Key Points
A recurrent gain of a region of chromosome 11 (11q24.3) occurs in up to one-quarter of cases of diffuse large B-cell lymphoma.
ETS1 and FLI1 genes are overexpressed and determine proliferation, survival, and differentiation arrest of the lymphoma cells.
Abstract
Diffuse large B-cell lymphoma (DLBCL) is the most common form of human lymphoma. DLBCL is a heterogeneous disease characterized by different genetic lesions. We herein report the functional characterization of a recurrent gain mapping on chromosome 11q24.3, found in 23% of 166 DLBCL cases analyzed. The transcription factors ETS1 and FLI1, located within the 11q24.3 region, had significantly higher expression in clinical samples carrying the gain. Functional studies on cell lines showed that ETS1 and FLI1 cooperate in sustaining DLBCL proliferation and viability and regulate genes involved in germinal center differentiation. Taken together, these data identify the 11q24.3 gain as a recurrent lesion in DLBCL leading to ETS1 and FLI1 deregulated expression, which can contribute to the pathogenesis of this disease.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma, accounting for 25% to 30% of adult cases in western countries, and it is characterized by heterogeneous histopathological, biological, and clinical features.1
DLBCL has been molecularly classified in at least 3 subtypes, reflecting different stages of B-cell differentiation: germinal center B-cell–like DLBCL (GCB) from germinal center (GC) centroblasts, activated B-cell–like DLBCL (ABC) from GC cells undergoing plasmacytic differentiation, and an intermediate type 3 DLBCL.2 Physiological GC B-cell differentiation requires a complex transcriptional program and DLBCL displays recurrent genetic abnormalities that typically circumvent this normal genetic program, allowing the neoplastic B cells to avoid plasmacytic differentiation and evade apoptosis.2 Briefly, chromosomal translocations involving the BCL2 oncogene and mutations of the EZH2 methyltransferase are almost exclusively observed in GCB DLBCL,3 whereas ABC DLBCL are preferentially associated with a disruption of terminal B-cell differen-tiation program (ie, PRDM1 inactivation and BCL6 deregulation)4,5 and constitutive activation of the nuclear factor κB (NF-κB) transcription pathway (most commonly via somatic mutations of TNFAIP3, CARD11, CD79B, and MYD88).6-8 Other recurrent lesions can be found in all the DLBCL subtypes, including inactivation of histone and/or chromatin modifying genes CREBBP, EP300, and MLL29,10 and of genes involved in immune recognition, such as β2-microglobulin and CD58.11 It is likely that additional, undescribed genetic defects may also contribute to the pathogenesis of DLBCL.
In the present study, aimed to identify new recurrent genetic defects in DLBCL, we performed genomic profiling analysis on 166 DLBCL samples, which enabled us to characterize a new recurrent lesion on chromosome 11q24.3 occurring in up to one-quarter of cases. The region contains 2 ETS transcription factors, ETS1 and FLI1. Known to be involved in the pathogenesis of multiple human cancers,12,13 both ETS1 and FLI1 have an important role in the development of lymphoid tissues and immune system control,14-16 regulating the activity of important players in B-cell growth and maturation such as the B-cell–specific activator protein PAX517,18 and the master regulator of plasma cell differentiation, PRDM1.19 Notably, Ets1 and Fli1 are repressed during the late phases of B-cell differentiation in an in vitro murine system.20 Here, we show that the 11q24.3 genomic lesion correlates with high levels of ETS1 and FLI1 expression in human DLBCL primary samples. Moreover, we demonstrated, in an in vitro model, that both ETS1 and FLI1 are critical for cell viability and that they regulate genes that are controlling the normal GC B-cell differentiation.
Methods
Tumor panel
DNA profiles, obtained using the GeneChip Human Mapping 250K NspI (Affymetrix, Santa Clara, CA), of 166 DLBCL samples from a previously published series were investigated (GSE15127).21 Genomic profiles of DLBCL cell lines were obtained using the 250K or single nucleotide polymorphism 6.0 array, as previously described.21,22 Gain was validated by fluorescence in situ hybridization using FLI1/EWSR1 probe (Cytocell, Cambridge, UK). Gene expression data of DLBCL clinical specimens were obtained using the Affymetrix Genechip U133 plus 2.0 (Affymetrix) (GSE10846). CEL files were imported and normalized using robust multiarray average algorithm in Partek Genomics Suite 6.4 (Partek, St. Louis, MO). Box-plots were created with Stata/SE v.12.1 (StataCorp, College Station, TX), and differences of expression between cases with and without the 11q24.3 gain were evaluated with a 1-sided t test for unpaired data with unequal variance. The distinction of DLBCL cases in GCB or in ABC was available in 134 cases: assessed in 77 cases by immunohistochemistry according to the algorithm of Hans et al23 (28 GCB, 47 non-GCB) and in 57 by GEP24 (30 GCB, 27 non-GCB/ABC), as previously described.21 All patients provided informed consent in accordance with the hospital’s institutional review board and the Declaration of Helsinki.
Immunohistochemistry
Formalin-fixed, paraffin-embedded, 4-µm-thick sections from 12 cases of DLBCL underwent immunohistochemical characterization with antibodies against ETS1 (1G11, Novocastra, Nunningen, Switzerland) and FLI1 (G146-222, BD Pharmingen, Buccinasco, Italy).
Short hairpin RNAs and plasmid constructs
The short hairpin RNA (shRNA) molecules were obtained from the Expression Arrest TRC library (Sigma-Aldrich, St. Louis, MO). Human pLKO.1 lentiviral shETS1 used were TRCN0000005588 and TRCN0000005591, shFLI1: TRCN0000005324 and TRCN0000005326. pLKO.1 GFP expressing vector was constructed as previously described.25 ETS1 expressing vector was cloned in pCMV6-Entry vector (Origene, Rockville, MD).
Cell culture and lentiviral infection
HEK293T cells were cultured under standard conditions (37°C in humidified atmosphere, with 5% CO2) in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum (FCS). DLBCL cell lines (OCI-Ly7, SU-DHL4, Val, U-2932, SU-DHL2, and OCI-Ly10) were obtained from Dalla Favera and Staudt laboratories. OCI-Ly7, SU-DHL4, Val, and U-2932 were maintained in RPMI1640 supplemented with 10% FCS and 1% penicillin/streptomycin and 2 mM glutamine. OCI-Ly7 medium was additioned with nonessential amino acids (100 μM), Na-pyruvate (1 nM), and β-mercaptoetanol (50 μM). SU-DHL2 and OCI-Ly10 were maintained in Iscove modified Dulbecco medium supplemented with 10% FCS, 1% penicillin/streptomycin, and 2 mM glutamine.
Transfections of HEK-293T were performed with JetPrime reagent (Polyplus Transfection, Illkirch, France) according to the manufacturer’s instructions. HEK293T were transfected with pLKO.1 lentiviral vector in combination with third-generation helper plasmids. Self-inactivating lentiviral particles were produced as previously described.25 Exponentially growing cells (3 × 105/mL) were infected with viruses and puromycin added to the medium 48 hours after infection. Alive cells were recovered after 72 hours and collected at different days.
Electroporation
OCI-Ly7 cells were transiently transfected with Amaxa Nucleofector methodology (Amaxa Inc., Gaithersburg, MD). Briefly, 5 × 106 cells per nucleofection sample were resuspended in 100 μL of nucleofector solution V. Two micrograms of pCMV6-ETS1 or control pCMV6-EV plasmid DNA was added to each cell suspension and transferred to an Amaxa-certified cuvette. Nucleofection was performed using the O-17 program.
Real-time polymerase chain reaction (PCR)
RNA was extracted using the RNA easy kit (Qiagen AG, Hombrechtikon, Switzerland) and reverse-transcribed using the Superscript First-Strand Synthesis System for real-time PCR kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer’s instructions.
Real-time PCR amplification was performed using Fast SYBR Green Master Mix on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). Primer sets (supplemental Table 1) were designed using Primer3.26 All samples were analyzed in triplicates.
Western blotting analysis
Cells were solubilized in hot SDS lysis buffer (2.5% SDS, Tris-HCl pH 7.4) and sonicated for 15 s. The following primary antibodies were used: anti-ETS1 (C-20, Santa Cruz Biotechnology, Santa Cruz, CA), anti-FLI1 (C-19, Santa Cruz Biotechnology), anti-PRDM1α (6D3, Santa Cruz Biotechnology) and anti-GAPDH (MAB374, Millipore, Zug, Switzerland), anti-IRF4 (no. 4964, Cell Signaling technology), and anti-XBP1s (clone 143F, Biolegend, CNIO, Spain).
Proliferation assays
Cells infected with pLKO.1shGFP, shETS1, or shFLI1 were seeded in triplicate in 24-well plates after puromycin selection and counted daily for 5 days.
For EdU labeling, 106 OCI-Ly7 cells were pulsed with EdU (10 μM) for 2 hours at 37°C in 5% CO2. EdU incorporation was determined by flow cytometry using the Click-iT EdU Alexa488 Kit (Invitrogen) according to the manufacturer’s instructions.
For competition assay, OCI-Ly7 cells infected with pLKO.1shGFP, shETS1, or shFLI1 (GFP negative) were mixed with an equal number of OCI-Ly7 cells infected with pLKO.1 GFP-expressing vector (GFP positive) and the percentage of GFP positive cells was evaluated by fluorescence-activated cell sorter analysis at days 3, 7, 10, 17, and 24.
Analysis of apoptosis by flow cytometry
Apoptosis was measured by flow cytometry after staining with annexin V and propidium iodide. Annexin V binding was revealed by incubation with FITC. Cells were analyzed by FACScan using CellQuest Program.
Chromatin immunoprecipitation
OCI-Ly7 cells infected with shGFP and shETS1 were collected, cross-linked with formaldehyde, and processed as previously described.27 After sonication, chromatin was immunoprecipitated with antibody for ETS1 (C-20 X, Santa Cruz Biotechnology). DNA-protein cross-links were reversed and DNA purified from total cell lysates (input) and immunoprecipitated fractions. Quantitative real-time PCR was performed using SYBR Green qPCR and the following primers: PRDM1 FW 5- GAGAAGCAGGAATGCAAGGT-3′ and PRDM1 REV 5′-AGCGGTCGGAGGCAGTAAT-3′. The amount of immunoprecipitated DNA was calculated in reference to a standard curve and normalized to input DNA.
Gene expression profiling
Gene expression profiling was performed after single ETS1 and FLI1 down-regulation using 2 shRNAs against each gene in 2 independent experiments. Total RNA was isolated using Trizol (Invitrogen Life Technologies). The concentration and the quality of total RNA were assessed as previously performed.28 Samples were processed using the HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA), according to the manufacturer’s protocol. Arrays were read on an Illumina HiScanSQ system. Data were first extracted with the Illumina GenomeStudio software and then imported in Partek Genomics Suite 6.4 and quantile normalized. Transcripts with differences in expression were identified by ANOVA using both shRNA for each gene against shGFP. Genes were considered significant for absolute fold change >1.2 and false discovery rate <0.20. Functional annotation was performed using the Gene Set Enrichment Analysis,29 and the enrichment was calculated for “chemical and genetic perturbations” and “canonical pathways” gene sets. Gene Set Enrichment Analysis default settings were used to define enrichment of gene sets. Raw data will be available at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) database.
Results
A recurrent 11q24.3 gain is associated with overexpression of ETS1 and FLI1
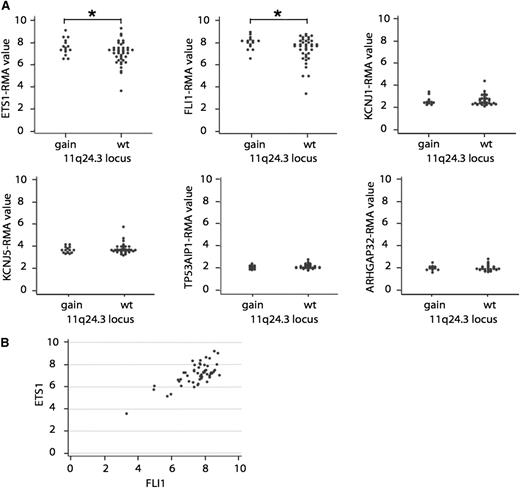
The analysis of the genomic profiles of 166 cases of DLBCL by high-density genome-wide single nucleotide polymorphism-based array21 had originally identified a minimal recurrent region of gain, not previously functionally characterized, mapping at 11q24.3 (GRCh37/hg19chr: 128 137,400-128 994,334) in 23% (38/166) of the cases. The region contained 6 genes: ETS1, FLI1, KCNJ1, KCNJ5, TP53AIP1, and ARHGAP32. Integration of genomic profiles with gene expression profiles in a set of 54 cases (16 bearing 11q24.3 gain) showed that, among these 6 genes, only the ETS1 and FLI1 genes were expressed (Figure 1A). Cases with 11q24.3 gain presented significantly higher RNA levels of ETS1 (fold change: 1.4, P = .0129) and FLI1 (fold change: 1.5, P = .0082) than wild-type cases (Figure 1A). There was also a positive correlation between the 2 mRNA levels across all DLBCL cases (correlation coefficient 0.73; Figure 1B). Expression was also confirmed at the protein level on 12 DLBCL cases, 4 bearing the gain (supplemental Figure 1). There was no significant association between 11q24.3 gain and the DLBCL cell of origin. As assessed on the GSE10846 dataset24 (167 ABC DLBCL, 183 GCB DLBCL, 66 unclassified DLBCL), ETS1 levels were higher in ABC and GCB DLBCL cases than in 9 samples of benign hyperplasia (P = .013 and P = .035, respectively) and in ABC than in GCB DLBCL (P < .001). FLI1 expression levels did not show any difference between the 2 DLBCL subtypes or in respect to the benign hyperplasia.
11q24.3 gain is a recurrent event in DLBCL and it is associated with high levels of ETS1 and FLI1. (A) Patients with the 11q24.3 gain express high levels of ETS1 and FLI1 mRNA. RNA levels of ETS1, FLI1, KCNJ1, KCNJ5, TP53AIP1, and ARHGAP32 in patients with 11q24.3 gain (gain) compared with patients without the gain (wt). *P value < .05. (B) Correlation between ETS1 and FLI1 mRNA levels in DLBCL samples.
11q24.3 gain is a recurrent event in DLBCL and it is associated with high levels of ETS1 and FLI1. (A) Patients with the 11q24.3 gain express high levels of ETS1 and FLI1 mRNA. RNA levels of ETS1, FLI1, KCNJ1, KCNJ5, TP53AIP1, and ARHGAP32 in patients with 11q24.3 gain (gain) compared with patients without the gain (wt). *P value < .05. (B) Correlation between ETS1 and FLI1 mRNA levels in DLBCL samples.
ETS1 and FLI1 sustain cell viability of a DLBCL cell line
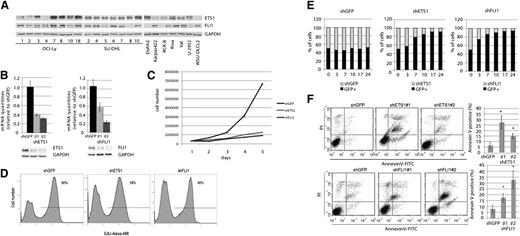
To study a putative pathogenetic role of ETS1 and FLI1 in DLBCL, we first screened 22 DLBCL cell lines, encompassing GCB and ABC subtypes, for ETS1 and FLI1 protein levels and found that most of them expressed ETS1 and FLI1, albeit at different levels. By combining these data with genome-wide DNA profiling, we found that among the evaluated cell lines, the OCI-Ly7 DLBCL cell line presented both the 11q24.3 gain observed in clinical specimens and the highest protein levels of ETS1 and FLI1 (Figure 2A; supplemental Figure 2). Thus, we considered the OCI-Ly7 DLBCL cell line a suitable model to in vitro study the pathogenetic roles of ETS1 and FLI1 in DLBCL.
ETS1 and FLI1 affect proliferation and viability in OCI-Ly7. (A) ETS1 and FLI1 protein levels in 22 DLBCL cell lines. Western blot analysis was carried out on whole-cell lysates with antibodies against ETS1, FLI1, and GAPDH as loading control. (B) ETS1 and FLI1 down-regulation in OCI-Ly7 cell line. Real-time PCR and western blot analysis of ETS1 (left) and FLI1 (right) levels after shRNA (day 5). As a control, β2-microglobulin mRNA and GAPDH protein expression were documented. (C) For the growth curve, OCI-Ly7 cells infected with control (shGFP) lentivirus or a lentivirus expressing shRNA for ETS1 and FLI1 (shETS1 and shFLI1, respectively), were seeded on 24-well plates at a density of 3 × 105 cells/well. Cultures were harvested every day, and the number of cells was determined. The numbers refer to mean values of triplicate determinations. (D) At day 3 of the growth curve, levels of EdU incorporation were determined by fluorescence-activated cell sorter analysis. (E) ETS1 and FLI1 down-regulation causes proliferation impairment. Y-axis: percentage of GFP positive and negative cells. X-axis: days. Histograms are representative of 1 experiment of 2. (F) ETS1 (upper) and FLI1 (lower) down-regulation induces apoptosis. Percentage of apoptotic cells was determined by ANNEXINV staining at day 5. Dot plots are representative of 1 experiment; histogram graphs represent the average of at least 3 independent experiments. *P value < .05.
ETS1 and FLI1 affect proliferation and viability in OCI-Ly7. (A) ETS1 and FLI1 protein levels in 22 DLBCL cell lines. Western blot analysis was carried out on whole-cell lysates with antibodies against ETS1, FLI1, and GAPDH as loading control. (B) ETS1 and FLI1 down-regulation in OCI-Ly7 cell line. Real-time PCR and western blot analysis of ETS1 (left) and FLI1 (right) levels after shRNA (day 5). As a control, β2-microglobulin mRNA and GAPDH protein expression were documented. (C) For the growth curve, OCI-Ly7 cells infected with control (shGFP) lentivirus or a lentivirus expressing shRNA for ETS1 and FLI1 (shETS1 and shFLI1, respectively), were seeded on 24-well plates at a density of 3 × 105 cells/well. Cultures were harvested every day, and the number of cells was determined. The numbers refer to mean values of triplicate determinations. (D) At day 3 of the growth curve, levels of EdU incorporation were determined by fluorescence-activated cell sorter analysis. (E) ETS1 and FLI1 down-regulation causes proliferation impairment. Y-axis: percentage of GFP positive and negative cells. X-axis: days. Histograms are representative of 1 experiment of 2. (F) ETS1 (upper) and FLI1 (lower) down-regulation induces apoptosis. Percentage of apoptotic cells was determined by ANNEXINV staining at day 5. Dot plots are representative of 1 experiment; histogram graphs represent the average of at least 3 independent experiments. *P value < .05.
We next down-regulated the ETS1 and FLI1 expression in OCI-Ly7 cells through lentiviral shRNA, utilizing 2 different hairpins for each gene (nos. 1 and 2). Both real-time PCR and western blotting analyses revealed significant reductions of the levels of the 2 ETS factors with respect to the shGFP control vector (Figure 2B). We then examined the effects of ETS1 and FLI1 down-regulation on cell proliferation and viability in the OCI-Ly7 cell line. We found that both ETS1 and FLI1 down-regulation induced a lower proliferation rate compared with the shGFP control (Figure 2C), and this phenotype was associated with a reduced percentage of cells in S-phase (Figure 2D). To evaluate the biological effects of ETS1 and FLI1 down-regulation at longer time points, we added cells infected with shRNA against ETS1 or FLI1 (GFP negative) to an equal amount of cells infected with a vector carrying the GFP marker (GFP positive). Fluorescence-activated cell sorter analysis of the percentage of the 2 cell populations at days 3, 7, 10, 17, and 24 following infection demonstrated that OCI-Ly7 cells with reduced levels of ETS1 or FLI1 proliferated less efficiently and were outgrown by GFP positive cells, at a variance with cells infected with the control vector (shGFP), thereby indicating an impairment of cell proliferation as a result of the ETS1 or FLI1 down-regulation (Figure 2E). Control shRNA transduced cells did not display this readout. In comparison with shETS1 cells, shFLI1 cells were more rapidly overcome by GFP-positive cells despite a similar percentage of regulation for both proteins, suggesting a more prominent role for FLI1 in regulating cell proliferation. We then investigated whether this phenotype was associated with cell death and found that down-regulation of ETS1 or FLI1 induced apoptosis in a significant percentage of OCI-Ly7, indicating that these 2 genes are critical for cell viability in this DLBCL cell line (Figure 2F).
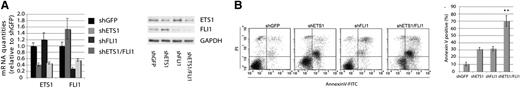
To investigate whether ETS1 and FLI1 in 11q24.3 gain positive patients could cooperate in regulating cell viability; we concomitantly down-regulated both genes in OCI-Ly7 (Figure 3A). Concomitant down-regulation of ETS1 and FLI1 clearly induced apoptosis in a percentage of cells that was significantly higher (70%) than those observed after individual down-regulation of ETS1 or FLI1. This strongly indicated that ETS1 and FLI1 can cooperatively sustain cell viability in DLBCL (Figure 3B).
ETS1 and FLI1 cooperate in sustaining cell viability in OCI-Ly7. (A) ETS1 and FLI1 concomitant down-regulation in OCI-Ly7 cell line. Real-time PCR (left) and western blot (right) analysis of ETS1 and FLI1 levels after shRNA (day 4) after infection. (B) ETS1 and FLI1 down-regulation induce apoptosis in a cooperative manner. Dot plots are representative of 1 experiment; histogram graphs represent the average of 3 independent experiments. **P value < .05.
ETS1 and FLI1 cooperate in sustaining cell viability in OCI-Ly7. (A) ETS1 and FLI1 concomitant down-regulation in OCI-Ly7 cell line. Real-time PCR (left) and western blot (right) analysis of ETS1 and FLI1 levels after shRNA (day 4) after infection. (B) ETS1 and FLI1 down-regulation induce apoptosis in a cooperative manner. Dot plots are representative of 1 experiment; histogram graphs represent the average of 3 independent experiments. **P value < .05.
ETS1 and FLI1 affect genes involved in B-cell terminal differentiation
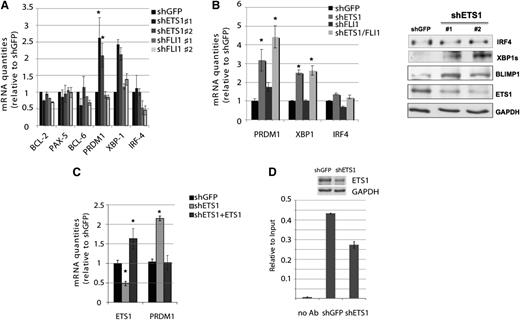
Because ETS factors are involved in B-cell differentiation and in the regulation of important players in GC differentiation,17,19 we then evaluated whether ETS1 and FLI1 regulate the expression of known genes playing a key role in GC B-cell development in the OCI-Ly7 DLBCL cell line in order to elucidate the mechanisms through which ETS1 and FLI1 might exert their role in DLBCL. We analyzed the mRNA levels of BCL2, BCL6, PAX5, IRF4, PRDM1, and XBP1 following knocking-down of ETS1 and FLI1. ETS1 down-regulation resulted in a significant up-regulation of PRDM1 and this effect was maintained, even when both ETS factors were down-regulated (Figure 4A-B). Similarly, silencing of ETS1 determined a XBP1 up-regulation, whereas no effect on IRF4 was observed, suggesting that ETS1 might act up-stream or independently from the NF-κB pathway (Figure 4A-B). These data were also confirmed at the protein level (Figure 4B). We then reinduced ETS1 expression in OCI-Ly7 cells previously infected with shETS1 and observed that ETS1 reintroduction restored PRDM1 mRNA levels to values similar to the control cells (shGFP) (Figure 4C), indicating that the observed PRDM1 up-regulation was induced by ETS1 silencing. In order to evaluate whether PRDM1 was directly regulated by ETS1, we looked for the presence of ETS binding site (EBS) on PRDM1 promoter and found a canonical EBS (−1035 bp). We performed chromatin immunoprecipitation by using antibody against ETS1 on OCI-Ly7 infected with the control shGFP vector and with shETS1. We detected an enrichment of ETS1 in the PRDM1 promoter region containing the EBS in OCI-Ly7 cells that was reduced when ETS1 expression was down-regulated (Figure 4D). We can thus conclude that ETS1 negatively regulates PRDM1 transcription.
ETS1 regulates genes involved in B-cell differentiation. (A) RNA levels of BCL2, PAX5, BCL6, PRDM1, XBP1, and IRF4 and β2-microglobulin transcripts after ETS1 or FLI1 down-regulation were determined by real-time PCR (day 5). Histogram graphs represent the average of at least 3 independent experiments. *P value < .05. (B) Left: RNA levels of PRDM1, XBP1, and IRF4 and β2-microglobulin transcripts were determined by real-time PCR after concomitant ETS1 and FLI1 down-regulation (day 3). *P value <.05. Right: western blot analysis with antibodies against ETS1, PRDM1, IRF4, active splice form XBP1s, and GAPDH as loading control after shETS1 (day 5). (C) ETS1 reintroduction restores PRDM1 mRNA levels in OCI-Ly7. RNA levels of ETS1 and PRDM1 were determined by real-time PCR at 24 hours after ETS1 reexpression in cells previously interfered for ETS1 expression. *P value < .05. (D) Chromatin immunoprecipitation analysis of PRDM1 promoter performed with antibody against ETS1 in OCI-Ly7 infected with the control shGFP or shETS1 lentivirus. Input and immunoprecipitated DNA was amplified by quantitative real-time PCR using primers amplifying the Δ-1100/-939 bp region of the PRDM1 promoter. Enrichments are presented as percentage of total input DNA. Upper: western blot analysis of ETS1 after shRNA.
ETS1 regulates genes involved in B-cell differentiation. (A) RNA levels of BCL2, PAX5, BCL6, PRDM1, XBP1, and IRF4 and β2-microglobulin transcripts after ETS1 or FLI1 down-regulation were determined by real-time PCR (day 5). Histogram graphs represent the average of at least 3 independent experiments. *P value < .05. (B) Left: RNA levels of PRDM1, XBP1, and IRF4 and β2-microglobulin transcripts were determined by real-time PCR after concomitant ETS1 and FLI1 down-regulation (day 3). *P value <.05. Right: western blot analysis with antibodies against ETS1, PRDM1, IRF4, active splice form XBP1s, and GAPDH as loading control after shETS1 (day 5). (C) ETS1 reintroduction restores PRDM1 mRNA levels in OCI-Ly7. RNA levels of ETS1 and PRDM1 were determined by real-time PCR at 24 hours after ETS1 reexpression in cells previously interfered for ETS1 expression. *P value < .05. (D) Chromatin immunoprecipitation analysis of PRDM1 promoter performed with antibody against ETS1 in OCI-Ly7 infected with the control shGFP or shETS1 lentivirus. Input and immunoprecipitated DNA was amplified by quantitative real-time PCR using primers amplifying the Δ-1100/-939 bp region of the PRDM1 promoter. Enrichments are presented as percentage of total input DNA. Upper: western blot analysis of ETS1 after shRNA.
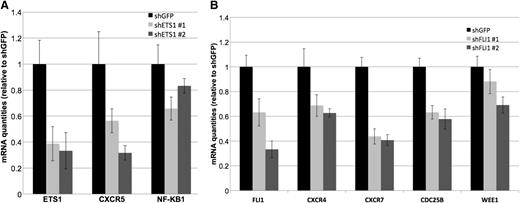
To further understand the potential mechanism of action of ETS1 and FLI1 in our DLBCL model, we performed gene expression profiling in OCI-Ly7 after individual ETS1 and FLI1 down-regulation using 2 shRNAs against each gene in 2 independent experiments (supplemental Figure 3). We looked for the individual genes significantly affected by silencing the 2 ETS factors and also for the more general effect on regulatory pathways. We determined the transcripts significantly affected by the individual shRNA (fold change >1.2, false discovery rate <0.20), and then we considered only those commonly regulated by both shRNA pairs for ETS1 and by both shRNA pairs for FLI1. ETS1 affected 331 transcripts (150 up- and 181 down-regulated) and FLI1 affected 213 transcripts (111 up- and 102 down-regulated) (supplemental Tables 2-3). Both visual inspection of the gene lists as well as the functional analysis of the changes affecting the transcriptome in its integrity highlighted that the genes affected by ETS1 and/or FLI1 silencing code for proteins involved in cell cycle regulation, BCR signaling, plasma cell differentiation, and chemotaxis (supplemental Tables 8-9), such as CXCR5, NFKB1 (ETS1-regulated), CXCR4, CXCR7, CDC25B, and WEE1 (FLI1-regulated) (Figure 5).
ETS1 and FLI1 affect genes involved in GC reaction. (A) Real-time PCR validation of down-regulation of CXCR5 and NFKB1 observed by gene expression profiling after ETS1 silencing. Histogram graphs represent the average of 4 independent silencing experiments. All down-regulation had a P value < .05. (B) Real-time PCR validation of down-regulation of CXCR4, CXCR7, CDC25B, and WEE1 observed by gene expression profiling after FLI1 silencing. Histogram graphs represent the average of 4 independent silencing experiments. All down-regulation had a P value < .05.
ETS1 and FLI1 affect genes involved in GC reaction. (A) Real-time PCR validation of down-regulation of CXCR5 and NFKB1 observed by gene expression profiling after ETS1 silencing. Histogram graphs represent the average of 4 independent silencing experiments. All down-regulation had a P value < .05. (B) Real-time PCR validation of down-regulation of CXCR4, CXCR7, CDC25B, and WEE1 observed by gene expression profiling after FLI1 silencing. Histogram graphs represent the average of 4 independent silencing experiments. All down-regulation had a P value < .05.
Only FLI1 is essential for cell viability of DLBCL cells without 11q24.3 gain
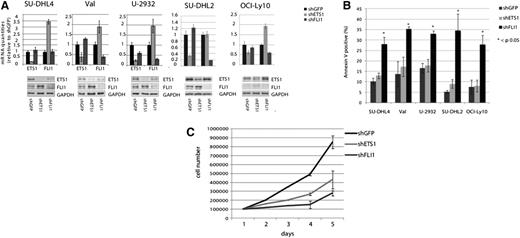
To assess whether ETS1 and FLI1 are required for cell viability in general in primary DLBCL independently on the presence of the 11q24.3 gain and high levels of ETS1 and FLI1 expression, we down-regulated the expression of these 2 molecules in a set of GCB (SU-DHL4, Val) and ABC (OCI-Ly10, SU-DHL2, and U-2932) DLBCL cell lines lacking the 11q24.3 gain (Figure 6A). Only FLI1 down-regulation significantly affected cell viability in a way similar to what observed in OCI-Ly7 cell line. Conversely, ETS1 down-regulation did not induce apoptosis (Figure 6B) although it did decrease cell proliferation (Figure 6C). These data suggested a different role of the 2 genes in DLBCL models (Figure 6B). Notably, in the majority of the DLBCL cell lines, ETS1 down-regulation induced a significant FLI1 up-regulation in line with previous reports in normal T cells,30 suggesting that elevated FLI1 levels might compensate for ETS1 down-regulation.17 To further investigate the molecular effects of the down-regulation of ETS1 and FLI1 in DLBCL cell lines without 11q24.3 gain, we performed the same GEP experiment performed in the OCI-LY7 cell line on 2 other DLBCL models, SU-DHL4 and SU-DHL2, and GCB- and ABC- models, respectively (supplemental Figure 3). Also in these cell lines, ETS1 and FLI1 silencing affected the transcription program. ETS1 silencing affected 1056 transcripts in SU-DHL4 (supplemental Table 4) and 400 transcripts in SU-DHL2 (supplemental Table 5), while FLI1 silencing affected 3051 transcripts in SU-DHL4 (supplemental Table 6) and 1040 transcripts in SU-DHL2 (supplemental Table 7). In these cell lines lacking the 11q24.3, ETS1 and FLI1 silencing did not appear to affect genes involved in B-cell maturation pathways (only on pathways regulating cell proliferation), confirming the phenotypic data obtained in these cells (supplemental Tables 10-13).
ETS1 and FLI1 down-regulation in other DLBCL cell lines. (A) ETS1 and FLI1 down-regulation in SU-DHL4, Val, U-2932, SU-DHL2, and OCI-Ly10 DLBCL cell lines. Real-time PCR and western blot analysis of ETS1 and FLI1 levels after shRNA (day 5). As a control, β2-microglobulin mRNA and GAPDH protein expression was documented. (B) Apoptosis in 5 different DLBCL cell lines after ETS1 and FLI1 down-regulation. Percentage of apoptotic cells was determined by annexinV staining at day 5. Histogram graphs represent the average of at least 2 independent experiments.*P value < .05. (C) For the growth curve, SU-DHL2 cells infected with control (shGFP) lentivirus or a lentivirus expressing shRNA for ETS1 and FLI1 (shETS1 and shFLI1, respectively) were seeded on 24-well plates at a density of 105 cells/well. Cultures were harvested every day and the number of cells was determined. The numbers refer to mean values of triplicate determinations.
ETS1 and FLI1 down-regulation in other DLBCL cell lines. (A) ETS1 and FLI1 down-regulation in SU-DHL4, Val, U-2932, SU-DHL2, and OCI-Ly10 DLBCL cell lines. Real-time PCR and western blot analysis of ETS1 and FLI1 levels after shRNA (day 5). As a control, β2-microglobulin mRNA and GAPDH protein expression was documented. (B) Apoptosis in 5 different DLBCL cell lines after ETS1 and FLI1 down-regulation. Percentage of apoptotic cells was determined by annexinV staining at day 5. Histogram graphs represent the average of at least 2 independent experiments.*P value < .05. (C) For the growth curve, SU-DHL2 cells infected with control (shGFP) lentivirus or a lentivirus expressing shRNA for ETS1 and FLI1 (shETS1 and shFLI1, respectively) were seeded on 24-well plates at a density of 105 cells/well. Cultures were harvested every day and the number of cells was determined. The numbers refer to mean values of triplicate determinations.
Discussion
In our genome-wide DNA profiling study, 23% of DLBCL samples carried a recurrent gain on chromosome 11q24.3. Among the 6 genes encompassed in the minimal common region, only the genes coding for the transcription factors ETS1 and FLI1 resulted to be expressed, with levels significantly higher in cases with the gain in comparison with those without. Functional studies on an in vitro model of DLBCL carrying the 11q24.3 gain and expressing high levels of ETS1 and FLI1 showed that these transcription factors cooperate in sustaining neoplastic B-cell proliferation and viability and affect the regulation of genes involved in the maturation process from GC B cell to plasma cell.
ETS1 and FLI1 are 2 ETS family members known to be involved in tumorigenesis and B-cell development.12-16 ETS1 overexpression is observed in a variety of human tumors.31 Recently, nonsynonymous mutations of ETS1 gene32 and copy number gains10 have been reported in a subgroup of DLBCL, suggesting a putative oncogenic role. FLI1 is involved in the oncogenic fusion-protein EWS-FLI1 of Ewing sarcoma, in which its transcriptional activity is pivotal.33 In mice, FLI1 regulates B-cell differentiation, proliferation, and apoptosis and contributes to the control of marginal zone and follicular B-cell development.34
In our study, the 11q24.3 gain was observed in both GCB- and ABC-DLBCL. The OCI-Ly7 cell line, the in vitro model bearing the 11q24.3 gain, is usually considered of GCB origin but is more likely representative of a type of DLBCL intermediate between GCB and ABC DLBCL.35 Indeed, also in our hands, the cell line expressed markers typical of both GCB (BCL6, PAX5) and non-GCB DLBCL (IRF4, PRDM1, XBP1) (data not shown). These observations suggest that the lesion might be relevant in a subgroup of DLBCL derived from the non-GC phase in accordance with the observed higher ETS1 RNA levels in ABC than in GCB DLBCL. Although both ETS1 and FLI1 were critical for the cell line carrying the 11q24.3 gain, ETS1 down-regulation did not affect the survival of other DLBCL cells even though it affected cell proliferation; only FLI1 was critical for the viability of additional DLBCL models, indicating that the latter gene regulates genes involved in cell proliferation and apoptosis independently of its levels of expression.
The observed up-regulation of PRDM1 after ETS1 silencing (likely due to a direct transcriptional regulation) in the cells carrying the 11q24.3 gain could explain the critical role of ETS1 on the viability of the OCI-Ly7 cell line but not of other cell lines. Indeed, SU-DHL4, bearing the classical features of GC-derived cells, did not express PRDM1 at mRNA or protein levels (data not shown). ETS1 down-regulation was not followed by PRDM1 mRNA up-regulation in Val and U-2932 cell lines in which the constitutively deregulated BCL6 represses the PRDM1 gene.36,37 Accordingly, SU-DHL2 and OCI-Ly10 also carry PRDM1 inactivating mutations.4,38 In accordance with these observations, genetic silencing of ETS1 and FLI1 followed by GEP showed that the 2 transcription factors were involved in the regulation of genes coding for proteins involved in cell cycle and B-cell development in the cell line bearing the 11q23 gain, while mainly genes involved in cell cycle regulation appeared differentially expressed in the remaining analyzed DLBCL cell lines. These results suggest that ETS1-mediated suppression of PRDM1 expression, as observed in the presence of 11q24.3 gain, would act as an alternative mechanism contributing to the pathogenesis of a subgroup of DLBCL in which constitutively high levels of ETS1 and FLI1 would deregulate the GC/plasma cell expression program.
An interesting finding of our study was that both ETS1 and FLI1 genes appeared to regulate the expression of chemokine receptors (ETS1, CXCR5, FLI1, CXCR4, and CXCR7), which can participate to the neoplastic process.39-42 Although less is known regarding the role of CXCR7,43,44 CXCR5 and CXCR4 regulate the development and the trafficking of B cells within the GC.45 In normal lymphoid tissue, CXCR4 is highly expressed in the dark zone of the GC, attracting the centroblasts, whereas CXCR5 expression is in the light zone and contributes to attracting B cells to the area and is then down-regulated during plasmacytic differentiation.46 CXCR4 signaling also has a pro-survival role in B cells,47 and, in a subset of DLBCL, FLI1 might promote B-cell survival through a positive regulation of CXCR4.
The effect of ETS1 and FLI1 on different genes involved in similar pathways was in accordance with other important observations, all indicating that the 2 ETS factors might contribute to DLBCL growth in a cooperative manner. First, the 2 genes are closely mapped, indicating that they might arise by gene duplication from a common ancestral gene48,49 and thus be coaffected by the 11q24.3 gain. Second, when both genes were down-regulated in the OCI-Ly7 cell line, the observed effects were much more severe than with single gene silencing. Third, ETS1 and FLI1 RNA expressions were correlated across DLBCL samples.
In conclusion, the combination of genomic and expression profiling suggested a new pathogenetic mechanism for a subgroup of DLBCL derived from the transformation of late GC B cells. A model could be proposed in which the acquisition of the 11q24.3 gain causes a constitutive high expression of ETS1 and FLI1 genes that would deregulate genes involved in the GC expression program and on cell proliferation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the San Salvatore Foundation (Lugano, Switzerland), Oncosuisse OCS-1939-8-2006 (Bern, Switzerland), Nelia et Amadeo Barletta Foundation (Lausanne, Switzerland), and Fondazione per la Ricerca e la Cura sui Linfomi in Ticino (Bellinzona, Switzerland). M.T. and M.S. are enrolled in the program in Pharmaceutical Sciences, University of Geneva, Switzerland.
Authorship
Contribution: P.B. designed and performed experiments, interpreted data, and co-wrote the manuscript; M.T. performed experiments, interpreted data, and helped write the manuscript; M.S. performed experiments and genomic profiling; M.P. performed immunohistochemistry, provided advice, and collected and characterized DLBCL samples; R.P. provided advice; A.A.M. performed experiments and provided advice; M.G.T. performed fluorescence in situ hybridization analyses; A.R. performed genomic and gene expression profiling; I.K. performed statistical analysis; J.I. interpreted gene expression profiling data; T.C.G., W.-C.C., G.G, M.A.P., F.C., and E.Z. provided advice and collected and characterized DLBCL samples; G.I. provided advice and collected and characterized DLBCL samples; and F.B. designed the study, interpreted data, and co-wrote the manuscript. All authors have approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for P.B. is Italian Institute of Technology, Center for Genomic Sciences, Milan, Italy.
Correspondence: Francesco Bertoni, Lymphoma and Genomics Research Program, IOR Institute of Oncology Research, via Vincenzo Vela 6, 6500 Bellinzona, Switzerland; e-mail: frbertoni@mac.com.
References
Author notes
P.B. and M.T. contributed equally to this study.