change in the frequency of these cells over the course of treatment.
CD161++/MAIT cells fail to recover with HAART. Patients with chronic-stage HIV infection were followed for 2 years of HAART, with samples taken before the start of treatment (T0), then at 1 year (T1) and 2 years (T2) into treatment (n = 29). PBMCs were stained for markers of interest including CD161, TCR Vα7.2 and CCR6, and a random selection were stimulated with PMA and ionomycin to analyze the production of IFNγ, IL17A and IL22 (n = 13). PBMCs from healthy controls were stained for surface markers of interest including CD161 (n = 23), TCR Vα7.2 (n = 15), and CCR6 (n = 15) or were stimulated with PMA and ionomycin to analyze the production of IFNγ, IL17A and IL22 (n = 23). (A) All HIV+ patients showed complete suppression of viral load and recovery of CD4+ T cells over the course of treatment. (B) There were no changes in the frequency of CD161++CD8+ T cells or CD161+CD8+ T cells during treatment. (C) CD161++Vα7.2+CD8+ T cells and IL17A-producing CD8+ T cells failed to recover over the course of treatment. Note data points in plots (B) and (C) lie on the x-axis (values of 0% were arbitrarily ascribed a value of 0.001% or 0.0001% so as to appear on the log scale). (D) There was a lower frequency of CCR6+CD8+ T cells in the HIV+ cohort at all treatment time points compared with healthy controls, and this population of cells failed to recover over the course of treatment. There was no difference in the frequency of CCR6+CD4+ T cells between healthy controls and the HIV+ cohort at any of the treatment time points, and no change in the frequency of these cells over the course of treatment.
We confirmed the loss of CD161++CD8+ T cells (1.4% versus 10.74%; P < .0001), CCR6+CD8+ T cells (6.85% versus 11.32%; P < .05), and Tc17 cells (0.05% versus 0.41%; P < .05) in the pretreatment samples versus healthy controls (Figure 4B-D). We found similar losses when gating on the Vα7.2+CD161++CD8+ T-cell subset (Figure 4C). Of note, normal or near normal frequencies were again seen in a minority of HIV-infected individuals.
Analysis over the 2-year period of treatment showed no changes in the frequency of classic Vα7.2+MAIT cells, CD161++CD8+ T cells, CCR6+CD8+ T cells, or Tc17 cells which remained significantly lower than those of the healthy controls at all time points (P < .05; Figure 4). Therefore CD161++/MAIT cells are not recovered by HAART.
HIV does not preferentially infect CD161++/MAIT cells
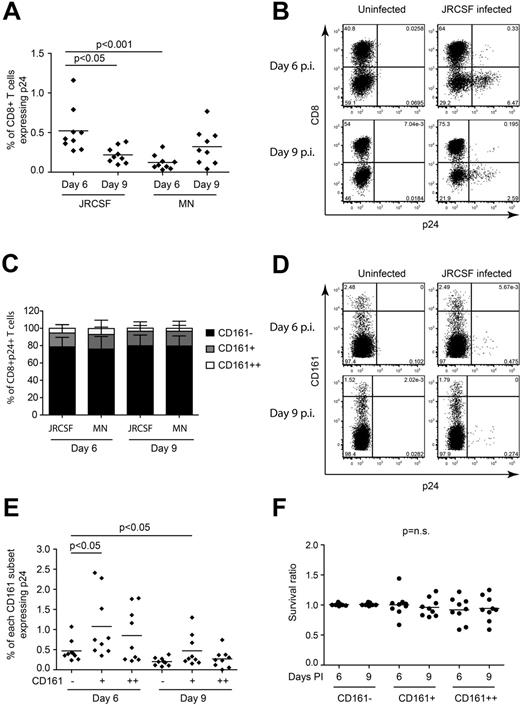
Infection of CD8+ T cells by HIV was previously reported.24,33-35 Given the expression of the entry coreceptor CCR5, we hypothesized that infection could contribute to CD161++/MAIT cell depletion. PBMCs from healthy donors were infected with either a CCR5-tropic (JR-CSF) or CXCR4-tropic (MN) strain of HIV in vitro. At both days 6 and 9 after infection, we observed infection of CD8+ T cells (Figure 5A-B). With both virus strains at both time points the considerable majority of infected CD8+ T cells were CD161−, with the fewest infected cells found in the CD161++ subset (Figure 5C-D). When considering the frequency of infection within each of the CD161 populations, it was not higher in the CD161++/MAIT cell population than the CD161− or CD161+ populations (Figure 5D-E). No loss of CD161++/MAIT cells occurred in the infected cultures relative to the uninfected control (Figure 5F). Direct exposure of PBMCs to HIV for 20 hours in vitro failed to activate CD161++/MAIT cells (data not shown).
CD161++/MAIT cells are not preferentially infected in vitro. PBMCs from healthy subjects (n = 9) were activated with PHA, cultured in rhIL2 and rhIL7, and infected with a CCR5-tropic virus (JR-CSF) or CXCR4 tropic virus (MN) at an MOI of 10. FACS analysis was performed on days 6 and 9 after infection as described in “Methods.” The gating strategy is shown in supplemental Figure 9B through D. (A) CD8+ T cells were infected at a low frequency by both viruses, with the greatest frequency of p24+ cells observed on day 6 after infection with JR-CSF. (B) Representative FACS plots of CD3+ lymphocytes from day 6 after infection with JR-CSF. (C) When gating on the total p24+CD8+ T cells, the majority of the infected CD8+ T cells were found in the non-MAIT CD161−CD8+ T-cell population on both day 6 and day 9 after infection with JR-CSF virus. (D) Representative FACS plots of p24 staining of CD8+ T cells mock infected or infected with JR-CSF. (E) Comparison of the frequency of infection within the CD161++/MAIT, and non-MAIT CD161+ and CD161−CD8+ T-cell populations. CD161++/MAIT cells were infected, but not more frequently than either the non-MAIT CD161− or CD161+CD8+ T-cell populations on either day 6 or day 9 after infection with JR-CSF virus. (F) There was no significant difference in the survival of the cells in the HIV infected cultures compared with the uninfected cultures. Cell survival ratio was calculated by normalizing the frequency of CD161++/MAIT cells in the infected culture to that of the uninfected culture.
CD161++/MAIT cells are not preferentially infected in vitro. PBMCs from healthy subjects (n = 9) were activated with PHA, cultured in rhIL2 and rhIL7, and infected with a CCR5-tropic virus (JR-CSF) or CXCR4 tropic virus (MN) at an MOI of 10. FACS analysis was performed on days 6 and 9 after infection as described in “Methods.” The gating strategy is shown in supplemental Figure 9B through D. (A) CD8+ T cells were infected at a low frequency by both viruses, with the greatest frequency of p24+ cells observed on day 6 after infection with JR-CSF. (B) Representative FACS plots of CD3+ lymphocytes from day 6 after infection with JR-CSF. (C) When gating on the total p24+CD8+ T cells, the majority of the infected CD8+ T cells were found in the non-MAIT CD161−CD8+ T-cell population on both day 6 and day 9 after infection with JR-CSF virus. (D) Representative FACS plots of p24 staining of CD8+ T cells mock infected or infected with JR-CSF. (E) Comparison of the frequency of infection within the CD161++/MAIT, and non-MAIT CD161+ and CD161−CD8+ T-cell populations. CD161++/MAIT cells were infected, but not more frequently than either the non-MAIT CD161− or CD161+CD8+ T-cell populations on either day 6 or day 9 after infection with JR-CSF virus. (F) There was no significant difference in the survival of the cells in the HIV infected cultures compared with the uninfected cultures. Cell survival ratio was calculated by normalizing the frequency of CD161++/MAIT cells in the infected culture to that of the uninfected culture.
E coli exposure triggers apoptosis of CD161++/MAIT cells
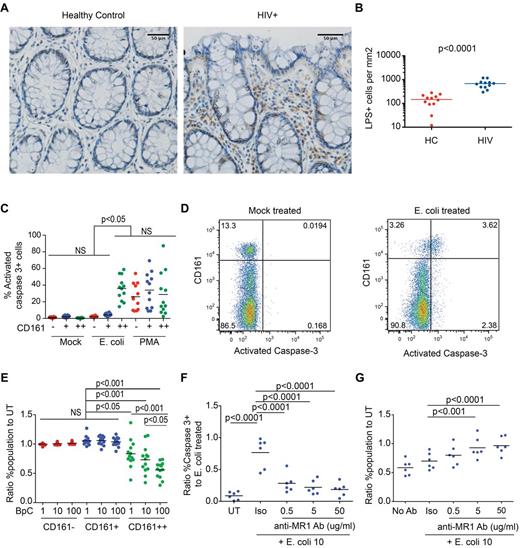
Microbial translocation has been suggested to occur in HIV infection after compromise of intestinal epithelial integrity.25,36,37 Given that CD161++CD8+ T cells are specifically activated by bacteria, and are present in gut tissue (Figures 1,Figure 2–3 and supplemental Figure 4) we tested whether bacterial antigen was present in gut tissue in HIV infection in vivo, and what the impact of antigen encounter on CD161++/MAIT cells was in vitro. Staining of colon sections from HIV+ patients for LPS demonstrated frequent LPS+ cells in the lamina propria and, in some sections, cell-free LPS (Figure 6A-B, supplemental Figure 6A-B); this is consistent with recent findings in SIV-infected rhesus macaques.25
E coli exposure triggers apoptosis and destruction of CD161++/MAIT cells in vitro. (A-B) Sections of colon from HIV-infected patients (n = 12) or controls (n = 12) were stained for lipopolysaccharide (LPS). (A) Representative images from control (left) and HIV-infected tissue (right) are shown. Scale bars, 50 μm. (B) Significantly more LPS+ cells were seen in the lamina propria in HIV infection. The number of LPS+ cells in the lamina propria was determined by manual counting as outlined in “Methods.” (C-G) PBMCs from healthy subjects (n = 12) were activated with PFA-fixed E coli at a bacteria per cell (BpC) ratio of 1, 10, or 100 or mock-treated and were analyzed after 20 hours incubation. (C) E coli–exposed CD161++CD8+ cells (BpC of 10) had higher frequencies of activated caspase-3–positive cells versus mock-treated cells or E coli–exposed CD161− and CD161+ cell populations. (D) Representative FACS plots, gated on CD8+ T cells, comparing activated caspase-3 expression in mock-treated and E coli–exposed PBMC cultures. (E) At all BpC of E coli, there was a reduction in survival of the CD161++CD8+ T-cell population. No significant differences were noted in the CD161+ or the CD161−CD8+ T-cell populations. Cell survival was determined by normalizing the frequency of the cell population of interest in the E coli–exposed culture to that in the mock-treated culture. A value of 1 is equivalent to 100% survival. (F) Anti-MR1–blocking antibody reduced the frequency of activated caspase-3–positive CD161++CD8+ T cells compared with the isotype control. Blocking of E coli–induced apoptosis was determined by normalizing the frequency of activated caspase 3+ CD161++C8+ T cells treated as indicated to that of the culture exposed to E coli alone. (G) Anti-MR1–blocking antibody increased the survival of CD161++CD8+ T cells in the E coli–exposed culture. Cell survival was determined by normalizing the frequency of CD161++CD8+ T cells in the antibody treated, E coli–exposed culture to that of the mock-treated culture.
E coli exposure triggers apoptosis and destruction of CD161++/MAIT cells in vitro. (A-B) Sections of colon from HIV-infected patients (n = 12) or controls (n = 12) were stained for lipopolysaccharide (LPS). (A) Representative images from control (left) and HIV-infected tissue (right) are shown. Scale bars, 50 μm. (B) Significantly more LPS+ cells were seen in the lamina propria in HIV infection. The number of LPS+ cells in the lamina propria was determined by manual counting as outlined in “Methods.” (C-G) PBMCs from healthy subjects (n = 12) were activated with PFA-fixed E coli at a bacteria per cell (BpC) ratio of 1, 10, or 100 or mock-treated and were analyzed after 20 hours incubation. (C) E coli–exposed CD161++CD8+ cells (BpC of 10) had higher frequencies of activated caspase-3–positive cells versus mock-treated cells or E coli–exposed CD161− and CD161+ cell populations. (D) Representative FACS plots, gated on CD8+ T cells, comparing activated caspase-3 expression in mock-treated and E coli–exposed PBMC cultures. (E) At all BpC of E coli, there was a reduction in survival of the CD161++CD8+ T-cell population. No significant differences were noted in the CD161+ or the CD161−CD8+ T-cell populations. Cell survival was determined by normalizing the frequency of the cell population of interest in the E coli–exposed culture to that in the mock-treated culture. A value of 1 is equivalent to 100% survival. (F) Anti-MR1–blocking antibody reduced the frequency of activated caspase-3–positive CD161++CD8+ T cells compared with the isotype control. Blocking of E coli–induced apoptosis was determined by normalizing the frequency of activated caspase 3+ CD161++C8+ T cells treated as indicated to that of the culture exposed to E coli alone. (G) Anti-MR1–blocking antibody increased the survival of CD161++CD8+ T cells in the E coli–exposed culture. Cell survival was determined by normalizing the frequency of CD161++CD8+ T cells in the antibody treated, E coli–exposed culture to that of the mock-treated culture.
E coli is known to activate CD161++/MAIT cells, but whether this might lead on to activation-induced cell death has not been explored. Although CD161++CD8+ T cells proliferated in vitro on exposure to E coli (supplemental Figure 6C-D), apoptosis was also induced in dose-dependent manner as measured by expression of activated caspase 3 at 20 hours (36.05% versus 0.45% in mock-treated controls, P < .001; Figure 6C-D), surface expression of phosphatidylserine at 44 hours (25.44% versus 5.49%, P = .004; supplemental Figure 6I), and selective loss of CD161++CD8+ T cells from the culture at 20 and 44 hours and 6 days (Figure 6E, supplemental Figure 6E-J). CD161++CD8+ T cells from patients with HIV also underwent apoptosis on exposure to increasing doses of E coli (supplemental Figure 6K-L). Although it is unclear to what extent such proliferation and apoptosis may occur in vivo, these data suggest that antigen/activation-induced cell death could plausibly contribute to depletion of CD161++/MAIT cells in HIV infection.
Consistent with the specific activation of CD161++CD8+ T cells by E coli (Figure 1), induction of apoptosis by E coli was also specific to CD161++CD8+ T cells as no differences in expression of either activated caspase-3, phosphatidylserine, or relative cell number were seen in the CD161− or CD161+CD8+ T-cell populations (Figure 6C-E, supplemental Figure 6I-J). In contrast, stimulation with PMA/ionomycin or large numbers of anti-CD2/CD3/CD28 microbeads induced expression of activated caspase 3 and phosphatidylserine expression in all subsets (Figure 6C, supplemental Figure 6M-N). Apoptosis in response to E coli was activation-induced, as blocking of MR1 significantly reduced the proportion of CD161++CD8+ T cells expressing activated caspase-3 compared with PBMCs treated with E coli alone (P < .0001; Figure 6F), abrogated the loss of CD161++CD8+ T cells relative to the mock-treated control (Figure 6G, supplemental Figure 6G), and inhibited E coli–induced proliferation (supplemental Figure 6F-H).
To investigate the mechanism of activation-induced apoptosis, we assessed changes in death receptor expression and the anti-apoptotic factor Bcl-2 after exposure to E coli. Marked up-regulation of Fas (CD95) and TNFRII was seen; small increases in the expression of DR4, which binds TRAIL, and Bcl-2 were also noted (supplemental Figure 7A-L). Combined blockade of both CD95 and TNFα reduced the percentage of phosphatidylserine+ CD161++CD8+ T cells but had no effect on the expression of activated caspase 3 or the loss of cells (supplemental Figure 7M-O).
Discussion
In this study, we demonstrated a specific decrease in the size of the CD161++/MAIT cell population in the blood of early and late-stage untreated HIV+ patients. Interestingly, these populations are not reconstituted in the blood during 2 years of HAART. We addressed potential mechanisms of loss from the bloodstream in HIV infection, including redistribution to tissues, and death via either direct viral infection or activation-induced apoptosis on encounter with translocated bacteria.
It should be noted that in healthy individuals the size of the CD161++/MAIT cell population varies considerably, both between individuals and with age.13 In some healthy individuals the frequency of CD161++/MAIT cells is as low as that observed in HIV infected patients (Figure 2A-C). Furthermore, normal or near normal frequencies of CD161++/MAIT cells were seen in a minority of HIV-infected individuals, The reason for this variation and the impact of low CD161++/MAIT cell numbers in health is unclear.
The reduced frequency of CD161++/MAIT cells in blood in HIV infection is unlikely to be because of dilution by HIV or herpesvirus-specific CD8+ T cells. Firstly, no difference was observed in the frequency of CD161+CD8+ T cells, despite the majority of HIV, CMV, and EBV-specific CD8+ T cells being CD161 negative (data not shown).38 Secondly, despite the decrease in frequency of HIV-specific CD8+ T cells reported with HAART,39,40 we observed no change in the frequency of CD161++/MAIT cells. Although loss secondary to HIV infection is the probable explanation for our findings, it is not possible to exclude that low CD161++/MAIT cell frequency may predispose to HIV infection.
Several groups have now consistently found that CD161++/MAIT cells produce a variety of cytokines, including IL17A and IL22,11,12 cytokines known to be important in mucosal immunity16,18 and to play a central role in the control of some HIV-associated infections.20,22,41,42 We showed a specific loss of the IL17A-producing CD161++/MAIT cell population from the blood of HIV-infected patients. The loss of Tc17 cells from the bloodstream has recently been reported in SIV-infected rhesus macaques with end-stage disease, but not in sooty mangabeys.43 This may suggest a role for Tc17 cells in disease pathology. Furthermore, the authors found similar losses of Tc17 cells in the colorectal mucosa, liver, spleen, and axillary lymph nodes of SIV-infected macaques, suggesting a system-wide loss of these cells in chronic infection.
CD161++/MAIT cells express the mucosal tissue homing markers CCR2, CXCR6, and CCR6, but not the lymph node homing markers CD62L and CCR7.11,12 CCL20, the ligand for CCR6, is expressed by healthy and inflamed intestinal mucosa in humans.44,45 Gut-associated lymphoid tissue (GALT) is a major site of infection and inflammation in early HIV infection1,2,26,46 and CCL20 has been shown in the colon and jejunum of SIV-infected macaques.47 We therefore sought to identify CD161++/MAIT cells in gut tissue in healthy controls and in subjects chronically infected with HIV. We were limited in the analysis of gut tissue because many relevant markers of CD161++/MAIT cells could not be reliably stained in formalin-fixed paraffin-embedded tissue, and we used high-level expression of the transporter protein MDR1 as a surrogate for high-level expression of CD161. Given these limitations, we did not see specific enrichment of CD161++/MAIT cells in the colon, although, an absolute increase in the numbers of CD161++/MAIT cells, which resolves with HAART, or changes in Vα7.2-expressing cells within the CD161++ subset cannot be excluded. Interestingly, there was no specific loss of CCR6+CD4+ T cells from the blood, which argues against CCL20-mediated T-cell retention in tissue.
One possible explanation for our data are that after recruitment to tissues, CD161++/MAIT cells die through a variety of mechanisms, including antigen-induced cell death (AICD), bystander activation, or infection. Regarding AICD, recent work has shown that MAIT cells are activated by a range of bacteria and fungi in an MR1-dependent manner.9,10 Given the increased permeability of the gut in HIV infection and the associated microbial translocation,25,36,37,48-50 we first analyzed gut samples to seek evidence for local accumulation of bacterial antigen. Immunohistochemical staining of colon biopsies demonstrated significant numbers of LPS+ cells, suggesting a high antigen burden. On in vitro activation of PBMCs by paraformaldehyde-fixed E coli, CD161++/MAIT cells specifically underwent apoptosis in a dose and MR1-dependent manner. Some specific proliferation in vitro in response to E coli was also observed, although again this was reduced at the highest dose, suggesting that there is a balance between proliferation and cell death. Up-regulation of CD95 and TNFRII expression by E coli–exposed CD161++/MAIT cells and the partial inhibition of apoptosis with dual blockade suggest a possible mechanism for cell death. Although these in vitro experiments may not accurately reflect what is occurring in vivo, the accumulation of apoptotic CCR6+CD8+ T cells in the spleens of HIV infected patients51 and the observed loss of Tc17 cells in macaques in all tissues studied43 are consistent with the idea that apoptosis in tissues contributes to the observed depletion of CD161++/MAIT cells in the blood.
Given the majority of the CD161++/MAIT cell population express CCR5 and that R5-tropic viruses are dominant in early HIV infection52,53 we investigated whether the loss of this subset could be because of preferential infection by HIV. Previous studies have shown infection of CD8+ T cells by HIV or SIV ex vivo and in vitro, albeit at low frequencies.24,33-35 Although we were able to show that both CCR5 and CXCR4-tropic viruses could infect CD8+ T cells in vitro, the frequency of infection was low despite a high viral MOI. Considering the potential of the CD161++/MAIT cells to traffic to sites of inflammation11 in early HIV infection, and the high viral titers present in such locations,46 infection of these cells in vivo may be more efficient than in our in vitro system. However, that CD161++MAIT cells were not preferentially infected, and that their numbers were not adversely affected by incubation with HIV argues against either infection or bystander cell death being solely responsible for the specific depletion of such a substantial population of cells.
No recovery of CD161++/MAIT cells was seen during 2 years of HAART. This may be linked to incomplete repair of the bacterial translocation defect and/or ongoing tissue inflammation, leading to continued CD161++/MAIT cell redistribution with or without AICD, failure of the precursor pool to proliferate, lack of generation of new cells from the thymus, or loss of an essential antigen presenting cell pool or site required for maintenance and/or expansion. Future studies in different patient groups, for example pediatric HIV infection, and in different tissues (eg, the liver) may help dissect these possible mechanisms. The mechanisms leading to loss of CD161++/MAIT cell populations in early infection and their failure to recover with HAART are integrated into a model (supplemental Figure 8).
The loss of the CD161++/MAIT cell population in HIV may have several important consequences. Firstly, it may contribute to increased susceptibility to M tuberculosis infection. The risk of tuberculosis doubles within a year of HIV infection, whereas the CD4 T-cell count remains relatively preserved.54 Recent studies have provided evidence of a role for MAIT cells in the immune response to M tuberculosis.9,10 Therefore, in addition to the early loss of M tuberculosis–specific memory CD4+ T cells,7 the loss of CD161++/MAIT cells in acute HIV infection may also contribute to the increased risk of tuberculosis associated with HIV infection. Secondly, CD161++/MAIT cell loss may also contribute to the risk of other bacterial and fungal infections. Indeed IL17 has recently been implicated in the control of mucocutaneous candiasis20 and invasive nontyphoidal Salmonella enterica infection.22 Finally, further compromise of the IL17A/IL22 axis, already disrupted through loss of Th17 cells, may result in increased impairment of mucosal immunity and contribute to reduced barrier function in HIV/AIDS, including increased bacterial translocation.
Overall the CD161++/MAIT cell population is a highly prominent cell population in the human immune system, and the negative impact of HIV infection occurs early and is not reversed by HAART. These data not only provide a novel paradigm for the immunopathogenesis of HIV but also suggest that future strategies to rescue or amplify this functional subset could help restore host defense, including against M tuberculosis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Wellcome Trust (WT091663MA); The Medical Research Council; The National Institute for Health Research, Biomedical Research Center, Oxford; the Oxford Martin School; the National Institutes for Health, National Institute of Allergy and Infectious Diseases (5U19A082630-04); and the Oxford Dominions Trust. The Swiss HIV cohort study is funded by the Swiss National Science Foundation (SNF grant No. 33CS30-134277) and by the SHCS research foundation. The London Infectious Diseases Biobank provided the untreated chronic-stage HIV+ PBMC samples. The Thames Valley Cohort provided the untreated chronic-stage HIV+ whole blood samples. The Swiss HIV Study Cohort provided the HIV+ samples for the longitudinal treatment cohort; the complete list of SCHS membership appears in supplemental Appendix 1. The early stage HIV+ PBMC samples were provided by the SPARTAC Trial; the complete list of membership appears in supplemental Appendix 2. HIV+ colon samples were provided by St Stephen's Center, Chelsea and Westminster Hospital. The HIV− tissue samples were provided by the Oxford GI Biobank.
National Institutes of Health
Wellcome Trust
Authorship
Contribution: C.C. and J.E.U. designed and performed experiments, analyzed data, and wrote the paper; K.G., A.K., M.H.H., K.A., J.R.F., and Y.-H.K. designed and performed experiments; H.U. provided technical advice; P.G., J.F., B.G., A.S., A.R., F.P., H.U., and the SHCS investigators A.R., H.F.G., and N.K. provided samples; T.H. provided a critical reagent and advice; P.K., C.C., and J.E.U. developed the concept and designed the study; R.E.P., J.F., A.R., and P.S. provided conceptual advice; and all authors commented on the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cormac Cosgrove, Ragon Institute of MGH, MIT and Harvard, Bldg 149, 13th St, Charlestown, MA 02129; e-mail: pcosgrove@partners.org.
References
Author notes
C.C. and J.E.U. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal