Key Points
Relapsed AML with NPM1 mutation is genetically related to the primary leukemia and characterized by an increase in high-risk aberrations.
DNMT3A mutations show the highest stability and thus may precede NPM1 mutations.
Abstract
Mutations in the nucleophosmin 1 (NPM1) gene are considered a founder event in the pathogenesis of acute myeloid leukemia (AML). To address the role of clonal evolution in relapsed NPM1-mutated (NPM1mut) AML, we applied high-resolution, genome-wide, single-nucleotide polymorphism array profiling to detect copy number alterations (CNAs) and uniparental disomies (UPDs) and performed comprehensive gene mutation screening in 53 paired bone marrow/peripheral blood samples obtained at diagnosis and relapse. At diagnosis, 15 aberrations (CNAs, n = 10; UPDs, n = 5) were identified in 13 patients (25%), whereas at relapse, 56 genomic alterations (CNAs, n = 46; UPDs, n = 10) were detected in 29 patients (55%) indicating an increase in genomic complexity. Recurrent aberrations acquired at relapse included deletions affecting tumor suppressor genes (ETV6 [n = 3], TP53 [n = 2], NF1 [n = 2], WT1 [n = 3], FHIT [n = 2]) and homozygous FLT3 mutations acquired via UPD13q (n = 7). DNMT3A mutations (DNMT3Amut) showed the highest stability (97%). Persistence of DNMT3Amut in 5 patients who lost NPM1mut at relapse suggests that DNMT3Amut may precede NPM1mut in AML pathogenesis. Of note, all relapse samples shared at least 1 genetic aberration with the matched primary AML sample, implying common ancestral clones. In conclusion, our study reveals novel insights into clonal evolution in NPM1mut AML.
Introduction
Acute myeloid leukemia (AML) is a clonal disease characterized by the presence of a variety of genetic alterations. Mutations in the nucleophosmin 1 (NPM1) gene represent one of the most common gene mutations (25%-30%) in AML.1 Approximately 85% of AML with NPM1 mutation (NPM1mut) display a cytogenetically normal (CN) karyotype if it is determined by chromosome banding analysis. High-resolution techniques like single-nucleotide polymorphism (SNP) arrays revealed only a few submicroscopic copy number alterations (CNAs) in NPM1mut AML, whereas uniparental disomies (UPDs) that result in loss of heterozygosity (LOH) were detected in 10% to 20% of the patients.2,3 NPM1mut co-occurs with known AML-associated mutations like FLT3, DNMT3A, IDH1, IDH2, and NRAS.1,4-7 These mutations seem to not accumulate in a random order but instead can be allocated to early and late events in the transformation process from a normal hematopoietic stem cell (HSC) or progenitor cell to leukemia.8,9
A high proportion of NPM1mut AML patients achieve complete remission (CR) with intensive chemotherapy; however, approximately 50% of these patients have relapse during the first 3 years after diagnosis, in particular patients with concurrent internal tandem duplications (ITDs) of the FLT3 gene.1,10-12 Relapse is generally less responsive to chemotherapy and a major cause of death in patients with AML. Other characteristics of NPM1mut AML include low expression or absence of the hematopoietic cell surface marker CD34, multilineage involvement, and a unique gene-expression and miRNA signature.13 On the basis of these clinical and biological characteristics, NPM1mut AML was included as a provisional entity in the current World Health Organization classification.14 In addition to the distinct properties of NPM1mut AML, NPM1mut is considered founder event in the pathogenesis of AML, based mainly on the observation that the mutation is stable over time and in general is maintained at relapse, even occurring after a long latency.15,16 However, we and other groups observed loss of NPM1mut in ∼10% of relapsed patients, and this loss was generally accompanied by further chromosomal and molecular changes.17-19 To date, it is still unclear whether these cases can be explained by clonal evolution from a common preleukemic ancestor or whether these patients developed therapy-related AML (t-AML) that evolved from an independent HSC or progenitor cell.
Clonal evolution is a hallmark of cancer.20 This process is highly affected by cytotoxic therapy because it selects for resistant clones that are the basis for reoccurrence of the disease. Previous studies on relapsed AML estimated clonal evolution in ∼40% of the cases based on chromosomal banding analysis.21-24 In a study by Raghavan and colleagues, SNP genotyping in 27 patients revealed the acquisition of UPDs, mainly UPD13q, in ∼40% of relapses.25 In addition, sequential analysis for AML-associated gene mutations such as FLT3, TP53, NRAS, DNMT3A, IDH1, and IDH2 showed a high variance of their stability.26-29
Thus far, most studies were restricted to small cohorts of AML patients and low-resolution chromosome banding analysis. Kühn et al published a SNP profiling study in adult and pediatric core-binding–factor AML that included 39 paired samples obtained at the time of diagnosis and at relapse.30 Although two-thirds of the relapses were identical to diagnosis or simply acquired additional aberrations, one-third arose from an ancestral clone because they lost one or more aberrations present at diagnosis. In a recent study, Parkin et al integrated cytogenetics, high-resolution SNP profiling, and mutational screening to assess clonal evolution in diagnosis and relapse samples from 28 unselected AML cases, corroborating a genetic relationship in all pairs as well as a high stability of somatic CNAs and UPDs.31 The most comprehensive approach to assess clonal evolution in relapsed AML was performed by Ding et al, who applied whole-genome sequencing in 8 primary/relapse AML pairs. In addition to identification of novel gene mutations in AML, this study unequivocally demonstrated that relapse either emerged from the predominant founding clone or a smaller subclone that shared some but not all mutations of the predominant clone.32 Furthermore, their data suggest that chemotherapy directly causes new gene mutations.
To assess clonal evolution in the genetically defined subgroup “AML with NPM1mut,” we analyzed paired leukemia samples obtained at the time of diagnosis and relapse from 53 NPM1mut AML patients by using high-resolution SNP-array profiling and mutation analysis of 10 AML-associated genes.
Methods
Patients
Patient characteristics for the 53 adult AML patients (24-66 years of age) are given in Table 1. All patients gave informed consent for both treatment and genetic analysis according to the Declaration of Helsinki. Approval was obtained from the institutional review boards of the participating AMLSG institutions.
Clinical characteristics of 53 NPM1mut patients at diagnosis (n = 53)
| Variable . | No. . | % . |
|---|---|---|
| Age, years | ||
| Median | 51 | |
| Range | 24-66 | |
| Sex | ||
| Female | 26 | 49 |
| Male | 27 | 51 |
| AML history | ||
| De novo | 51 | 96 |
| t-AML | 2 | 4 |
| Cytogenetics (banding analysis) | ||
| Normal karyotype | 44 | 83 |
| Deletion 9q | 4 | 8 |
| Not available | 5 | 9 |
| NPM1 mutation type | ||
| A | 40 | 75 |
| B | 5 | 9 |
| D | 4 | 8 |
| Other | 4 | 8 |
| WBC count, ×109/L | ||
| Median | 23 | |
| Range | 1-253 | |
| BM blasts, % | ||
| Median | 90 | |
| Range | 22-100 | |
| Induction cycles, n | ||
| 1 | 12 | 23 |
| 2 | 41 | 77 |
| Remission status after first induction cycle (n = 53) | ||
| CR | 38 | 72 |
| PR | 14 | 26 |
| RD | 1 | 2 |
| Remission status after second induction cycle (n = 41) | ||
| CR | 40 | 98 |
| Relapse | 1 | 2 |
| Consolidation therapy (n = 52) | ||
| High-dose Cytarabin | 44 | 83 |
| Allogeneic SCT | 4 | 8 |
| Autologous SCT | 4 | 8 |
| CIR, months | ||
| Median | 8.8 | |
| Range | 1.3-110.5 | |
| Overall survival after diagnosis, months | ||
| Median | 17.3 | |
| Overall survival after relapse, months | ||
| Median | 7.3 | |
| Variable . | No. . | % . |
|---|---|---|
| Age, years | ||
| Median | 51 | |
| Range | 24-66 | |
| Sex | ||
| Female | 26 | 49 |
| Male | 27 | 51 |
| AML history | ||
| De novo | 51 | 96 |
| t-AML | 2 | 4 |
| Cytogenetics (banding analysis) | ||
| Normal karyotype | 44 | 83 |
| Deletion 9q | 4 | 8 |
| Not available | 5 | 9 |
| NPM1 mutation type | ||
| A | 40 | 75 |
| B | 5 | 9 |
| D | 4 | 8 |
| Other | 4 | 8 |
| WBC count, ×109/L | ||
| Median | 23 | |
| Range | 1-253 | |
| BM blasts, % | ||
| Median | 90 | |
| Range | 22-100 | |
| Induction cycles, n | ||
| 1 | 12 | 23 |
| 2 | 41 | 77 |
| Remission status after first induction cycle (n = 53) | ||
| CR | 38 | 72 |
| PR | 14 | 26 |
| RD | 1 | 2 |
| Remission status after second induction cycle (n = 41) | ||
| CR | 40 | 98 |
| Relapse | 1 | 2 |
| Consolidation therapy (n = 52) | ||
| High-dose Cytarabin | 44 | 83 |
| Allogeneic SCT | 4 | 8 |
| Autologous SCT | 4 | 8 |
| CIR, months | ||
| Median | 8.8 | |
| Range | 1.3-110.5 | |
| Overall survival after diagnosis, months | ||
| Median | 17.3 | |
| Overall survival after relapse, months | ||
| Median | 7.3 | |
WBC, white blood cell; BM, bone marrow; SCT, stem cell transplantation; PR, partial remission; RD, refractory disease; CIR, cumulative incidence of relapse.
SNP microarray-based genotyping analysis
SNP profiling was performed as described recently.3,30 Microarray raw data were made publicly available at NCBI’s Gene Expression Omnibus (GEO-accession number: GSE46951).
Further details are described in the supplemental Methods.
Results
Cytogenetics, CNAs, and UPDs at diagnosis and relapse
At the time of diagnosis.
Data from chromosome banding analysis were available in 48 patients. Among the 43 patients (90%) with normal karyotype, SNP profiling identified 4 CNAs in 3 patients (Unique patient number [UPN] #8, #11, #17) including 3 deletions (13q21.1 [size, 0.02 Mb], 16p13.3-p13.13 [10.8 Mb], 9q21.32 [0.89 Mb]) and one gain [17q (11.1 Mb)]; in 4 patients (UPN #5, #13, #21, #47), cytogenetic analysis revealed a 9q deletion that was confirmed by SNP-array profiling. One patient (UPN#4) had a 47,XYY karyotype that was also present in the remission and relapse sample, confirming the constitutional nature (XYY syndrome). In this patient, a deletion at 9q21.32-q21.33 (2.4 Mb) was identified by SNP-array genotyping. Among the 5 patients without available cytogenetics, SNP profiling detected one gain at 4p16.3 (0.17 Mb). In total, 10 somatic CNAs (gain, n = 2; deletions n = 8) were identified by SNP profiling at the time of diagnosis. Moreover, SNP profiling allowed the detection of UPDs in 5 patients (as a sole abnormality in 4 patients, in combination with a CNA in 1 patient); in 4 cases UPD affected chromosomal arm 13q (UPN#10, #11, #23, #24), all resulting in homozygous FLT3-ITD mutations, and in one case chromosomal band 1p (UPN#1) was affected (supplementary Tables 1 and 2).
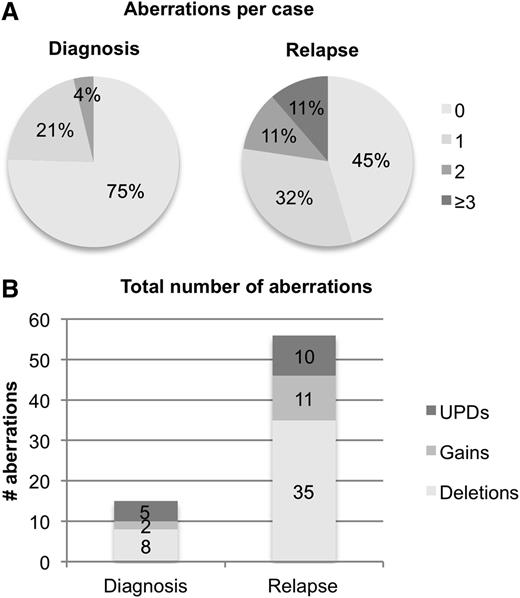
Taken together, we identified one CNA or UPD in 11 patients (21%) and 2 aberrations in 2 patients (4%) at diagnosis (Figure 1, Table 2), with del(9q) and UPD13q being the only recurrent aberrations.
Frequencies of somatic CNA and UPDs identified by SNP array. (A) Number of SNP findings (combined CNA and UPD) per patient at diagnosis and relapse. (B) Absolute number of gains (bright gray), deletions (white), and UPDs (dark gray) at diagnosis and relapse.
Frequencies of somatic CNA and UPDs identified by SNP array. (A) Number of SNP findings (combined CNA and UPD) per patient at diagnosis and relapse. (B) Absolute number of gains (bright gray), deletions (white), and UPDs (dark gray) at diagnosis and relapse.
CNAs and UPDs identified at diagnosis and relapse by SNP profiling
| Aberration . | Candidate genes in CDR . | Diagnosis . | Lost . | Acquired . | Relapse . |
|---|---|---|---|---|---|
| del(9q21) | HNPRK, NTRK2 | 6 | –3 | 3 | |
| UPD13q | FLT3 | 4 | –1 | +7 | 10 |
| UPD1p | ? | 1 | –1 | 0 | |
| Nonrecurrent CNAs at diagnosis | 4 | –2 | 2 | ||
| gain(11q23.3) | MLL | 0 | +3 | 3 | |
| del(12p13) | ETV6 | 0 | +3 | 3 | |
| del(11p13) | WT1 | 0 | +3 | 3 | |
| del(17p) | TP53 | 0 | +2 | 2 | |
| del(17q11.2) | NF1 | 0 | +2 | 2 | |
| del(3p14.2) | FHIT | 0 | +2 | 2 | |
| del(4q22.1) | ? | 0 | +2 | 2 | |
| Trisomy 5 | ? | 0 | +1 | 1 | |
| Monosomy 7 | ? | 0 | +1 | 1 | |
| Nonrecurrent CNAs at relapse | 0 | +22 | 22 | ||
| Total | 15 | –7 | +48 | 56 |
| Aberration . | Candidate genes in CDR . | Diagnosis . | Lost . | Acquired . | Relapse . |
|---|---|---|---|---|---|
| del(9q21) | HNPRK, NTRK2 | 6 | –3 | 3 | |
| UPD13q | FLT3 | 4 | –1 | +7 | 10 |
| UPD1p | ? | 1 | –1 | 0 | |
| Nonrecurrent CNAs at diagnosis | 4 | –2 | 2 | ||
| gain(11q23.3) | MLL | 0 | +3 | 3 | |
| del(12p13) | ETV6 | 0 | +3 | 3 | |
| del(11p13) | WT1 | 0 | +3 | 3 | |
| del(17p) | TP53 | 0 | +2 | 2 | |
| del(17q11.2) | NF1 | 0 | +2 | 2 | |
| del(3p14.2) | FHIT | 0 | +2 | 2 | |
| del(4q22.1) | ? | 0 | +2 | 2 | |
| Trisomy 5 | ? | 0 | +1 | 1 | |
| Monosomy 7 | ? | 0 | +1 | 1 | |
| Nonrecurrent CNAs at relapse | 0 | +22 | 22 | ||
| Total | 15 | –7 | +48 | 56 |
For the complete list, see supplementary Tables 1 to 3. CDR, commonly deleted region.
At the time of relapse.
At relapse, cytogenetic analysis was available for 14 patients and revealed a normal karyotype in 6 patients (all with a normal karyotype at diagnosis) and an aberrant karyotype in 8 patients (6 with normal karyotype, 2 with no available metaphases at diagnosis) (supplementary Table 3). The only recurrent abnormality detected by banding analysis in 2 cases (UPN#10, #26) was an unbalanced translocation derivative chromosome (12;17)(q10;q10), with loss of 17p comprising TP53 and 12p comprising ETV6, which were both confirmed by SNP profiling (supplementary Figure 1 A-D). One case (UPN#22) had a balanced t(2;17)(q35;q11), and SNP profiling detected a 1.6 Mb deletion at the 17q11 breakpoint comprising the tumor suppressor NF1 (supplementary Figure 1 E-F).
SNP-array analysis of the 53 relapse samples revealed that the total number of CNAs increased from 10 at diagnosis to 46 (35 deletions and 11 gains), and the total number of UPDs from 5 to 10 (Figure 1). In 29 patients (55%), we identified at least 1 CNA or UPD: 17 patients (32%) had one, 6 patients (11%) had two, and 6 patients (11%) had three or more CNAs and/or UPDs. The mean number of aberrations (CNAs and UPDs) increased from 0.28 per case at diagnosis to 1.06 per case at relapse (paired Student t test, P = .002).
Five of 10 CNAs detected by SNP profiling at diagnosis were also present at the time of relapse [del(9q) (3 of 6), gain(4p16.3), del(13q12.1)], whereas 5 [del(9q) (3 of 6), del(16q), gain(17q)] were no longer detectable (Figure 2A).
Visualization of recurrent deletions on 9q and UPD on 13q that were present at diagnosis (indicated as D) and/or relapse (indicated as R) by dCHIP. (A) Log2 ratios of 6 patients with del(9q) at the time of diagnosis. Patients #8, #13, and #47 maintained the same deletions at both time points, whereas in patients #4, #5, and #21, del(9q) was lost. Blue indicates deleted and red indicates gained chromosomal segments. (B) Cases with UPD13q at diagnosis and relapse (n = 3), at diagnosis only (n = 1), or at relapse only (n = 7). Mutation status is indicated below the corresponding FLT3-ITD mutation: −/− no mutation; −/+, heterozygous mutation; +/+ homozygous mutation. Blue indicates homozygous (= LOH) and yellow heterozygous regions of chromosome 13 as determined by SNP profiling.
Visualization of recurrent deletions on 9q and UPD on 13q that were present at diagnosis (indicated as D) and/or relapse (indicated as R) by dCHIP. (A) Log2 ratios of 6 patients with del(9q) at the time of diagnosis. Patients #8, #13, and #47 maintained the same deletions at both time points, whereas in patients #4, #5, and #21, del(9q) was lost. Blue indicates deleted and red indicates gained chromosomal segments. (B) Cases with UPD13q at diagnosis and relapse (n = 3), at diagnosis only (n = 1), or at relapse only (n = 7). Mutation status is indicated below the corresponding FLT3-ITD mutation: −/− no mutation; −/+, heterozygous mutation; +/+ homozygous mutation. Blue indicates homozygous (= LOH) and yellow heterozygous regions of chromosome 13 as determined by SNP profiling.
Newly acquired recurrent alterations comprised deletions of tumor suppressor genes at 12p13 (ETV6, n = 3), 11p13 (WT1, n = 3), 17p (TP53, n = 2), 17q11.2 (NF1, n = 2), and 3p14.2 (FHIT, n = 2), as well as a gain of 11q23.3 (MLL, n = 3). Nonrecurrent aberrations comprised trisomy 5 and monosomy 7 (Table 2, supplementary Tables 1 and 2).
The majority of the altered genomic regions at relapse (76%) were exclusively affected in relapse samples. CNAs in these regions could be detected in neither the 53 diagnostic samples nor in an independent cohort of 140 newly diagnosed NPM1mut AML analyzed on the same platform (data not shown).
Two of the 5 patients with UPD at diagnosis lost UPD at relapse (UPD13q in UPN#23 and UPD1p in UPN#1). Conversely, 7 patients acquired a new UPD13q with homozygous FLT3-ITD mutation at relapse (5 with heterozygous and 2 without FLT3-ITD mutation at diagnosis) (Figure 2B).
Gene mutation pattern at diagnosis and relapse
At the time of diagnosis, we observed a typical pattern for concurrent gene mutations in NPM1mut AML (Figure 3). Heterozygous DNMT3A mutations (DNMT3Amut) were detected in 32 patients (60%). The majority of mutations (n = 19, 59%) resulted in an amino acid substitution altering the arginine residue at codon 882. Twenty-four patients (45%) had FLT3-ITD mutations, of which 5 were homozygous defined by an FLT3-ITD to FLT3–wild-type ratio >1; 4 of these cases had UPD13q. The tyrosine kinase domain (TKD) of FLT3 was mutated in 11 cases (21%). Fourteen patients (26%) harbored NRAS mutations, affecting amino acid positions at G12 (n = 7), G13 (n = 6), and G175 (n = 1). Heterozygous IDH1/IDH2 mutations (all affecting R132 and R140, respectively) were detected in 8 (15%) and 12 (25%) patients, respectively. No MLL-PTD, TP53, and ASXL1 mutations were detected in the samples obtained at diagnosis.
Mutation pattern in paired (diagnosis [D]/relapse [R]) samples of 53 NPM1mut patients. Each column represents an individual patient (UPN numbers are indicated in the top row). Colored bars indicate the presence of a mutation, blank bars represent wild-type for the respective gene, and data not available are indicated by a gray bar. Bright and dark green bars represent heterozygous and homozygous FLT3-ITD mutations, respectively. Bars marked by “X” represent different mutation types found at diagnosis and relapse. “Stability” was calculated by the number of mutations that persisted in relapse divided by all mutations present at diagnosis. “Acquired at Relapse” was calculated by the number of cases that acquired a new mutation at relapse and were not detected at diagnosis divided by the number of cases that did not have the same mutation at diagnosis. Mutations in the same gene but of different type (FLT3-ITD, n = 3; NRAS, n = 1) in a diagnosis/relapse pair are considered as loss and new acquisition of a mutation. For MLL-PTD, TP53, and ASXL1, stability could not be determined because no mutations were detected at diagnosis. na, not applicable.
Mutation pattern in paired (diagnosis [D]/relapse [R]) samples of 53 NPM1mut patients. Each column represents an individual patient (UPN numbers are indicated in the top row). Colored bars indicate the presence of a mutation, blank bars represent wild-type for the respective gene, and data not available are indicated by a gray bar. Bright and dark green bars represent heterozygous and homozygous FLT3-ITD mutations, respectively. Bars marked by “X” represent different mutation types found at diagnosis and relapse. “Stability” was calculated by the number of mutations that persisted in relapse divided by all mutations present at diagnosis. “Acquired at Relapse” was calculated by the number of cases that acquired a new mutation at relapse and were not detected at diagnosis divided by the number of cases that did not have the same mutation at diagnosis. Mutations in the same gene but of different type (FLT3-ITD, n = 3; NRAS, n = 1) in a diagnosis/relapse pair are considered as loss and new acquisition of a mutation. For MLL-PTD, TP53, and ASXL1, stability could not be determined because no mutations were detected at diagnosis. na, not applicable.
At the time of relapse, the gene mutation pattern changed in 34 patients (64%): 5 patients (9%) lost NPM1mut at relapse, even when applying a highly-sensitive RNA-based NPM1mut-specific real-time quantitative polymerase chain reaction18 (see next subsection).
In 31 (97%) relapse samples, we detected the same DNMT3Amut type as determined at diagnosis, whereas in 1 patient the DNMT3Amut was lost. Only 1 (5%) of 22 patients with DNMT3A wild-type at diagnosis acquired a DNMT3Amut at relapse; 2 patients with heterozygous DNMT3Amut at diagnosis acquired a second DNMT3Amut at relapse. FLT3-ITD persisted in 18 of 24 patients (75%), 2 patients (8%) lost a heterozygous mutation, and 1 patient (4%) lost a homozygous mutation together with loss of UPD13q; in 6 cases, heterozygous FLT3-ITD evolved to a homozygous state, 5 of them by UPD13q. In addition, 3 of 24 patients (13%) with FLT3-ITD mutation at diagnosis had a different ITD length at relapse, and 7 patients lost 1 or more of the ITD clones present at diagnosis while maintaining at least one of the original ITD-clones (supplementary Figure 2). Conversely, 9 of 29 (31%) FLT3-ITD–negative patients acquired FLT3-ITD mutations (heterozygous, n = 7; homozygous, n = 2) at relapse. FLT3-TKD mutation was maintained at relapse in only 1 of 11 patients (9%), and none of the 42 patients without FLT3-TKD mutation at diagnosis acquired this mutation at relapse. Only 5 of 14 (36%) patients maintained the same NRAS mutation type, whereas 4 of 39 (10%) previously NRAS wild-type cases acquired an NRAS mutation at relapse; 1 patient showed a different mutation type at relapse. Two of 8 patients lost IDH1, and 1 of 12 cases lost IDH2 mutation at relapse, whereas the remaining 6 IDH1mut and 11 IDH2mut patients presented with the identical mutation types at relapse. Four (9%) of 45 IDH1 wild-type patients acquired a new IDH1mut. One (2%) TP53 mutation (G245S) was detected in a relapse sample with del(17p) on cytogenetic analysis. Of note, 3 of 5 patients who lost NPM1mut at relapse acquired an MLL-PTD mutation that corresponded to a small gain at 11q23.3 detected by SNP-array analysis. Two patients acquired an ASXL1 mutation at relapse.
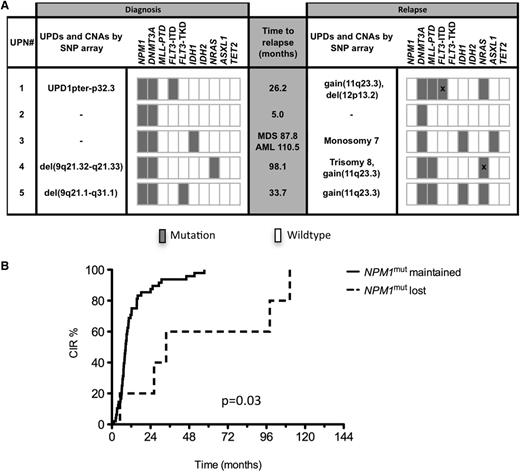
Loss of NPM1 mutation at relapse
Five patients lost NPM1mut at relapse; at diagnosis we detected genomic aberrations in 3 of these patients by SNP analysis [UPN#5, del(9q21.1-q31.1); UPN#1, UPD 1pter-32P.2; UPN#4, del(9q21.32-q21.33)] (Figure 4A), whereas no aberrations were identified in the other 2 patients (UPN#2, #3). At relapse, none of these genomic lesions remained detectable, whereas new aberrations occurred: patients UPN #5, #1, and #4 all acquired a small gain at chromosomal band 11q23 that corresponded to an MLL-PTD mutation; in addition, patient UPN#4 acquired trisomy 8, and patient UPN#1 a del(12p13.2) comprising the ETV6 gene. UPN#2 maintained a normal SNP profile at relapse, and patient UPN#3 acquired a monosomy 7 at relapse.
Genetic profiling and CIR in patients who lost or retained NPM1mut. (A) Genetic profiling by SNP array and gene mutation analyses of 5 patients with loss of NPM1mut at relapse. Bars marked by “X” represent different mutation types found at diagnosis and relapse. (B) CIR in patients with loss of NPM1mut and patients who retained NPM1mut.
Genetic profiling and CIR in patients who lost or retained NPM1mut. (A) Genetic profiling by SNP array and gene mutation analyses of 5 patients with loss of NPM1mut at relapse. Bars marked by “X” represent different mutation types found at diagnosis and relapse. (B) CIR in patients with loss of NPM1mut and patients who retained NPM1mut.
Characterization of concurrent gene mutation patterns revealed further insights into the clonal evolution of these 5 cases of AML: UPN#5 lost the FLT3-TKD mutation and acquired an IDH1 and NRAS mutation; UPN#4 changed its NRAS mutation type (G13V to G13D) at relapse, and UPN#3 still showed the same IDH1 mutation and acquired an ASXL1 mutation; UPN#1 had a heterozygous FLT3-ITD mutation at diagnosis with an ITD size of 15 bp. This ITD clone was lost at relapse and 2 new FLT3-ITD clones with a longer size (36 and 54 bp) were acquired. The only stable marker that was consistently present at both time points in all 5 cases was DNMT3Amut without any changes in mutation type. The median time interval between achievement of CR and relapse in these 5 AML cases was 33.7 months (range, 5.0-110.5 months) and thus significantly longer than in the 48 patients maintaining the NPM1mut at relapse (median, 8.6 months; range, 1.3-57.4; P = .03) (Figure 4B). UPN#3 developed peripheral blood cytopenia 87.8 months after AML diagnosis, and a bone marrow smear revealed dysplasias with 5% myeloid blasts, consistent with a diagnosis of myelodysplastic syndrome (MDS). At that time, cytogenetic analysis revealed monosomy 7 that was also detected after transformation in AML 22.7 months later. No history of MDS preceding AML relapse was reported in the remaining 4 patients.
None of the 5 patients with NPM1mut loss at relapse responded to intensive salvage therapy, whereas 66% of patients that maintained NPM1mut achieved a second CR (P = .009).
There was no striking difference with regard to clinical characteristics at diagnosis between patients who lost NPM1mut and patients who maintained NPM1mut (supplementary Table 4).
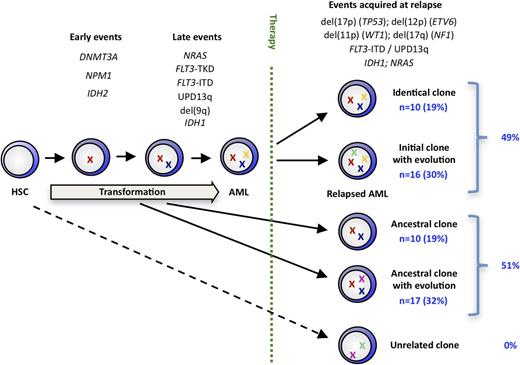
Clonal evolution in NPM1mut AML
We integrated our findings from high-resolution SNP profiling and mutation screening to assess the clonal relationship of the diagnosis/relapse pairs. Twenty-six relapses (49%) maintained all mutations, CNAs, and UPDs at relapse, with 10 relapses (19% of total cohort) being genetically identical and 16 (30%) having additional alterations. The remaining 27 relapse samples (51%) lost (with or without acquisition of a new aberration) 1 or more aberrations that were present in their matched diagnosis sample. Ten (19% of total cohort) of these cases simply lost aberrations or mutations present at diagnosis, whereas in 17 (32%) we found 1 or more newly acquired somatic genetic lesions. None of the 53 AML diagnosis/relapse pairs showed a completely unrelated aberration pattern because all diagnosis/relapse pairs shared at least 1 genetic lesion (Figure 5). We used the number of identical genetic aberrations (gene mutations, CNAs, UPDs) at diagnosis and relapse in an individual patient as a quantitative estimate for the genetic relationship between both AML samples. This number correlated inversely with the time to relapse, with primary/relapse pairs showing a high degree of genetic relationship relapsing earlier than pairs that were less genetically related (supplementary Figure 3).
A model of clonal evolution in NPM1mut AML based on the stability of each genetic marker evaluated in our study.
A model of clonal evolution in NPM1mut AML based on the stability of each genetic marker evaluated in our study.
Discussion
High-resolution SNP-array profiling of 53 relapsed NPM1mut AML patients revealed only few genomic aberrations in the primary AML samples (mean, 0.28/case) corroborating the chromosomal stability of leukemic cells in newly diagnosed NPM1mut AML. At the time of relapse, we observed a significant increase (mean, 1.06/case) in the number of genomic alterations identified by SNP profiling. This change to a higher genetic complexity at relapse is consistent with previous studies that compared diagnosis and relapse samples in unselected AML cohorts by chromosomal banding analysis.21-24 CNAs and UPDs detected at diagnosis were not necessarily present in the matched relapse sample, as exemplified by the recurrent del(9q) that was lost in 3 of 6 (50%) cases and therefore represent late events in NPM1mut AML. This in contrast to the study by Parkin et al,31 where all CNAs detected at diagnosis persisted at relapse in 28 cases of different cytogenetic and genetic AML subgroups and were therefore interpreted as early events.
Distinct chromosomal regions were recurrently affected by CNAs at relapse, like gain of 11q23 comprising MLL (n = 3) and deletions on 17p (n = 2), 17q (n = 2), 12p (n = 3), 11p (n = 3), and 3p14.2 (n = 2) affecting TP53, NF1, ETV6, WT1, and FHIT, respectively. Parallel cytogenetic and SNP-array–based analysis in 14 relapse samples revealed that deletions in part resulted from unbalanced translocations. The observation that most aberrations (76%) were exclusively found in relapse samples as opposed to NPM1mut AML at diagnosis suggests that they were induced by chemotherapy or were present in small subclones that were selected by therapy. Most of these aberrations found at the time of relapse are associated with high-risk AML and therefore might in part explain the poor outcome of relapsed AML.33,34
Although genomic aberrations as detected by chromosome banding analysis or SNP profiling are rare in newly diagnosed cases of NPM1mut AML, most cases harbor concurrent gene mutations. Comparative mutation analysis of primary and relapse samples revealed a high stability for mutations in DNMT3A (97%), IDH2 (92%), and NPM1 (91%), whereas FLT3-ITD (75%) and IDH1 (75%) were less stable than what is mostly in line with previous studies.16,27,28,35-38 Conversely, the majority of FLT3-TKD (91%) and NRAS mutations (64%) present in the primary leukemia were not retained at relapse. The loss of a somatic mutation or aberration at relapse implies that it is (i) a late event in the development of AML and (ii) cells with these mutations are more sensitive to chemotherapy. In this respect, previous studies did not find an impact of either FLT3-TKD or NRAS mutations on outcome.7,39 FLT3-ITD mutations are acquired in one-third of FLT3-ITD–negative patients at the time of relapse, highlighting that FLT3-ITD mutations occur late in the pathogenesis of AML as corroborated by other studies,40 but in contrast to FLT3-TKD and NRAS mutations, they represent important driver mutations for relapsed AML. Furthermore, the incidence of UPD13q with homozygous FLT3-ITD mutations increased from diagnosis (8%, n = 4) to relapse (19%, n = 10), similar to the study by Raghavan et al,25 which observed an acquisition of UPD13q at the time of relapse in 22% of patients. In our cohort, IDH1 mutations were less stable and more often acquired than in previous studies.37,38 Therefore, IDH1 mutations might represent later events in NPM1mut AML than in other AML subgroups. In relapsed NPM1mut AML, TP53 and ASXL1 mutations do not seem to play a major role because only 1 TP53 and 2 ASXL1 mutations were detected at this time point.
The limited sensitivity (∼5%) of the techniques used in this study did not allow us to determine whether newly acquired mutations or genomic alterations at relapse preexisted in small subclones at the time of diagnosis or whether they were acquired during or after chemotherapy. Deep sequencing of 8 paired diagnosis/relapse AML samples by Ding et al could not detect the majority of the relapse-specific mutations in the primary AML sample, suggesting that these mutations were acquired after therapy.32 However, even high-coverage targeted sequencing has a detection limit of ∼2% and therefore will not allow the detection of very small sublcones that expand under selection pressure with chemotherapy. We also cannot rule out the possibility that distinct mutations, in particular frequent ones like NPM1 mutation type A, were lost and reacquired at relapse. Within this context, one patient showed different NRAS mutation types at diagnosis and relapse, indicating that this is a possible mechanism of clonal evolution. Similarly, 3 of 24 patients showed different FLT3-ITD clones at relapse. However, this was not the case for DNMT3Amut, where all patients maintained the same nucleotide changes.
Loss of NPM1mut has been described to occur in ∼10% of relapsed AML cases.17-19 Because of the long latency between primary leukemia and relapse, together with a diverging pattern of genomic alterations and gene mutations, these cases were initially interpreted as t-AML rather than relapse.17,18 By extending the number of analyzed genes in these patients, we consistently found DNMT3Amut in the primary and relapse sample in 5 of 5 NPM1mut loss cases. For each patient, we found the same nucleotide changes in DNMT3A, making it more likely that a common ancestral DNMT3Amut but NPM1 wild-type clone gave rise to both primary and relapsed AML as opposed to a scenario in which 2 hematopoietic clones have independently acquired the same DNMT3Amut. The observation that at least in these 5 cases DNMT3Amut most likely preceded NPM1mut controverts the current concept that NPM1mut is generally the founder event in NPM1mut AML.13 However, 1 case in our cohort lost DNMT3Amut at relapse and maintained NPM1mut, highlighting that the sequential order in which they occur is not strictly determined. DNMT3A knockout in mouse HSCs strongly induces self-renewal, supporting its role in malignant transformation.41 However, even after several rounds of transplantation, these mice do not develop AML or other myeloid malignancies, suggesting the need for secondary genetic hits. This might explain the long latency between the primary leukemia and relapse observed in the 5 patients with NPM1mut loss. Although results are based on a very small patient number, we nevertheless find the observation intriguing because it supports the hypothesis that the persisting DNMT3Amut/NPM1wt clone again has to acquire additional mutations to transform to AML. Further studies including patients in continuous complete remission are warranted to assess the likelihood of persisting DNMT3Amut clones to evolve to MDS or AML.
In our study, combined SNP-array profiling and mutational analysis revealed that all 53 relapses shared at least 1 mutation or aberration with their matched primary sample, implicating a common cell of origin. Based on our findings, there is no evidence for t-AML that derived from a genetically unrelated hematopoietic clone. Similarly, previous combined genomic and molecular studies in AML31,32 and acute lymphoblastic leukemia42,43 have demonstrated that the primary leukemia and relapse in general are genetically related and derive directly from the dominant leukemic clone or from a common ancestral clone. In addition, the increase of specific genomic aberrations that we found in our NPM1mut cohort at the time of relapse suggests that DNA-damaging chemotherapy causes secondary genetic changes in persisting (pre-)leukemic clones. In this respect, some of the specific alterations we found exclusively at relapse, such as −7/del(7q) and derivative chromosome (12;17)(q10;q10), are frequent in patients who were previously treated with chemotherapy.44 This hypothesis is further supported by the whole-genome sequencing study that revealed a marked increase in the absolute number of a specific type of mutation (transversions) in the genomes of relapsed AML compared with de novo AML.32
In our study, 26 of the 53 relapses (49%) evolved directly from the dominant primary clone because it maintained all the mutations and genomic aberrations present in the primary AML sample. The other group of 27 (51%) relapse samples lost some of the genetic changes of the initial clone but shared at least one common genetic alteration, implying that they derived from an ancestral clone. Although limited by the markers studied, our data are in line with data from a whole-genome sequencing study in relapsed AML, where 3 of 8 relapses evolved from the dominant primary clone and the remaining 5 from a minor, genetically related subclone.32 Genetic characterization of fluorescence-activated cell–sorted HSCs and leukemic stem cells of AML patient samples directly demonstrated that these (pre-)leukemic subclones that comprise some but not all mutations (mostly FLT3-ITD) coexist with the dominant leukemic clone at diagnosis.40
In conclusion, although NPM1mut is generally considered a primary event in NPM1mut AML, our data suggest that mutations in DNMT3A can precede NPM1 mutations. Even after a long latency of up to 110 months, all relapses were genetically related to the initial diagnosis of AML, suggesting that persisting subclones or preleukemic HSCs acquire new secondary aberrations rather than being genetically unrelated t-AML. To achieve long-term leukemia-free survival, future therapies in AML need to eradicate all (pre-)leukemic clones present at diagnosis that persist during therapy and contribute to relapse.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all AMLSG institutions and investigators who contributed to this study as well as Karin Bezet and Juliane Grohlke for technical support with molecular analyses.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG; SFB 1074, project B3), Germany
Authorship
Contribution: J.K., L.B., V.T., F.T., V.I.G., and P.P. performed research; V.T. performed cytogenetic analysis; J.K., L.B., V.T., V.I.G., P.P., F.T., F.G.R., M.W.M.K., K.H., S.K.-S., B.S., and G.G., analyzed and interpreted data; J.K., D.S., and R.F.S. performed statistical analysis; L.B., V.I.G., P.P., F.G.R., M.W.M.K., T.K., M.S., J.K., A.G., R.F.S., H.D., and K.D. were involved directly or indirectly in care of patients and sample procurement; R.F.S., H.D., and K.D. provided administrative support; J.K., L.B., H.D., and K.D. designed the research; and J.K., L.B., H.D., and K.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.


![Figure 3. Mutation pattern in paired (diagnosis [D]/relapse [R]) samples of 53 NPM1mut patients. Each column represents an individual patient (UPN numbers are indicated in the top row). Colored bars indicate the presence of a mutation, blank bars represent wild-type for the respective gene, and data not available are indicated by a gray bar. Bright and dark green bars represent heterozygous and homozygous FLT3-ITD mutations, respectively. Bars marked by “X” represent different mutation types found at diagnosis and relapse. “Stability” was calculated by the number of mutations that persisted in relapse divided by all mutations present at diagnosis. “Acquired at Relapse” was calculated by the number of cases that acquired a new mutation at relapse and were not detected at diagnosis divided by the number of cases that did not have the same mutation at diagnosis. Mutations in the same gene but of different type (FLT3-ITD, n = 3; NRAS, n = 1) in a diagnosis/relapse pair are considered as loss and new acquisition of a mutation. For MLL-PTD, TP53, and ASXL1, stability could not be determined because no mutations were detected at diagnosis. na, not applicable.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/1/10.1182_blood-2013-01-479188/4/m_100f3.jpeg?Expires=1765936285&Signature=248KChGfF59LrSyRwZbtfYeHhuXRtRN9qOQkL1OokrAjgQ7Hv4YLrAbDo96~Tj5ZmZCBBl7ZO1VsaUJ3giNiqJyKHq8sheJH1xI~-SIYT5ZBvI3v2Tq6F9nEd9doHHo0lSxPQ4glIvwkA39sfT2Bn1z9ZBGJzBy9Rs7Kujt5zH7DrabrFiNmzaVK~NDQ6831IIwZgLHnlJa33DBSmPMhx044HLJJSUkchePaNlAIHhCxBN8YfwyspZwWgi6nD3S4xpBSrzrfKSNSr7AqRN23L8~ep~Gy85U1tiMhipXs6hcTT2s~6YlRNOY1F058qVDcU5czUWjx25F6f8Tx8avcdQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)