Key Points
PU.1 is a potent tumor suppressor in cHL cells and the induction of PU.1 is a possible therapeutic option for patients with cHL.
Abstract
PU.1 has previously been shown to be down-regulated in classical Hodgkin lymphoma (cHL) cells via promoter methylation. We performed bisulfite sequencing and proved that the promoter region and the −17 kb upstream regulatory element of the PU.1 gene were highly methylated. To evaluate whether down-regulation of PU.1 is essential for the growth of cHL cells, we conditionally expressed PU.1 in 2 cHL cell lines, L428 and KM-H2. Overexpression of PU.1 induced complete growth arrest and apoptosis in both cell lines. Furthermore, in a Hodgkin lymphoma tumor xenograft model using L428 and KM-H2 cell lines, overexpression of PU.1 led to tumor regression or stable disease. Lentiviral transduction of PU.1 into primary cHL cells also induced apoptosis. DNA microarray analysis revealed that among genes related to cell cycle and apoptosis, p21 (CDKN1A) was highly up-regulated in L428 cells after PU.1 induction. Stable knockdown of p21 rescued PU.1-induced growth arrest in L428 cells, suggesting that the growth arrest and apoptosis observed are at least partially dependent on p21 up-regulation. These data strongly suggest that PU.1 is a potent tumor suppressor in cHL and that induction of PU.1 with demethylation agents and/or histone deacetylase inhibitors is worth exploring as a possible therapeutic option for patients with cHL.
Introduction
Hodgkin lymphoma is a B-cell malignancy that occurs frequently in the white population, and is relatively rare within Japanese and other Asian populations.1 To date, the combination of chemotherapy and irradiation has led to a dramatic improvement in both progression-free survival and overall survival of stage I and II patients, which now exceeds 90%.2 In contrast, the prognosis of the remaining patients who relapse or fail to make complete remission, and in stage III and IV patients, is relatively poor.3-9 In addition, patients who achieve long-term disease-free survival frequently have infertility and secondary malignancies, including breast cancer and cardiac failure, which are related to chemotherapeutic agents and radiation therapies.10-12 Therefore, the development of new therapeutic strategies is necessary to improve clinical outcome and reduce the long-term side effects of current treatments in these patients. Nevertheless, our understanding of the mechanisms underlying the pathogenesis of Hodgkin lymphoma, which are necessary for the generation of novel, molecularly targeted agents, remains incomplete. It is known that both alleles of tumor necrosis factor, α-induced protein 3 (TNFAIP3)(A20) are deleted in a third of patients with Hodgkin lymphoma of nodular sclerosis histology and in the classic Hodgkin lymphoma (cHL) cell line, KM-H2.13
Hodgkin lymphoma is subdivided into cHL, which constitutes the majority of patients (95%), and nodular lymphocyte predominant Hodgkin lymphoma.1 In cHL, lymphoma cells do not express the B cell–specific surface antigens, CD19 and CD20, or the B cell–specific transcription factors, Bob.1, PU.1, and SpiB.14,15
PU.1 is an Ets family transcription factor that is essential for the differentiation of both myeloid and lymphoid cells.16,17 PU.1 is expressed in granulocytes, monocytes/macrophages, and B cells, but not in erythrocytes or T cells. The expression of PU.1 requires an upstream regulatory element (URE) located −14 kb and −17 kb upstream of the transcriptional start site of the murine and human PU.1 genes, respectively, in addition to promoter regulatory elements.18-20 In murine models, deletion of the −14 kb URE led to down-regulation of PU.1 expression to 20% of wild-type mice, and surprisingly, knockout mice developed acute myeloid leukemia and a B-cell chronic lymphocytic leukemia-like disease.21,22 These data suggest that PU.1 has tumor suppressor activity in myeloid cells and B cells. We recently reported that PU.1 is down-regulated in a subset of multiple myeloma cells and in most myeloma cell lines. Conditional expression of PU.1 induced complete growth arrest and apoptosis in myeloma cell lines, suggesting that PU.1 is a potent tumor suppressor in multiple myeloma.20
Previous studies have reported that PU.1 is also down-regulated in cHL cells via methylation of the PU.1 promoter.23 Therefore, in this study, we evaluated whether PU.1 is tumor suppressor in cHL cells.
Methods
Bisulfite sequencing
Genomic DNA was treated with sodium bisulfite as previously described20 and subjected to 35 cycles of PCR. A 149-bp PU.1 promoter-exon1 region, including a PU.1 binding site, was amplified with the primers 5′-GTAGTTTAGGGGGTAGGTTTGAGTT-3′and 5′-AAAAAAAACCCTTCCATTTTACAC-3′. PCR products were directly sequenced. A 244-bp product encompassing the −17 kb URE, including PU.1 and runt-related transcription factor 1 (RUNX1/AML1) binding sites, which are required for PU.1 expression, was amplified with the primers 5′-ATTTTTTTGAGGTTTGGTTTAGGTT-3′ and 5′-CTACAACTACCCCTATTTCCACATC-3′. Both regions were previously shown to contain CpG islands.20
Cell culture
Human Hodgkin lymphoma cell lines L428, KM-H2, L540, HDLM2, and HD-70 and their derivatives were grown in RPMI 1640 medium containing 10% (volume/volume) FBS at 37°C.
Constructs
pCAG20-1, pUHD-3 puromycin and pUHD-10-3 IRES-GFP plasmids were kind gifts from Dr Takumi Era (Division of Molecular Neurobiology, Institute of Molecular Embryology and Genetics, Kumamoto University, Japan).24 The human PU.1 cDNA was subcloned into the blunt-ended EcoRI site of pUHD10-3 IRES-GFP, resulting in pUHD10-3 PU.1-IRES-GFP.
Generation of stable transformants conditionally expressing PU.1
To obtain PU.1-inducible cHL cell lines using the tetracycline-off system, 1 × 107 L428 or KM-H2 cells were cotransfected with 10 μg each of ScaI-digested pCAG20-1 and pUHD-3 puromycin plasmids by electroporation. Cells were selected with 1 μg/mL puromycin, and subsequently transfected with 10 μg of ScaI-digested pUHD10-3 PU.1-IRES-GFP and 2 μg of HindIII-digested pPGKneo.20 After isolation of G418-resistant clones, GFP expression was evaluated after tetracycline removal. On confirmation of GFP expression, these cells were designated L428tetPU.1 and KM-H2tetPU.1.
Xenograft model
A total of 7 × 106 L428tetPU.1 or KM-H2tetPU.1 cells were injected subcutaneously into Rag2−/−Jak3−/−balb/c mice (n = 16 per group). Mice were given drinking water containing tetracycline (500 μg/mL) 3 days before injection. Subcutaneous tumors were grown to 1- to 2-cm in diameter (∼ 30-35 days after injection), and half of the cohort (n = 8) were maintained on tetracycline-treated water, whereas the remaining mice discontinued tetracycline and were given pure water. Tumor size was measured every 7 days.
Primary cHL cells
Primary cells were obtained from lymph node biopsy samples of 3 patients with cHL. Informed consent for sample collection was obtained according to protocols approved by the institutional review boards and in accordance with the Declaration of Helsinki. Primary cHL cells were purified by negative selection using an anti-CD3, CD14, CD16, CD19, CD20, and CD56 antibody cocktail (BioLegend) and antimouse IgG antibody-conjugated magnetic beads (Miltenyi Biotec). After negative selection, the purity of Hodgkin cells was confirmed by May-Giemsa staining and immunostaining with anti-CD30 antibody of cytospin samples or flow cytometry after staining with anti-CD30 antibody.
Lentiviral transduction
The CSII-EF-MCS-IRES-Venus lentiviral vector and packaging constructs, VSV-G Rev and CAG-HIVgp, were purchased from the RIKEN BioResource Center. The human PU.1 cDNA was subcloned into NotI site of CSII-EF-MCS-IRES-Venus. Virus was produced and cells were infected as previously described (http://www.brc.riken.jp/lab/cfm/Subteam_for_Manipulation_of_Cell_Fate/Home.html).25,26
DNA microarray analysis
Total RNA was extracted from L428tetPU.1 and KM-H2tetPU.1 cells using Trizol reagent at 3 different time points: days 0, 1, and 3 after tetracycline removal. RNA was hybridized to Illumina:Sentrix Human-6 Expression BeadChips or Illumina HumanHT-12 Version 4 Expression BeadChips, according to the manufacturer's instructions. Gene expression profiles were analyzed using GeneSpring Version 12.0 software.27 Data for L428tetPU.1 cells are available at the Gene Expression Omnibus under accession number GSE42437; data for KM-H2tetPU.1 cells are available under GSE42440.
Real-time PCR
Quantitative TaqMan PCR was performed with commercially available assay-on demand probe primer sets for p21 and β-actin (Applied Biosystems) and TaqMan Universal PCR Master Mix reagent according to the manufacturer's instructions. Reactions were performed using an Illumina Eco Real-Time PCR system. The expression levels of β-actin were used to normalize the relative expression levels of p21. The expression level of p21 in L428tetPU.1 cells before tetracycline removal was set to 100.
Western blot analysis
Cell lysates were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were incubated with anti-PU.1, anti-p21WAF/CIP1, and antiactin primary antibodies (Santa Cruz Biotechnology) for 3-12 hours. Membranes were then incubated with peroxidase-labeled secondary antibodies for 30 minutes and developed using an enhanced chemiluminescence system (GE Healthcare).
Generation of L428tetPU.1 cells stably expressing p21 siRNA
siRNA expression vectors were generated by insertion of annealed oligonucleotides targeting p21 and scrambled control siRNAs into the BamHI and HindIII sites of pRNA-U6.1/Zeo or pRNA-U6.1/ Hygro (GenScript).27 siRNA expression vectors were transfected into L428tetPU.1 cells by electroporation, and stable transformants were obtained by selection with zeocin (400 μg/mL) or hygromycin (200 μg/mL).
Detection of apoptosis
For detection of apoptosis, cHL cells were stained with an annexin V Phycoerythrin or Allophycocyanin Apoptosis Detection kit (Medical and Biologic Laboratories). Cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson).
Cell-cycle analysis
L428tetPU.1 and KM-H2tetPU.1 cell-cycle profiles were analyzed by staining with bromodeoxyuridine (BrdU) and 7-aminoactinomycin D (BrdU Flow Kits; BD Biosciences PharMingen),28 3 days after tetracycline withdrawal. Cells were analyzed by flow cytometry (FACSCalibur).
Results
PU.1 is down-regulated in Hodgkin lymphoma by methylation of the promoter and a −17 kb URE of the PU.1 gene
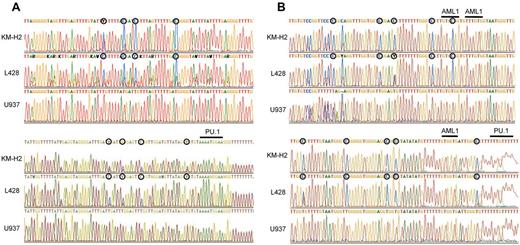
Previous studies have shown that PU.1 is generally highly down-regulated in both Hodgkin lymphoma cell lines and primary Hodgkin lymphoma cells.23 In these studies, methylation-specific PCR revealed that the PU.1 promoter is methylated, leading to down-regulation of PU.1 expression. We previously demonstrated that PU.1 is strongly down-regulated in myeloma cell lines and the PU.1 promoter was highly methylated as shown by bisulfite sequencing.20 To confirm this mechanism of PU.1 down-regulation in Hodgkin lymphoma, we conducted bisulfite sequencing of the PU.1 promoter region in the Hodgkin lymphoma cell lines, L428 and KM-H2. As shown in Figure 1A, the PU.1 promoter region was heavily methylated in both cell lines, and there was no C-T conversion in CpG islands after bisulfite treatment of genomic DNA. Gene expression often requires cis-elements located > 10 kb or, in some cases, more than several hundred kilobases upstream of the transcriptional start site or downstream of the transcriptional termination site.29-36 The expression of PU.1 also requires a URE located −14 kb and −17 kb upstream of its promoter in mice and humans, respectively.18,19,21,22 Previously, we reported that PU.1 is down-regulated in multiple myeloma cells via methylation of its promoter and the −17 kb URE.20 We therefore evaluated the methylation status of the −17 kb URE of PU.1 in L428 and KM-H2 cells. The −17 kb URE of PU.1 was highly methylated in both L428 and KM-H2 cells, indicating that PU.1 is silenced in cHL cells via methylation of both the promoter and the −17 kb URE (Figure 1B).
The promoter region and the −17 kb URE of the PU.1 gene are highly methylated in cHL cells. (A) Bisulfite sequencing confirmed that the promoter of PU.1 is highly methylated in L428 and KM-H2 Hodgkin lymphoma cell lines. ○ represents methylated cytosines. Underlining indicates the PU.1 binding site downstream of the translation initiation site. (B) Bisulfite sequencing revealed that the −17 kb URE was highly methylated in L428 and KM-H2 Hodgkin lymphoma cell lines. ○ represents methylated cytosines. Underlining indicates 3 RUNX1 (AML1) binding sites and a PU.1 binding site.
The promoter region and the −17 kb URE of the PU.1 gene are highly methylated in cHL cells. (A) Bisulfite sequencing confirmed that the promoter of PU.1 is highly methylated in L428 and KM-H2 Hodgkin lymphoma cell lines. ○ represents methylated cytosines. Underlining indicates the PU.1 binding site downstream of the translation initiation site. (B) Bisulfite sequencing revealed that the −17 kb URE was highly methylated in L428 and KM-H2 Hodgkin lymphoma cell lines. ○ represents methylated cytosines. Underlining indicates 3 RUNX1 (AML1) binding sites and a PU.1 binding site.
PU.1 induces growth arrest and apoptosis of Hodgkin lymphoma cell lines, L428 and KM-H2
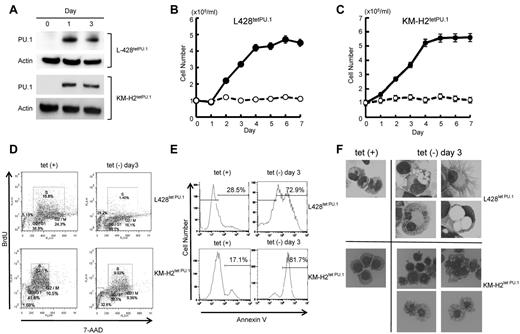
We next evaluated whether down-regulation of PU.1 may play a role in Hodgkin lymphoma cell growth. We generated Hodgkin lymphoma cell lines that conditionally express PU.1 using a tet-off system, designated L428tetPU.1 and KM-H2tetPU.1. After the removal of tetracycline from growth medium, PU.1 was highly up-regulated in both L428tetPU.1 and KM-H2tetPU.1 cell lines (Figure 2A). Conditional expression of PU.1 induced complete growth arrest of both L428tetPU.1 and KM-H2tetPU.1 cells over the course of 7 days (Figure 2B-C). Cell-cycle analysis using BrdU and 7-aminoactinomycin D staining revealed that induction of PU.1 led to a decrease in S phase cells in both L428tetPU.1 (15.8% vs 1.4%) and KM-H2tetPU.1 cells (32.1% vs 9.9%) after 3 days, suggesting that PU.1 induced G1 arrest in these cells (Figure 2D). We also observed an increase in the sub-G1 population in both L428tetPU.1 and KM-H2tetPU.1 cells. Consistent with these data, annexin V staining revealed that PU.1 expression led to a significant increase in apoptotic cells in L428tetPU.1 and KM-H2tetPU.1 cells at day 3 (28.5% vs 72.9% and 17.1% vs 81.7%, respectively; Figure 2E). Morphologically, L428tetPU.1 cells expressing PU.1 were enlarged and displayed numerous cell processes and vacuoles of various sizes, which in some cases occupied the majority of the cellular mass and nuclear compartment (Figure 2F). In addition, a number of L428tetPU.1 PU.1-expressing cells exhibited nuclear fragmentation, a typical feature of apoptosis (Figure 2F). In comparison, KM-H2tetPU.1 cells expressing PU.1 contained relatively smaller vacuoles, and many cells exhibited nuclear condensation, which is also a feature of apoptotic cells. These data demonstrate that PU.1 induces complete growth arrest and apoptosis in L428 and KM-H2 cells.
PU.1 induces growth arrest and apoptosis in L428 and KM-H2 cHL cell lines in vitro. (A) Western blot of PU.1 protein after tetracycline withdrawal. PU.1 was highly induced after tetracycline withdrawal in L428tetPU.1 and KM-H2tetPU.1 cells. PU.1 induced growth arrest in L428tetPU.1 (B) and KM-H2tetPU.1 cells (C) after tetracycline removal (○), whereas uninduced cells (●) grew comparably to wild-type parental cells. (D) Cell-cycle analysis was performed in L428tetPU.1 and KM-H2tetPU.1 cells by BrdU and 7-aminoactinomycin D staining. PU.1 induced G1 arrest and led to a significant decrease in S-phase cells in L428tetPU.1 and KM-H2tetPU.1 cell lines. (E) PU.1 induced apoptosis in L428tetPU.1 and KM-H2tetPU.1 cells as assessed by annexin V staining. (F) PU.1 induced morphologic changes in L428tetPU.1 and KM-H2tetPU.1 cells. L428tetPU.1 cells expressing PU.1 exhibited numerous different-sized cell processes and vacuoles compared with uninduced cells. A number of cells displayed nuclear fragmentation (Tet−, day 3, bottom left panel). The majority of KM-H2tetPU.1 cells expressing PU.1 also contained relatively small-sized vacuoles and nuclear condensation, indicative of apoptosis. Images were acquired using a BX60 microscope and DP70 digital camera with DP controller software (Olympus; ×400 magnification).
PU.1 induces growth arrest and apoptosis in L428 and KM-H2 cHL cell lines in vitro. (A) Western blot of PU.1 protein after tetracycline withdrawal. PU.1 was highly induced after tetracycline withdrawal in L428tetPU.1 and KM-H2tetPU.1 cells. PU.1 induced growth arrest in L428tetPU.1 (B) and KM-H2tetPU.1 cells (C) after tetracycline removal (○), whereas uninduced cells (●) grew comparably to wild-type parental cells. (D) Cell-cycle analysis was performed in L428tetPU.1 and KM-H2tetPU.1 cells by BrdU and 7-aminoactinomycin D staining. PU.1 induced G1 arrest and led to a significant decrease in S-phase cells in L428tetPU.1 and KM-H2tetPU.1 cell lines. (E) PU.1 induced apoptosis in L428tetPU.1 and KM-H2tetPU.1 cells as assessed by annexin V staining. (F) PU.1 induced morphologic changes in L428tetPU.1 and KM-H2tetPU.1 cells. L428tetPU.1 cells expressing PU.1 exhibited numerous different-sized cell processes and vacuoles compared with uninduced cells. A number of cells displayed nuclear fragmentation (Tet−, day 3, bottom left panel). The majority of KM-H2tetPU.1 cells expressing PU.1 also contained relatively small-sized vacuoles and nuclear condensation, indicative of apoptosis. Images were acquired using a BX60 microscope and DP70 digital camera with DP controller software (Olympus; ×400 magnification).
PU.1 induces growth arrest, regression of subcutaneous tumors, and prolonged survival in a Hodgkin lymphoma xenograft mouse model
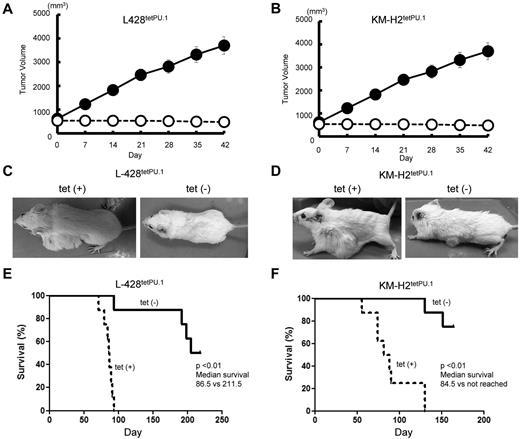
We next investigated the role of PU.1 in a xenograft model of Hodgkin lymphoma. A total of 7 × 106 L428tetPU.1 or KM-H2tetPU.1 cells were injected subcutaneously in Rag2−/−Jak3−/−balb/c mice, and tumors were grown to 1- to 2-cm in diameter. Mice were then divided into 2 treatment groups: one group continued drinking tetracycline-treated water (n = 8), whereas the other group was given nontreated water (n = 8). In mice taking tetracycline, tumors continued to grow and increase in size (Figure 3A-B), and a number of mice developed skin ulcers possibly induced by tumor necrosis. In contrast, tumors in mice given nontreated water ceased to grow and decreased in size in several cases (Figure 3C-D). All mice injected with L428tetPU.1 or KM-H2tetPU.1 cells, taking tetracycline, died within 96 days or 143 days, respectively. In contrast, more than 50% of mice without tetracycline survived > 200 days in both xenograft models. Furthermore, mice given nontreated water and harboring tumors < 1 cm in diameter had a 100% survival rate. In comparison, mice without tetracycline and harboring larger tumors tended to have skin ulcers at the site of the shrinking tumor and acquired lethal infections.
PU.1 induces growth arrest and apoptosis in both L428tetPU.1 and KM-H2tetPU.1 cHL cell lines in vivo. Tumor size in L428tetPU.1 (A) and KM-H2tetPU.1 (B) xenograft mouse models over the course of 7 days. Tumors in L428tetPU.1 and KM-H2tetPU.1 mice given tetracycline continued to grow (●), compared with tumors in mice without tetracycline, which ceased to grow and in some cases decreased in size (○). Tumors in L428tetPU.1 (C) and KM-H2tetPU.1 (D) xenografts did not grow after tetracycline withdrawal. Left panels: Tumors in a xenograft mouse taking tetracycline water. Right panel: Tumors after tetracycline withdrawal. Survival curves of L428tetPU.1 (E) and KM-H2tetPU.1 (F) xenograft mice. L428tetPU.1 xenograft mice taking tetracycline all died within 96 days, whereas 7 of 8 mice without tetracycline survived > 190 days. KM-H2tetPU.1 xenograft mice taking tetracycline all died within 143 days, whereas 6 of 8 mice without tetracycline survived > 155 days.
PU.1 induces growth arrest and apoptosis in both L428tetPU.1 and KM-H2tetPU.1 cHL cell lines in vivo. Tumor size in L428tetPU.1 (A) and KM-H2tetPU.1 (B) xenograft mouse models over the course of 7 days. Tumors in L428tetPU.1 and KM-H2tetPU.1 mice given tetracycline continued to grow (●), compared with tumors in mice without tetracycline, which ceased to grow and in some cases decreased in size (○). Tumors in L428tetPU.1 (C) and KM-H2tetPU.1 (D) xenografts did not grow after tetracycline withdrawal. Left panels: Tumors in a xenograft mouse taking tetracycline water. Right panel: Tumors after tetracycline withdrawal. Survival curves of L428tetPU.1 (E) and KM-H2tetPU.1 (F) xenograft mice. L428tetPU.1 xenograft mice taking tetracycline all died within 96 days, whereas 7 of 8 mice without tetracycline survived > 190 days. KM-H2tetPU.1 xenograft mice taking tetracycline all died within 143 days, whereas 6 of 8 mice without tetracycline survived > 155 days.
These data suggest that PU.1 may act as a tumor suppressor of cHL in vivo.
PU.1 induces apoptosis in primary cells purified from patients with Hodgkin lymphoma
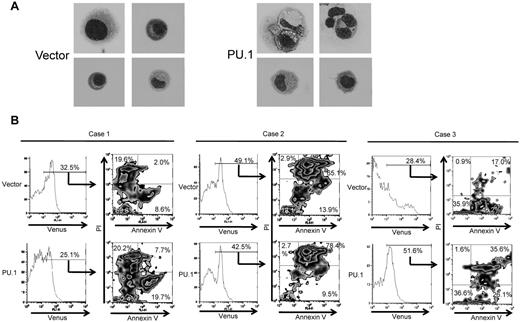
Next, to evaluate whether PU.1 induces apoptosis of primary cHL cells, we purified lymphoma cells from the lymph nodes of patients with cHL. Negative selection was performed with anti-CD3, -CD14, -CD16, -CD19, -CD20, and -CD56 antibodies, and purified cells consisted of > 90% Hodgkin and Reed Sternberg-cells as confirmed by May-Giemsa staining of cytospin samples (Figure 4A). These cells were also CD30-positive as confirmed by flow cytometry or immunohistochemistry (supplemental Figure 1A-B, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). After purification, primary Hodgkin lymphoma cells cultured alone survived for 1 day. However, coculture with cells obtained from patient lymph nodes prolonged survival to at least 3 days, as confirmed by microscopy of cytospin samples. Therefore, we infected cells obtained from whole lymph nodes, including Hodgkin cells, with lentivirus overexpressing human PU.1 or empty vector control. After 3 days in culture, primary Hodgkin lymphoma cells were purified by negative selection and subjected to annexin V staining. Cells transduced with lentivirus were recognized as Venus-positive cells and were analyzed by staining with propidium iodide (PI) and annexin V antibody. In case 1, overexpression of PU.1 led to a decrease in live cells (PI−/annexin V−) compared with empty vector (52.4% vs 69.8%), and a concomitant increase in PI−/annexin V+ preapoptotic cells (19.7% vs 8.6%; Figure 4B, left panel). In addition, primary cHL cells transduced with PU.1 exhibited various-sized vacuoles in cytosol (Figure 4A, right panels). In case 2, overexpression of PU.1 also led to a decrease in live cells (PI−/annexin V−) compared with control vector (9.4% vs 18.1%), and an increase in PI+ apoptotic cells (PI+/annexin V− and PI+/annexin V+) compared with vector only (81% vs 68%; Figure 4B, middle panel). In case 3, overexpression of PU.1 also led to a decrease in live cells (PI−) compared with control vector (62.7% vs 92.1%) and an increase in PI+ apoptotic cells (PI+/annexin V− and PI+/annexin V+) compared with vector only (37.2% vs 17.9%; Figure 4B right panel). These experiments were performed in biologic duplicate with similar results. These data suggest that PU.1 also induces apoptosis in primary Hodgkin lymphoma cells.
PU.1 induces apoptosis in primary cHL cells from patients. (A) Morphology of purified, primary cHL cells transduced with control lentivirus (Case 1, 4 left panels) or PU.1 expressing lentivirus (Case 1, 4 right panels). A number of primary classic Hodgkin cells expressing PU.1 contained various-sized vacuoles Images were acquired using a BX60 microscope and DP70 digital camera with DP controller software (Olympus; ×1000 magnification). (B) Primary cHL cells transduced with PU.1 lentivirus tended to undergo apoptosis compared with cells transduced with empty vector. Apoptosis was assessed in Venus-positive cells by flow cytometry after staining with PI and allophycocyanin-conjugated annexin V. Stable transduction of primary cHL cells from case 1 with PU.1 lentivirus led to a decrease in the percentage of live PI−/annexin V− cells and an increased percentage of PI−/annexin V+ preapoptotic cells, compared with control vector. Stable transduction of primary cHL cells from case 2 with PU.1 lentivirus also led to a decrease in the percentage of live PI−/annexin V− cells and an increased percentage of PI+ apoptotic cells. Stable transduction of primary cHL cells from case 3 with PU.1 lentivirus also led to a decrease in the percentage of live PI−/annexin V+ cells and an increased percentage of PI+ apoptotic cells.
PU.1 induces apoptosis in primary cHL cells from patients. (A) Morphology of purified, primary cHL cells transduced with control lentivirus (Case 1, 4 left panels) or PU.1 expressing lentivirus (Case 1, 4 right panels). A number of primary classic Hodgkin cells expressing PU.1 contained various-sized vacuoles Images were acquired using a BX60 microscope and DP70 digital camera with DP controller software (Olympus; ×1000 magnification). (B) Primary cHL cells transduced with PU.1 lentivirus tended to undergo apoptosis compared with cells transduced with empty vector. Apoptosis was assessed in Venus-positive cells by flow cytometry after staining with PI and allophycocyanin-conjugated annexin V. Stable transduction of primary cHL cells from case 1 with PU.1 lentivirus led to a decrease in the percentage of live PI−/annexin V− cells and an increased percentage of PI−/annexin V+ preapoptotic cells, compared with control vector. Stable transduction of primary cHL cells from case 2 with PU.1 lentivirus also led to a decrease in the percentage of live PI−/annexin V− cells and an increased percentage of PI+ apoptotic cells. Stable transduction of primary cHL cells from case 3 with PU.1 lentivirus also led to a decrease in the percentage of live PI−/annexin V+ cells and an increased percentage of PI+ apoptotic cells.
Growth arrest of L428tetPU.1 cells induced by PU.1 is at least partially dependent on up-regulation of p21
To elucidate the mechanisms underlying cell-cycle arrest and apoptosis induced by PU.1, we compared gene expression profiles of L428tetPU.1 and KM-H2tetPU.1 cells 0, 1, and 3 days after PU.1 induction, by DNA microarray. In the case of L428tetPU.1 cells, the top 30 genes up-regulated and down-regulated at day 1 and day 3 are shown in supplemental Tables 1 through 4. Genes up-regulated 24 hours after PU.1 induction (> 8-fold at day 1) are shown in Figure 5A. These genes contain IFN-stimulated genes (ISGs), including TRIM22, IFI44L, and OAS3. We previously reported that ISGs, including IFIT1, IFITM1, IFIT2, IFIT4, IFI27, ISG15, LY6E, and IFI6, were up-regulated in the multiple myeloma cell line, U266, after induction of PU.1 expression using the same tet-off system. IRF7, a key transcription factor involved in IFN signal transduction, was also highly up-regulated as previously shown in U266 cells expressing PU.1. Within the category of genes related to cell cycle or apoptosis, we observed that p21 (CDKN1A) was highly up-regulated at day 1 and day 3 after PU.1 induction, and this was confirmed by real-time PCR and Western blot (Figure 5B-C).
p21 is highly up-regulated in L428tetPU.1 cells after PU.1 induction. (A) Heatmap of DNA microarray analysis comparing the gene expression profiles of L428tetPU.1 cells after PU.1 induction at days 0, 1, and 3. p21 (CDKN1A) mRNA was highly up-regulated after PU.1 induction. (B) p21 mRNA was up-regulated in both L428tetPU.1 and KM-H2tetPU.1 cells after PU.1 induction. Real-time PCR was performed 1 and 3 days after PU.1 induction. (C) p21 protein was up-regulated in L428tetPU.1 but not in KM-H2tetPU.1 cells after PU.1 induction. Western blot for p21, PU.1, and actin was performed 1 and 3 days after PU.1 induction.
p21 is highly up-regulated in L428tetPU.1 cells after PU.1 induction. (A) Heatmap of DNA microarray analysis comparing the gene expression profiles of L428tetPU.1 cells after PU.1 induction at days 0, 1, and 3. p21 (CDKN1A) mRNA was highly up-regulated after PU.1 induction. (B) p21 mRNA was up-regulated in both L428tetPU.1 and KM-H2tetPU.1 cells after PU.1 induction. Real-time PCR was performed 1 and 3 days after PU.1 induction. (C) p21 protein was up-regulated in L428tetPU.1 but not in KM-H2tetPU.1 cells after PU.1 induction. Western blot for p21, PU.1, and actin was performed 1 and 3 days after PU.1 induction.
To clarify the role of p21 up-regulation in Hodgkin lymphoma cell growth, we stably silenced p21 in L428tetPU.1 cells using siRNA. p21 siRNA strongly suppressed p21 expression in L428tetPU.1 cells, before and after PU.1 induction (Figure 6A). Stable knockdown of p21 rescued L428tetPU.1 cells from growth arrest induced by PU.1 (Figure 6B). Taken together, these data suggest that L428tetPU.1 growth arrest induced by PU.1 is at least partially dependent on p21 up-regulation.
PU.1-induced cell growth arrest in L428tetPU.1 cells is dependent on p21 up-regulation. (A) Stable knockdown of p21 strongly suppressed p21 expression, even after PU.1 induction in L428tetPU.1 cells. (B) The growth arrest of L428tetPU.1 cells induced by PU.1 overexpression was reversed by targeted knockdown of p21. PU.1-induced growth arrest in L428tetPU.1 cells stably transduced with scrambled siRNA (○), whereas PU.1 failed to induce growth arrest in L428tetPU.1 cells stably transduced with p21 siRNA (▵).
PU.1-induced cell growth arrest in L428tetPU.1 cells is dependent on p21 up-regulation. (A) Stable knockdown of p21 strongly suppressed p21 expression, even after PU.1 induction in L428tetPU.1 cells. (B) The growth arrest of L428tetPU.1 cells induced by PU.1 overexpression was reversed by targeted knockdown of p21. PU.1-induced growth arrest in L428tetPU.1 cells stably transduced with scrambled siRNA (○), whereas PU.1 failed to induce growth arrest in L428tetPU.1 cells stably transduced with p21 siRNA (▵).
In the case of KM-H2tetPU.1 cells, genes up-regulated after PU.1 induction (> 10-fold at day 1) are shown in supplemental Figure 2. The top 30 genes up-regulated and down-regulated one or 3 days after PU.1 induction are shown in supplemental Tables 5 through 8. p21 was not included in these genes. Indeed, p21 was not up-regulated at the protein level after PU.1 induction, whereas p21 mRNA was up-regulated 6-fold (Figure 5B-C). Therefore, the mechanisms underlying cell-cycle arrest in KH-M2tetPU.1 cells expressing PU.1 may be distinct from those of L428tetPU.1 cells expressing PU.1. There were no highly up-regulated ISGs; however, IRF7 was highly up-regulated at both day 1 (13-fold) and day 3 (14.6-fold). Among genes related to the immune system, IL1RN, LILRA3, IGDCC3, and LILRA2 were highly up-regulated both at day 1 and day 3. Within the category of genes related to cell cycle or apoptosis, Aurora kinase C and FGFR4 were highly up-regulated one and 3 days after PU.1 induction. Among down-regulated genes that may be related to cell cycle or apoptosis, PRDM1 and TNFRSF18 were highly down-regulated at day 1 after PU.1 induction. Notably, PRDM1 is a key transcription factor involved in late B-cell differentiation; however, down-regulation of PRDM1 was not sustained (0.9-fold) 3 days after PU.1 induction. Further studies are required to elucidate the role of these genes in cell-cycle arrest and apoptosis observed in KM-H2 tetPU.1 cells after PU.1 induction.
5-aza-2′-deoxycytidine and/or trichostatin A induces up-regulation of PU.1 and apoptosis in cHL cells
Given that PU.1 is down-regulated in Hodgkin lymphoma cells by promoter and URE methylation, and that overexpression of PU.1 induced growth arrest and apoptosis of Hodgkin lymphoma cells, we hypothesized that up-regulation of PU.1 after treatment with demethylation agents and/or histone deacetylase (HDAC) inhibitors might represent a new therapeutic modality for Hodgkin lymphoma patients. We first evaluated the ability of 5-aza-2′-deoxycytidine to induce PU.1 expression. Treatment of Hodgkin lymphoma cell lines with 1μM 5-aza-2′-deoxycytidine induced PU.1 mRNA expression in HD-70, L540, HDLM2, and KM-H2 cells but not in L428 cells (Figure 7A).
Individual or combined treatment with demethylation agent and HDAC inhibitors is a possible therapeutic strategy for cHL. (A) 5-aza-2′-deoxycytidine and/or trichostatin A (TSA) induced PU.1 expression in cHL cell lines. Treatment of L540, HDLM2, HD-70, and KM-H2 cells with 1μM 5-aza-2′-deoxycytidine induced PU.1 expression after 3 days. In contrast, treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine failed to induce PU.1 expression, whereas the combined treatment with 1μM 5-aza-2′-deoxycytidine and 500nM TSA led to induction of PU.1 expression after 3 days. (B-E) Treatment of L540 (B), HD-70 (C), HDLM2 (D), and KM-H2 (E) cells with 1μM 5-aza-2′-deoxycytidine induced growth arrest. (F) Treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine failed to induce growth arrest. (G) The combined treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine and 500nM of TSA induced growth arrest.
Individual or combined treatment with demethylation agent and HDAC inhibitors is a possible therapeutic strategy for cHL. (A) 5-aza-2′-deoxycytidine and/or trichostatin A (TSA) induced PU.1 expression in cHL cell lines. Treatment of L540, HDLM2, HD-70, and KM-H2 cells with 1μM 5-aza-2′-deoxycytidine induced PU.1 expression after 3 days. In contrast, treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine failed to induce PU.1 expression, whereas the combined treatment with 1μM 5-aza-2′-deoxycytidine and 500nM TSA led to induction of PU.1 expression after 3 days. (B-E) Treatment of L540 (B), HD-70 (C), HDLM2 (D), and KM-H2 (E) cells with 1μM 5-aza-2′-deoxycytidine induced growth arrest. (F) Treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine failed to induce growth arrest. (G) The combined treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine and 500nM of TSA induced growth arrest.
We next evaluated the ability of HDAC inhibitors to induce PU.1 expression. Whereas treatment with SAHA failed to induce PU.1 expression in either L428, KM-H2 cells, trichostatin A induced expression of PU.1 mRNA in KM-H2 cells, but not in L428 cells (data not shown). We also evaluated the effect of combined treatment with both 5-aza-2′-deoxycytidine and trichostatin A on cell growth and apoptosis in Hodgkin lymphoma cell lines. Treatment of L428 cells with 1μM 5-aza-2′-deoxycytidine and 500nM trichostatin A led to induction of PU.1 mRNA expression (Figure 7A right panel). We next evaluated the effects of these agents on these Hodgkin lymphoma cell growth. Treatment with 1μM 5-aza-2′-deoxycytidine induced growth arrest in HD-70, L540, HDLM2, and KM-H2 cells (Figure 7B-E); however, L428 cells remained unaffected after treatment with 1μM 5-aza-2′-deoxycytidine (Figure 7F). In contrast, the combination of 1μM 5-aza-2′-deoxycytidine and 500nM trichostatin A induced growth arrest in L428 cells (Figure 7G). Given that the combination of 5-aza-2′-deoxycytidine and trichostatin A, but not treatment with 5-aza-2′-deoxycytidine alone, was capable of inducing PU.1 expression and growth arrest in L428 cells, these results indicate that PU.1 is at least partially responsible for inducing growth arrest of L428 cells in this setting. These data suggest that up-regulation of PU.1 by demethylation agents and HDAC inhibitors may represent a new therapeutic strategy for the treatment of cHL.
Discussion
In this study, we present data demonstrating that PU.1 is a potent tumor suppressor in cHL. First, using bisulfite sequencing, we showed that PU.1 is silenced by the methylation of its promoter and a −17 kb URE. Next, we demonstrated that PU.1 induced growth arrest and apoptosis in cHL cell lines, L428 and KM-H2, and primary cHL cells in vitro. In addition, using a xenograft mouse model harboring tumors of L428 or KM-H2 cells, we showed that PU.1 up-regulation induced tumor regression and promoted long-term survival, compared with mice without PU.1 induction. Finally, we showed that, in the case of L428 cells, growth arrest induced by PU.1 was mediated, at least in part, by the up-regulation of p21.
The oncogenic mechanisms underlying cHL are not well understood. Many B cell–specific genes are down-regulated in cHL cells, including B cell–specific transcription factors, Oct2, Bob1, and PU.1, and cell surface markers, CD19, CD20, and CD79.15 Down-regulation of these genes is caused by promoter methylation.23 In contrast, cHL cells express CD30, a marker expressed on activated B cells and T cells. However, to date, the significance of the down-regulation of B cell–specific genes and the expression of CD30 in cHL remain enigmatic.
One of the genetic lesions of cHL is constitutive activation of the canonical nuclear factor of κ light polypeptide gene enhancer in B-cells (NF-κB) pathway.37 A recent report showed that TNFAIP3 (A20), a negative regulator of the NFκB pathway, is inactivated by deletions and somatic mutations in 40% of cHL patients and in the cHL cell line, KM-H2.13 In addition to TNFAIP3, somatic mutations of NFκB1A, which also inhibits the NF-κB signaling pathway, have been identified in KM-H2 cells.37 NFκB1A, also known as IκB, is mutated in ∼ 20% of cHL patients.37-40 It is also well documented that ∼ 50% of cHL patients harbor amplification or gain of REL, which is a component of NFκB.41-43 Taken together, these data indicate that constitutive activation of NFκB is a key event underlying the pathogenesis of cHL. Our microarray analysis of L428tetPU.1 cells revealed that PU.1 induced a 2- to 3-fold up-regulation of TNFAIP3 at both 1 and 3 days (data not shown). Up-regulation of TNFAIP3 may block the NFκB pathway, therefore accounting for the observed cell-cycle arrest and apoptosis in L428 tetPU.1 cells after PU.1 induction. In contrast, we did not observe any significant change in the expression of NFκB composite proteins (> 2-fold) in L428tetPU.1 cells after PU.1 induction, indicating that PU.1 does not affect the expression of composite members of the NFκB protein complex.
In this study, we also elucidated the mechanisms of L428tetPU.1 and KM-H2tetPU.1 cell growth arrest after PU.1 induction. Gene expression profiling of L428tetPU.1 cells after induction of PU.1 revealed up-regulation of p21, and this accounted at least in part for the cell growth arrest. These results are very similar to our previous finding that p21 is up-regulated by PU.1, leading to cell-cycle arrest in the U266 myeloma cell line.27 Therefore, loss of p21 expression may be involved in the dysregulation of cell growth in some subsets of B-cell malignancies that have lost PU.1 expression. In contrast, the KM-H2 Hodgkin lymphoma cell line did not display p21 up-regulation at the protein level after PU.1 induction. Thus, up-regulation of p21 does not explain the growth arrest observed after PU.1 induction in all subsets of B-cell malignancies. In the case of apoptosis, we previously reported that PU.1 induces apoptosis in U266 myeloma cells via up-regulation of the TNF-related apoptosis inducing ligand.27 Nevertheless, TNF-related apoptosis inducing ligand was intrinsically highly expressed in L428tetPU.1 and KM-H2 tetPU.1 cells in the absence of PU.1 expression, and PU.1 did not further induce TNF-related apoptosis inducing ligand expression in either cell line. Therefore, the mechanism of apoptosis induced by PU.1 in these Hodgkin lymphoma cell lines is probably different from that observed in U266 myeloma cells. The identification of downstream effector proteins involved in PU.1-induced apoptosis in cHL cells may therefore provide new molecular targets for therapy in these patients.
In conclusion, we show that up-regulation of PU.1 in cHL cells induces cell-cycle arrest and apoptosis in vitro and in vivo, and demonstrate that cell-cycle arrest is accounted for, at least partially, by up-regulation of p21. Based on these data, it is possible that demethylation agents and HDAC inhibitors, which induce PU.1 up-regulation, may represent new molecular target modalities for treatment of patients with cHL.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Authorship
Contribution: H.Y. conducted the majority of experiments, generated KM-H2tetPU.1 cells and the xenograft model mice, and wrote the manuscript; S.U. isolated mRNA for DNA microarray analysis; H.T. designed the project; S.E. performed lentiviral transduction and flow cytometry; Y. Kawano analyzed survival curves of xenograft mice; H.N., T.I., and K.A. performed DNA microarray analysis; Y. Komohara and M.T. performed immunostaining of primary Hodgkin lymphoma cells; H.H., T.W., and H.M. gave useful advice; S.O. generated Rag2−/−Jak3−/−balb/c mice and gave useful advice to generate xenograft mice; and Y.O. designed the project, generated L428tetPU.1 cells, and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yutaka Okuno, Department of Hematology, Kumamoto University of Medicine, 1-1-1 Honjo, Kumamoto 860-8556, Japan; e-mail: yokuno@gpo.kumamoto-u.ac.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal