Key Points
Hypomorphic IFNGR2 alleles encode misfolded proteins with abnormal N-glycosylation.
Chemical modifiers of N-glycosylation effectively complemented the patients’ response to IFN-γ.
Abstract
We report a molecular study of the two known patients with autosomal recessive, partial interferon-γ receptor (IFN-γR)2 deficiency (homozygous for mutations R114C and G227R), and three novel, unrelated children, homozygous for S124F (P1) and G141R (P2 and P3). IFN-γR2 levels on the surface of the three latter patients’ cells are slightly lower than those on control cells. The patients’ cells also display impaired, but not abolished, response to IFN-γ. Moreover, the R114C, S124F, G141R and G227R IFNGR2 hypomorphic alleles all encode misfolded proteins with abnormal N-glycosylation. The mutants are largely retained in the endoplasmic reticulum, although a small proportion reach and function at the cell surface. Strikingly, the IFN-γ response of the patients’ cells is enhanced by chemical modifiers of N-glycosylation, as previously shown for patients with gain-of-glysosylation T168N and misfolding 382-387dup null mutations. All four in-frame IFNGR2 hypomorphic mutant alleles encoding surface-expressed receptors are thus deleterious by a mechanism involving abnormal N-glycosylation and misfolding of the IFN-γR2 protein. The diagnosis of partial IFN-γR2 deficiency is clinically useful, as affected patients should be treated with IFN-, unlike patients with complete IFN-γR2 deficiency. Moreover, inhibitors of glycosylation might be beneficial in patients with complete or partial IFN-γR2 deficiency due to misfolding or gain-of-glycosylation receptors.
Introduction
Mendelian susceptibility to mycobacterial disease (MSMD) is a rare syndrome characterized by the occurrence of infectious diseases due to weakly pathogenic mycobacteria, such as Bacillus Calmette-Guérin vaccine strains and environmental mycobacteria, in otherwise healthy subjects (MIM#209950).1-4 Infections generally occur in children, with a wide range of clinical symptoms, from localized to disseminated infections.1-3 The patients are also vulnerable to the more virulent Mycobacterium tuberculosis,5-7 and about half of them also suffer from nontyphoidal Salmonella infection.8,9 Other infections are increasingly documented, albeit typically in individual patients.10-12 The first genetic etiology of MSMD was discovered in 1996 and involved mutations affecting the ligand-binding chain of the interferon-γ receptor (IFN-γR1).13,14 To date, 7 autosomal (IFNGR1, IFNGR2, STAT1, IL12B, IL12RB1, IRF8, and ISG15) and 2 X-linked (CYBB and NEMO) morbid genes have been discovered.2,3,15-19 There is also allelic heterogeneity, resulting in the definition of up to 17 different genetic etiologies, based on mode of transmission (dominant or recessive), the expression of the mutated allele (eg, expressed on the cell surface or not for receptors), and the function affected (eg, phosphorylation or DNA binding for transcription factors). The products of the affected genes are all involved in IFN-γ–mediated immunity, controlling its induction (IL12B, IL12RB1, IRF8, ISG15, and NEMO) and/or its response (IFNGR1, IFNGR2, STAT1, IRF8, and CYBB).
There are several forms of IFN-γR2 deficiency. Autosomal dominant (AD) IFN-γR2 deficiency by haplo-insufficiency was reported in only 1 MSMD patient; the penetrance is low, as 17 other known heterozygous individuals have remained healthy.19 Another IFNGR2 mutation, also found in a healthy individual, was shown to be dominant-negative at the cellular level.20 Autosomal recessive (AR) IFN-γR2 deficiency is a more common cause of MSMD, reported thus far in 14 patients with complete penetrance.19-27 Two forms of complete IFN-γR2 deficiency have been reported, defined on the basis of cell surface expression levels. In 6 patients from 4 kindreds, a premature stop codon results in a lack of receptor expression at the cell surface.19-21 In 6 patients from 5 kindreds, the defect is characterized by the detectable expression of nonfunctional receptors at the cell surface.23-26 In 3 patients, a missense mutation (T168N) has been shown to create a new N-glycosylation site, which abolishes the cellular response to IFN-γ.23,24 In another patient, the mutation (382-387dup) is not gain-of-glycosylation but results in misfolded proteins that can also surprisingly be rescued with inhibitors of glycosylation.25 A partial form of AR IFN-γR2 deficiency has been reported in only 2 MSMD patients, homozygous for the R114C22 and G227R mutations.27 Both patients display impaired, but not abolished, cellular responses to IFN-γ. The molecular mechanism of disease in patients carrying these hypomorphic IFNGR2 missense alleles is unknown. This is an important question, because the severity of clinical disease in patients with IFN-γR2 (and IFN-γR1) deficiency is correlated with the cellular response to IFN-γ.28 We therefore attempted to identify novel patients with a partial form of AR IFN-γR2 deficiency and to investigate the underlying mechanism of disease.
Materials and methods
Ethics statement
This study was conducted in accordance with the Helsinki Declaration, with written informed consent obtained from the patient family. Approval for this study was obtained from the Comité de Protection des Personnes and Institut National de la Santé et de la Recherche Médicale in France and the Rockefeller Institutional Review Board (New York, NY).
Expression vectors and transfections
The wild-type (WT) sequence of IFNGR2 was inserted into the Topo-pcDNA3.1-tagged (at the C terminus; Invitrogen) V5 plasmid, and into the pEGFP-N1 vector (Clontech Laboratories) according to the manufacturer’s instructions. The R114C, S124F, G141R, G227R, 382-387dup, T168N, and 278delAG mutants were generated by site-directed mutagenesis (Quikchange site-directed mutagenesis kit; Stratagene), according to the kit manufacturer’s instructions. HEK293T cells and SV40 fibroblasts from a healthy control (WT/WT) and an IFN-γR2–deficient patient (278delAG/278delAG)21 were transiently transfected with a mock vector, WT IFNGR2, R114C, S124F, G141R, G227R, 382-387dup, T168N, and 278delAG mutant constructs in the presence of Lipofectamine Reagent (Invitrogen), as described by the manufacturer. The cells were analyzed 48 hours after transfection.
Chemical treatment, endoglycosidase H and peptide N-glycosidase F digestions, and western blotting
After 6 hours of transfection, HEK293T cells were left untreated or were treated with 166 µM kifunensine (Toronto Research Chemicals). After 48 hours of transfection, cells were then lysed with 1% NP-40 lysis buffer, and the proteins were recovered and left untreated or digested overnight at 37°C with endoglycosidase-H (EndoH; Biolabs) or peptide N-glycoside-F (PNGaseF; Biolabs) in the appropriate buffer. The products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blotting. The polyvinylidene difluoride membrane was probed with horseradish perioxidase (HRP)-conjugated anti-V5 antibody (Invitrogen) and anti–glyceraldehyde-3-phosphate dehydrogenase antibody (Santa Cruz).
Confocal fluorescence microscopy
SV40 fibroblasts from an IFN-γR2–deficient patient were transfected with enhanced green fluorescent protein (EGFP)-tagged constructs for 48 hours. Then, cells were stained with rabbit anti–protein disulfide-isomerase (PDI) (Abcam ab3672) as an endoplasmic reticulum (ER) marker, followed by Alexa Fluor 546 goat anti-rabbit immunoglobulin G (H+L) (Molecular Probes A-11010) and mouse anti–golgin-97 as a trans-golgi marker (Molecular Probes CDF4), followed by Alexa Fluor 647 goat anti-mouse immunoglobulin G (H+L) (Molecular Probes A21236). Images were acquired with DeltaVision Image Restoration Microscope (Applied Precision), and the Pearson’s coefficient and colocalization analysis were performed with IMARIS software (Bitplane).
Results
Novel IFNGR2 mutations in patients with MSMD
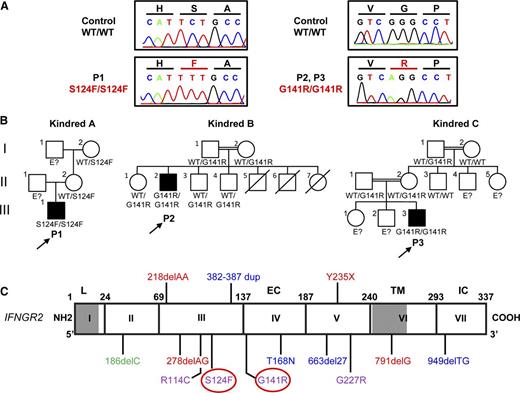
We studied 3 unrelated patients with MSMD from Mexico (P1) and Turkey (P2 and P3). We sequenced by the Sanger method the 7 exons and flanking intron regions of IFNGR1 and IFNGR2 on P1 leukocyte gDNA. P1 carries a homozygous mutation in exon 3 of IFNGR2, in which a C was replaced with a T at nucleotide position 371, leading to the replacement of a serine (S) with a phenylalanine (F) in position 124 (S124F) (Figure 1A,C). The mother and maternal grandmother were heterozygous (Figure 1B, kindred A). No DNA sample was available for the father or grandfather. Whole exome sequencing was carried out in P2 and P3 (supplemental Table 1 on the Blood Web site). Both patients carry a homozygous mutation in exon 4 of IFNGR2, in which a G was replaced with an A at nucleotide position 421, leading to the replacement of a glycine (G) with an arginine (R) at position 141 (G141R; Figure 1A,C). The G141R mutation was confirmed by Sanger sequencing in P2 and P3. The parents and 3 siblings of P2 were heterozygous for G141R. The parents of P3 were heterozygous for G141R (Figure 1B, kindreds B and C). An analysis of single nucleotide polymorphisms (SNPs) derived from the exome data of P2 and P3 showed that they had a common homozygous haplotype surrounding the IFNGR2 gene, encompassing 0.9 Mb (corresponding to 10 SNPs with a mean intermarker distance of 120 kb, ranging from 0.3 to 277 kb). The ESTIAGE program29 estimated the age of the most recent common ancestor to 103 generations (95% confidence interval [CI]: 33-491 generations). Assuming a generation time of 25 years, the most recent common ancestor of the patients therefore lived 2575 years ago (95% CI: 825-12 275 years). Both S124F and G141R are located in the extracellular domain of IFN-γR2. The variants were not found in the 1000 genomes project and dbSNP 134 databases. The S124F mutation was not found in 50 white healthy controls, and the G141R mutation was not found in 200 Turkish healthy controls that were sequenced. The S124 and G141 residues of IFN-γR2 have been conserved through evolution, and both Polyphen II and sorting intolerant from tolerant predicted both mutations to be “probably damaging” (supplemental Materials). In addition, the 3 patients display high levels of IFN-γ in plasma.30 Thus, the homozygous S124F and G141R variants probably underlie AR MSMD.
Identification of 3 new patients with recessive partial IFN-γR2 deficiency and MSMD. (A) Electropherogram showing the TCT-TTT mutation in P1 and the GGG-AGG mutation in P2 and P3 (indicated in red). (B) Familial segregation of the S124F and G141R mutations. Family A is from Mexico. Families B and C are from Turkey. E?, not genotyped. Healthy individuals are shown in white. Solid black shapes indicate patients with MSMD. The probands are indicated by arrows. Each kindred is designated by a capital letter (A-C), each generation by a roman numeral (I-III), and each individual by an Arabic number. (C) Schematic diagram of the IFNGR2 gene with all previously described mutations and the S124F and G141R mutations described here (highlighted by a red circle). Coding exons are numbered with roman numerals and delimited by a vertical bar. Regions corresponding to the leader sequence (L, 1-22), extracellular domain (EC, 23-248), transmembrane domain (TM, 249-272), and intracellular domain (IC, 273-337) are indicated. Mutations marked in red cause AR complete IFN-γR2 deficiency with no detectable expression of IFN-γR2 at the cell surface. The mutations marked in blue cause AR complete IFN-γR2 deficiency with detectable surface expression of a nonfunctional IFN-γR2. With the antibody now available, EBV-B cells from patients carrying the 663del27 mutations23 and 382-387dup25 were shown to have impaired and normal surface expression of IFN-γR2, respectively. The mutation marked in green causes AD partial IFN-γR2 deficiency. The mutations marked in purple cause AR partial IFN-γR2 deficiency.
Identification of 3 new patients with recessive partial IFN-γR2 deficiency and MSMD. (A) Electropherogram showing the TCT-TTT mutation in P1 and the GGG-AGG mutation in P2 and P3 (indicated in red). (B) Familial segregation of the S124F and G141R mutations. Family A is from Mexico. Families B and C are from Turkey. E?, not genotyped. Healthy individuals are shown in white. Solid black shapes indicate patients with MSMD. The probands are indicated by arrows. Each kindred is designated by a capital letter (A-C), each generation by a roman numeral (I-III), and each individual by an Arabic number. (C) Schematic diagram of the IFNGR2 gene with all previously described mutations and the S124F and G141R mutations described here (highlighted by a red circle). Coding exons are numbered with roman numerals and delimited by a vertical bar. Regions corresponding to the leader sequence (L, 1-22), extracellular domain (EC, 23-248), transmembrane domain (TM, 249-272), and intracellular domain (IC, 273-337) are indicated. Mutations marked in red cause AR complete IFN-γR2 deficiency with no detectable expression of IFN-γR2 at the cell surface. The mutations marked in blue cause AR complete IFN-γR2 deficiency with detectable surface expression of a nonfunctional IFN-γR2. With the antibody now available, EBV-B cells from patients carrying the 663del27 mutations23 and 382-387dup25 were shown to have impaired and normal surface expression of IFN-γR2, respectively. The mutation marked in green causes AD partial IFN-γR2 deficiency. The mutations marked in purple cause AR partial IFN-γR2 deficiency.
Expression of IFN-γR1 and IFN-γR2 in the patients’ cells
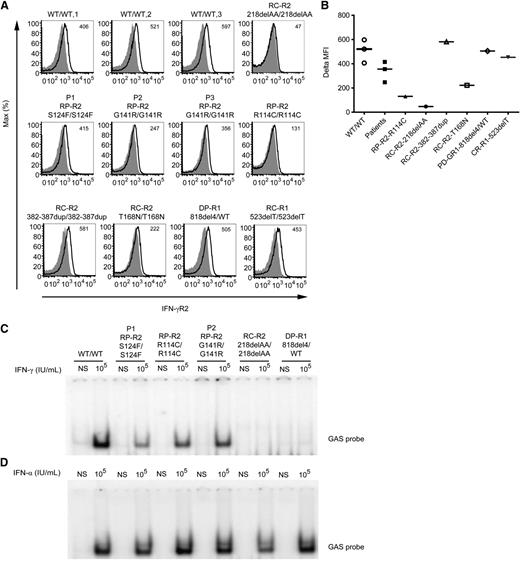
We used flow cytometry to investigate the expression of IFN-γR2 and of IFN-γR1 (as a control) on the surface of Epstein-Barr virus–transformed B cells (EBV-B cells) from 3 healthy controls, P1, P2, P3, the previously described patient with AR partial IFN-γR2 deficiency (RP-R2, mutation R114C/R114C),22 a patient with complete IFN-γR2 deficiency without receptor expression at the cell surface (RC-R2, 218delAA/218delAA),19 2 patients with complete IFN-γR2 deficiency but detectable expression of nonfunctional receptors at the cell surface (T168N and 382-387dup),23-25 a patient with AD partial IFN-γR1 deficiency (DP-R1, 818del4/WT),31 and a patient with complete IFN-γR1 deficiency (RC-R1, 523delT/523delT).32 Levels of IFN-γR1 expression at the cell surface were similar for all cell lines evaluated with the GIR-94 antibody (recognizing an epitope in the extracellular part of IFN-γR1), except for the AD partial IFN-γR1 mutant protein, for which IFN-γR1 expression levels were higher, as expected,31 and cells with complete IFN-γR1 deficiency were used as a negative control32 (supplemental Figure 1A). We then used a novel commercially available antibody (FAB773A; R&D Systems) that recognizes the extracellular part of IFN-γR2 and works well in flow cytometry.19,33 IFN-γR2 expression in the patients’ cells was found to be slightly higher than in R114C cells and slightly lower than that in cells from the healthy controls (Figure 2A-B). With the antibody now available, EBV-B cells from the patient carrying T168N mutation showed detectable surface expression of IFN-γR2. The 382-387dup cells25 also showed a normal surface expression of IFN-γR2. Cells with complete IFN-γR2 deficiency due to a frameshift mutation (RC-R2, 218delAA/218delAA)19 served as negative control (Figure 2A-B). Altogether, suboptimal levels of IFN-γR2 were detected on the surface of cells from the new patients.
IFN-γR2 expression and impaired STAT1-DNA-binding activity in response to IFN-γ stimulation of the patients’ cells. (A) Flow cytometry analysis of IFN-γR2 expression on the surface of EBV-B cells from 3 healthy controls (WT/WT-1, WT/WT-2, and WT/WT-3), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA), patient P1 with the recessive S124F mutation conferring partial deficiency (RP-R2, S124F/S124F), patients P2 and P3 with the recessive G141R mutation (RP-R2, G141R/G141R), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), 2 patients with complete recessive IFN-γR2 deficiency and surface expression (RC-R2, 382-387dup/382-387dup and RC-R2, T168N/T168N), a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/WT), and a patient with complete recessive IFN-γR1 deficiency (RC-R1, 523delT/523delT). These results are reproducible and representative of ≥3 independent experiments. The histograms represent the expression of IFN-γR2 (bold line) and the isotype control (gray filled). (B) Difference in mean fluorescence intensity (ΔMFI) values between specific and isotype antibody are indicated. Response of EBV-B cells to (C) IFN-γ and (D) IFN-α (105 IU/mL for 20 minutes) as determined by EMSA of GAS probe-binding nuclear proteins from a healthy control (WT/WT), patients P1 with the S124F mutation (RP-R2, S124F/S124F) and P2 (RP-R2, G141R/G141R) studied here, a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA), and a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/ WT). These results are reproducible and representative of ≥3 independent experiments.
IFN-γR2 expression and impaired STAT1-DNA-binding activity in response to IFN-γ stimulation of the patients’ cells. (A) Flow cytometry analysis of IFN-γR2 expression on the surface of EBV-B cells from 3 healthy controls (WT/WT-1, WT/WT-2, and WT/WT-3), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA), patient P1 with the recessive S124F mutation conferring partial deficiency (RP-R2, S124F/S124F), patients P2 and P3 with the recessive G141R mutation (RP-R2, G141R/G141R), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), 2 patients with complete recessive IFN-γR2 deficiency and surface expression (RC-R2, 382-387dup/382-387dup and RC-R2, T168N/T168N), a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/WT), and a patient with complete recessive IFN-γR1 deficiency (RC-R1, 523delT/523delT). These results are reproducible and representative of ≥3 independent experiments. The histograms represent the expression of IFN-γR2 (bold line) and the isotype control (gray filled). (B) Difference in mean fluorescence intensity (ΔMFI) values between specific and isotype antibody are indicated. Response of EBV-B cells to (C) IFN-γ and (D) IFN-α (105 IU/mL for 20 minutes) as determined by EMSA of GAS probe-binding nuclear proteins from a healthy control (WT/WT), patients P1 with the S124F mutation (RP-R2, S124F/S124F) and P2 (RP-R2, G141R/G141R) studied here, a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA), and a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/ WT). These results are reproducible and representative of ≥3 independent experiments.
Impaired DNA-binding activity in response to IFN-γ in the patients’ cells
We assessed the responses to IFN-γ and IFN-α (as a control) of P1, P2, and P3 EBV-B cells and compared these responses with those of healthy control, R114C/R114C IFNGR2,22 218delAA/218delAA,19 IFNGR2, and 818del4/WT IFNGR1 cells.31 EBV-B cells were either left unstimulated or stimulated with IFN-γ and IFN-α (105 IU/mL) for 20 minutes at 37°C. Nuclear proteins were extracted, and electrophoretic mobility shift assays (EMSAs) were performed with a γ-activating sequence (GAS) probe. As expected, cells from the patient with complete IFN-γR2 deficiency displayed no γ-activating factor (GAF) DNA-binding activity when stimulated with IFN-γ (Figure 2C). Cells from P1, P2, and P3 presented an impaired, but not abolished, response to IFN-γ in terms of GAS-DNA binding, compared with the WT (Figure 2C; supplemental Figure 1B). As a control, the response to IFN-α stimulation was assessed and found to be similar in all cell lines (Figure 2D). Competition and supershift experiments in the presence of excess unlabeled probe and antibodies specific for signal transducer and activator of transcription (STAT)1, STAT2, STAT3, and interferon regulatory factor 9/p48 demonstrated that GAS-binding activity of GAF in WT and P1 complexes was mediated exclusively by STAT1/STAT1 homodimers (supplemental Figure 2A-B). These results suggested that the S124F and G141R mutations were responsible for partial IFN-γR2 deficiency with an impaired, but not abolished, response to IFN-γ.
Impaired induction of IFN-γ target genes in the patient’s cells
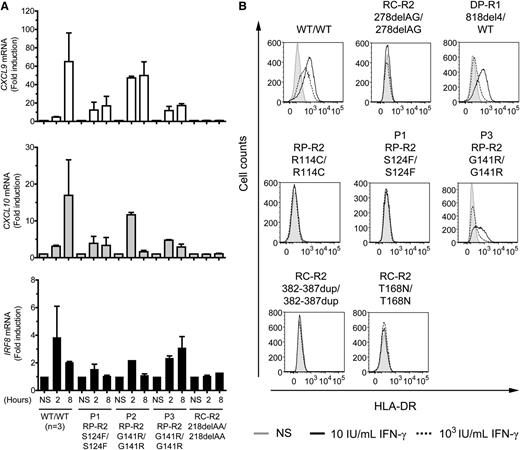
We then investigated later events in the IFN-γ signaling pathway. The induction of CXCL9, CXCL10, and IRF8 after 2 and 8 hours of stimulation with 103 IU/mL IFN-γ was similar in EBV-B cells from P1, P3, and the other patient with partial recessive IFN-γR2 deficiency (R114C), but weaker than in EBV-B cells from healthy controls (Figure 3A; supplemental Figure 3A-C). P2 showed higher induction of these genes than P1 and P3, specifically after 2 hours of stimulation, but lower than in EBV-B cells from healthy controls (Figure 3A). We also performed whole blood activation assays for the patients. As expected, the production of IFN-γ in response to BCG plus interleukin (IL)-12 was similar to that in controls (data not shown). In response to BCG alone and BCG plus IFN-γ, the cells of P1, P2, and P3 produced normal amounts of IL-12p40. However, the production of IL-12p70 was significantly impaired (supplemental Figure 4A-B). We then used flow cytometry to assess the induction of human leukocyte antigen, DR subregion (HLA-DR) at the surface of SV40 fibroblasts in response to stimulation for 48 hours with IFN-γ. As previously reported,31 HLA-DR induction was normal in 818del4/WT IFN-γR1 cells at high concentrations and impaired at low doses of IFN-γ (Figure 3B). By contrast, no induction was observed in cells from P1 and the previously reported patient with partial IFN-γR2 deficiency (R114C)22 or the patients with complete IFN-γR2 deficiency (278delAG and 382-387dup).21,25 SV40 fibroblasts from P2 were not available. However, P3 SV40 fibroblasts showed impaired HLA-DR induction (Figure 3B). The defect was not as pronounced in P3 as in P1. Thus, both early and late events in the IFN-γ signaling pathway are impaired in the cells of P1 and P3.
Impaired induction of CXCL9, CXCL10, IRF8, and surface HLA-DR expression in response to IFN-γ in the patients’ cells. (A) Quantitative reverse transcription-polymerase chain reaction was used to assess the induction of the CXCL9, CXCL10, and IRF8 mRNA after stimulation with IFN-γ (103 IU/mL for 2 and 8 hours) in EBV-B cells from healthy controls (n = 3), patient P1 with the S124F mutation (RP-R2, S124F/S124F), patients P2 and P3 with the G141R mutation (RP-R2,G141R/G141R), and a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA). The values shown are mean values ± standard deviation, calculated from 2 independent experiments. (B) SV40 fibroblasts from a healthy control (WT/WT), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), patients P1 (RP-R2, S124F/S124F) and P3 (RP-R2, G141R/G141R), 3 patients with recessive complete IFN-γR2 deficiency (RC-R2, 278delAG/278delAG; RC-R2, 382-387dup/382-387dup; and RC-R2-T168N/T168N), and a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/WT) were stimulated with the indicated doses of IFN-γ for 48 hours. HLA-DR induction was determined by flow cytometry. NS, no stimulation. These results are reproducible and representative of 2 independent experiments.
Impaired induction of CXCL9, CXCL10, IRF8, and surface HLA-DR expression in response to IFN-γ in the patients’ cells. (A) Quantitative reverse transcription-polymerase chain reaction was used to assess the induction of the CXCL9, CXCL10, and IRF8 mRNA after stimulation with IFN-γ (103 IU/mL for 2 and 8 hours) in EBV-B cells from healthy controls (n = 3), patient P1 with the S124F mutation (RP-R2, S124F/S124F), patients P2 and P3 with the G141R mutation (RP-R2,G141R/G141R), and a patient with recessive complete IFN-γR2 deficiency (RC-R2, 218delAA/218delAA). The values shown are mean values ± standard deviation, calculated from 2 independent experiments. (B) SV40 fibroblasts from a healthy control (WT/WT), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), patients P1 (RP-R2, S124F/S124F) and P3 (RP-R2, G141R/G141R), 3 patients with recessive complete IFN-γR2 deficiency (RC-R2, 278delAG/278delAG; RC-R2, 382-387dup/382-387dup; and RC-R2-T168N/T168N), and a patient with dominant partial IFN-γR1 deficiency (DP-R1, 818del4/WT) were stimulated with the indicated doses of IFN-γ for 48 hours. HLA-DR induction was determined by flow cytometry. NS, no stimulation. These results are reproducible and representative of 2 independent experiments.
Complementation of IFN-γ responsiveness with WT IFNGR2
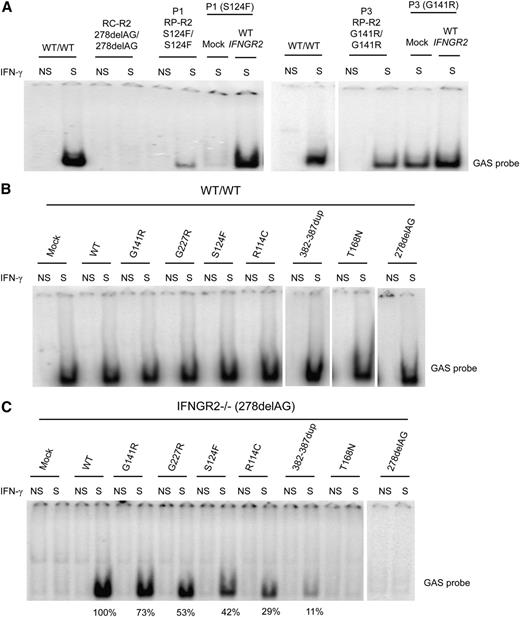
We then transfected SV40 fibroblasts from P1 and P3 with WT IFNGR2, and the response to IFN-γ was evaluated by EMSA. GAF-DNA binding activity was restored in the transfected cells, as shown by comparison with mock-transfected and untransfected cells (Figure 4A). These results strongly suggested that the S124F and G141R mutations of IFNGR2 were responsible for the impaired response to IFN-γ in the patients. We also transfected SV40 fibroblasts from a healthy control (WT) with a mock vector, WT, R114C, S124F, G141R, G227R, 382-387dup, T168N, and 278delAG mutant constructs. No detectable dominant-negative effect of any IFNGR2 mutant allele was observed in the WT-transfected cells, as the IFN-γ–dependent binding activity was similar for all the alleles tested (Figure 4B). SV40 fibroblasts from an IFN-γR2–deficient patient (278delAG/278delAG)21 were also transfected with the WT and mutant IFNGR2 alleles. Transfection with the R114C and S124F alleles resulted in responses to IFN-γ that were weaker than those observed after transfection with the WT allele (∼29% and 42%, respectively; Figure 4C). Transfection with G141R and G227R showed responses to IFN-γ that were stronger (∼73% and 53% of the WT allele, respectively) than that observed with R114C and S124F but weaker than that observed after transfection with the WT allele. Overexpression of 382-387dup showed mild activity. As a control, we also evaluated the surface expression of IFN-γR1 and IFN-γR2 by flow cytometry in unstimulated cells after transfection. The levels of IFN-γR1 expression at the cell surface of WT and IFN-γR2–deficient cells (278delAG/278delAG)21 were similar for all transfectants, whereas stronger IFN-γR2 expression was observed in cells transfected with the IFNGR2 WT, R114C, S124F, and 382-387dup alleles (supplemental Figure 5A-B). Collectively, these data suggested that the S124F and G141R mutant alleles are hypomorphic, as were the 2 previously reported alleles R114C22 and G227R.27
Complementation of the IFN-γ response with WT IFNGR2. (A) SV40 fibroblasts from P1 and P3 were transiently transfected with a mock vector and WT IFNGR2 and stimulated with 104 IU/mL IFN-γ. DNA-binding activity was then analyzed by EMSA with a GAS probe. Untransfected SV40 fibroblasts from a healthy control (WT/WT), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 278delAG/278delAG), and patients P1 and P3 studied here with the S124F (RP-R2, S124F/S124F) and G141R (RP-R2, G141R/G141R) mutations were used as controls. Vertical lines have been inserted to indicate a repositioned gel lane. (B) SV40 fibroblasts from a healthy control (WT/WT) and (C) an IFN-γR2–deficient patient (278delAG/278delAG) were transiently transfected with mock vector, WT, R114C, S124F, G141R, G227R, 382-387dup, T168N, and 278delAG IFNGR2-tagged V5 constructs. Transfected cells were either left unstimulated or were stimulated with 104 IU/mL of IFN-γ for 20 minutes. GAS-binding activity was evaluated by EMSA. The results shown are representative of 2 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
Complementation of the IFN-γ response with WT IFNGR2. (A) SV40 fibroblasts from P1 and P3 were transiently transfected with a mock vector and WT IFNGR2 and stimulated with 104 IU/mL IFN-γ. DNA-binding activity was then analyzed by EMSA with a GAS probe. Untransfected SV40 fibroblasts from a healthy control (WT/WT), a patient with recessive complete IFN-γR2 deficiency (RC-R2, 278delAG/278delAG), and patients P1 and P3 studied here with the S124F (RP-R2, S124F/S124F) and G141R (RP-R2, G141R/G141R) mutations were used as controls. Vertical lines have been inserted to indicate a repositioned gel lane. (B) SV40 fibroblasts from a healthy control (WT/WT) and (C) an IFN-γR2–deficient patient (278delAG/278delAG) were transiently transfected with mock vector, WT, R114C, S124F, G141R, G227R, 382-387dup, T168N, and 278delAG IFNGR2-tagged V5 constructs. Transfected cells were either left unstimulated or were stimulated with 104 IU/mL of IFN-γ for 20 minutes. GAS-binding activity was evaluated by EMSA. The results shown are representative of 2 independent experiments. Vertical lines have been inserted to indicate a repositioned gel lane.
Four hypomorphic IFNGR2 missense alleles encode misfolded proteins with abnormal N-glycolysation
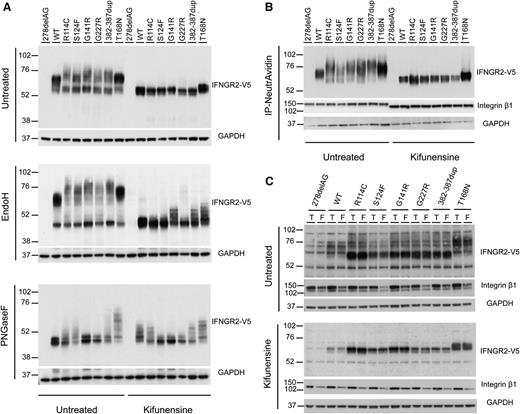
Next, we investigated the biochemical properties of the S124F, G141R, G227R, and R114C hypomorphic IFNGR2 alleles, in line with our previous study of T168N and 382-387dup null alleles.23-25 HEK293T cells were transiently transfected with 278delAG, WT, R114C, S124F, G141R, G227R, 382-387dup, and T168N IFN-γR2-tagged V5 constructs. IFN-γR2 was detected in whole cell extracts by western blotting with the V5-HRP antibody. The WT IFN-γR2 had a molecular weight (MW) of ∼60 kDa (Figure 5A, top panel). Most of the detectable R114C, S124F, G141R, and G227R proteins had an apparent MW of ∼55 kDa. However, proteins with a higher MW (∼75 kDa) were also detected in these same samples (Figure 5A). A similar migration pattern was observed for the product of the previously described 382-387dup allele.25 By contrast, the gain of glycosylation mutant T168N showed a larger MW (∼76 kDa), as previously described.23 All of the 55-kDa mutant proteins were sensitive to treatment with endoglycosidase H (Endo H) (Figure 5A, middle panel), which removes high-mannose glycans (immature oligosaccharides), strongly suggesting an abnormal pattern of N-glycosylation and abnormal folding of the proteins in the ER. As previously described, the heavier T168N proteins were resistant to Endo H treatment23 (Figure 5A, middle panel). In addition, treatment with PNGaseF, which removes all N-glycans (immature and mature), restored the MW of all the mutant proteins to that of the WT protein (Figure 5A, bottom panel). It has been shown that the use of modifiers of N-glycosylation (eg, kifunensine) can improve the maturation of misfolded proteins.25,34-37 We therefore treated the transfected HEK293T cells with kifunensine before western blotting. This treatment impaired the processing of the WT and all mutant proteins, which were also sensitive to Endo H treatment (Figure 5A). These results strongly suggest that S124F, G141R, G227R, and R114C alleles encode misfolded proteins with abnormal N-glycolysation.
Biochemical properties of IFN-γR2 after various chemical treatments. (A) HEK293T cells were transiently transfected with 278delAG, WT, R114C, S124F, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged V5 constructs. They were then incubated alone or with 166 μM kifunensine for 48 hours. Whole cell extracts were generated and left untreated or digested with Endo-H and PNGaseF overnight at 37°C. They were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with antibodies against V5 and glyceraldehyde-3-phosphate dehydrogenase. (B-C) Cell surface expression of IFN-γR2 was evaluated in a surface biotinylation assay in SV40 fibroblasts from the IFN-γR2–deficient patient (278delAG/278delAG) transfected with 278delAG, WT, S124F, R114C, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged V5 constructs. These cells were then incubated alone or with 166 μM kifunensine for 48 hours. (B) Surface receptors isolated by precipitation with NeutroAvidin agarose beads. (C) Total extracts (T) or flow-through (F) fractions were immunoblotted with antibodies against V5, integrin β1, and glyceraldehyde-3-phosphate dehydrogenase.
Biochemical properties of IFN-γR2 after various chemical treatments. (A) HEK293T cells were transiently transfected with 278delAG, WT, R114C, S124F, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged V5 constructs. They were then incubated alone or with 166 μM kifunensine for 48 hours. Whole cell extracts were generated and left untreated or digested with Endo-H and PNGaseF overnight at 37°C. They were then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with antibodies against V5 and glyceraldehyde-3-phosphate dehydrogenase. (B-C) Cell surface expression of IFN-γR2 was evaluated in a surface biotinylation assay in SV40 fibroblasts from the IFN-γR2–deficient patient (278delAG/278delAG) transfected with 278delAG, WT, S124F, R114C, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged V5 constructs. These cells were then incubated alone or with 166 μM kifunensine for 48 hours. (B) Surface receptors isolated by precipitation with NeutroAvidin agarose beads. (C) Total extracts (T) or flow-through (F) fractions were immunoblotted with antibodies against V5, integrin β1, and glyceraldehyde-3-phosphate dehydrogenase.
Characterization of the IFN-γR2 molecules expressed at the cell surface
We further characterized the IFN-γR2 molecules expressed at the cell surface. We transfected IFN-γR2–deficient SV40 fibroblasts with WT, 278delAG, R114C, S124F, G141R, G227R, 382-387dup, and T168N IFN-γR2–tagged V5 constructs and biotinylated the cell surface proteins. The cell lysates were subjected to immunoprecipitation with NeutrAvidin agarose, and the total extract, the flow through, and the elution fractions were analyzed by western blotting with the V5-HRP antibody. WT IFN-γR2 (∼60 kDa) and T168N (∼75 kDa)23 were detected at the cell surface (Figure 5B) and, to a lesser extent, in the cytoplasmic fraction (Figure 5C, flow-through). By contrast, most of the R114C, S124F, G141R, G227R, and 382-387dup mutant proteins were located in the cytoplasmic fraction and had a lower apparent MW (∼55 kDa) (Figure 5C). All of the mutant IFN-γR2 proteins, except T168N, displayed lower levels of surface expression than the WT protein (normalized with integrin β1, a 135-kDa cell surface protein). These data suggest that the antibody used in the flow cytometry experiments shown previously does not recognize all forms of surface-expressed IFN-γR2 molecules (Figure 2A-B; supplemental Figure 5A-B). Moreover, the R114C, S124F, G141R, and G227R proteins had a slightly higher MW than the WT (Figure 5B). However, the MW of the R114C, S124F, G141R, and G227R expressed at the cell surface was lower than that of the 382-386dup proteins. After treatment with kifunensine, all the mutant IFN-γR2 molecules expressed on the cell surface had a MW similar to that of the WT protein (∼60 kDa). Thus, R114C, S124F, G141R, and G227R IFN-γR2 molecules with a higher MW than WT IFN-γR2 molecules are transported to the cell surface, allowing residual cellular responses to IFN-γ in the patients. The defect may be partial because of the higher MW of the surface-expressed molecules, the smaller number of molecules at the surface, or both.
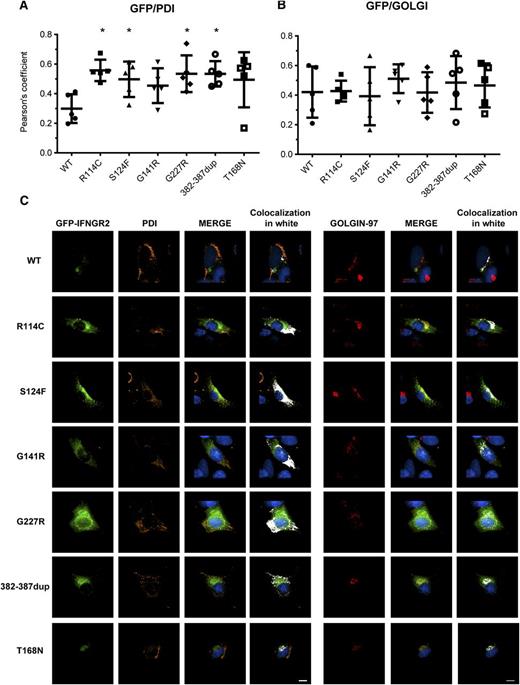
Mutant IFN-γR2 proteins are largely retained in the ER
We then evaluated the subcellular localization of the mutant IFN-γR2 proteins by confocal microscopy using transfected SV40 fibroblasts from the IFN-γR2–deficient patient (278delAG/278delAG).21 The variability of EGFP IFN-γR2 expression after transient transfection was taken into account when comparing cells from WT and each of the 4 hypomorphic mutants. The colocalization with PDI (ER marker) and golgin-97 (trans-Golgi marker) in each cell was quantified using the Pearson’s coefficient (Figure 6A-B). The visualized results of localization of 1 representative cell are shown (Figure 6C). We found that the colocalization of PDI with WT IFN-γR2 was much less than with the R114C, S124F, G141R, G227R, and 382-387dup mutant proteins and also surprisingly for the T168N mutant, suggesting their retention in the ER (Figure 6A,C). No significant differences in the colocalization with golgin-97 were observed between WT and mutant proteins (Figure 6B-C). These results are consistent with the mutant proteins being Endo H sensitive by western blotting (Figure 5A, middle panel) and confirm that the misfolded proteins are retained in the ER. We then evaluated if the accumulation of the mutant IFN-γR2 proteins in the ER activate the unfolded protein response (UPR) pathway. We used 2 approaches: the induction of the mRNA encoding the chaperone binding immunoglobulin protein and the splicing of the mRNA encoding the transcription factor x-box binding protein 1. There was no detectable difference in the induction of BiP mRNA levels or splicing of XBP1 mRNAs between control and patients’ SV40 fibroblasts. Serving as a control, all cell lines activated the UPR pathway normally in response to brefeldin-A (supplemental Figure 6A-B). We further investigated the activation of the UPR by transfecting HEK293 cells with WT and mutant IFNGR2 alleles. We used WT, C194X, and G185R-ELA2 alleles as controls. The mutant ELA2 alleles induced UPR, unlike the mutant IFNGR2 alleles (supplemental Figure 6C).
Subcellular localization of mutant IFN-γR2 proteins. SV40 fibroblasts from the IFN-γR2–deficient patient (278delAG/278delAG) were transiently transfected for 48 hours with WT, S124F, R114C, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged EGFP constructs. Cells were fixed and permeabilized, and intracellular double staining with PDI (orange) and golgin-97 (red) was performed. Images were acquired with DeltaVision Image Restoration Microscope with the 60× objective. Colocalization of GFP with (A) PDI and (B) golgin-97 was measured as Pearson’s coefficients in dataset volumes. The values shown are the mean ± standard deviation of 5 cells analyzed. Differences between WT and each mutant were analyzed using the Student t test. P < .05 was considered significant and is indicated by an asterisk. (C) Colocalization of WT and mutant proteins with PDI or golgin-97 were visualized by merging the images, and then a colocalization channel was created using IMARIS software. Colocalized pixels are show in white. Nuclei were stained with 4,6 diamidino-2-phenylindole (blue). Scale bars, 5 μm.
Subcellular localization of mutant IFN-γR2 proteins. SV40 fibroblasts from the IFN-γR2–deficient patient (278delAG/278delAG) were transiently transfected for 48 hours with WT, S124F, R114C, G141R, G227R, 382-387dup, and T168N IFNGR2-tagged EGFP constructs. Cells were fixed and permeabilized, and intracellular double staining with PDI (orange) and golgin-97 (red) was performed. Images were acquired with DeltaVision Image Restoration Microscope with the 60× objective. Colocalization of GFP with (A) PDI and (B) golgin-97 was measured as Pearson’s coefficients in dataset volumes. The values shown are the mean ± standard deviation of 5 cells analyzed. Differences between WT and each mutant were analyzed using the Student t test. P < .05 was considered significant and is indicated by an asterisk. (C) Colocalization of WT and mutant proteins with PDI or golgin-97 were visualized by merging the images, and then a colocalization channel was created using IMARIS software. Colocalized pixels are show in white. Nuclei were stained with 4,6 diamidino-2-phenylindole (blue). Scale bars, 5 μm.
Chemical complementation of the hypomorphic IFNGR2 alleles with modifiers of N-glycosylation
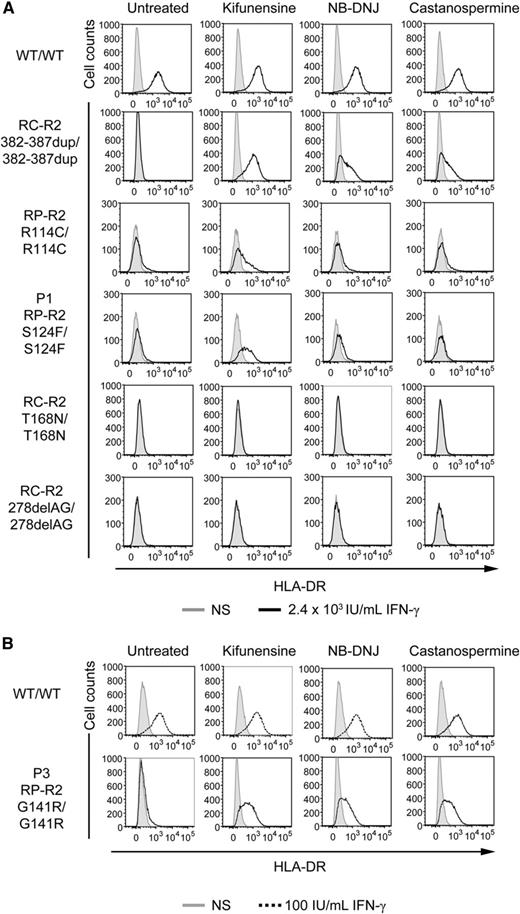
Finally, we investigated whether the cells from the patients could be functionally complemented with kifunensine, N-butyldeoxynojirimycin hydrochloride (NB-DNJ), and castanospermine, as previously shown in a patient with complete IFN-γR2 deficiency due to the mutation 382-387dup (misfolding).25 SV40 fibroblasts were cultured in the presence or absence of these drugs for 16 hours and were then stimulated with IFN-γ for 48 hours. HLA-DR induction was evaluated by flow cytometry. The responsiveness of WT cells was not affected by the treatment with any of these drugs (Figure 7). As previously reported,25 the cells from the patient homozygous for the 382-387dup in-frame mutation responded normally to IFN-γ after all these treatments. Cells from the patient homozygous for the T168N mutation are not complemented with these drugs; this mutation is not misfolding but gain of glycosylation and can be complemented by preventing or removing N-glycosylation with tunicamycin or PNGaseF, respectively, because the gain of glycosylation itself abolishes the receptor function23,24 (Figure 7). Kifunensine treatment also significantly improved the response to IFN-γ in P1, P3, and R114C cells, but not in cells lacking IFN-γR2 expression due to homozygosity for the 278delAG frameshift allele, used as a negative control (Figure 7). Cells homozygous for the G227R mutation were not available. The lack of full complementation with kifunensine in S124F and R114C cells, unlike for G141R, suggests that the serine 124 and arginine 114 residues may be important for IFN-γR2 function, regardless of their impact on glycosylation-related folding and cell traffic. Overall, these results indicate that the S124F and G141R IFNGR2 alleles are responsible for the impaired response to IFN-γ in the patients' cells and that this defect can be rescued by the introduction of the WT IFNGR2 allele or by treatment with modifiers of N-glycosylation. Similar conclusions were reached for the R114C allele and may apply to the G227R allele.
Chemical complementation with kifunensine of the response of the patients’ SV40 fibroblasts to IFN-γ. SV40 fibroblasts from a healthy control (WT/WT), 2 patients with recessive complete IFN-γR2 deficiency with detectable surface expression of a nonfunctional IFN-γR2 (RC-R2, 382-387dup/382-387dup and RC-R2-T168N/T168N), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), patient P1 with the S124F mutation (RP-R2, S124F/S124F), patient P3 with the G141R mutation (RP-R2, G141R/G141R), and a patient with recessive complete IFN-γR2 deficiency and no expression of the receptor at the cell surface (RC-R2, 278delAG/278delAG) were incubated for 72 hours in complete culture medium without (gray filled) stimulation, or with (A) 2.4 × 104 IU/mL IFN-γ (bold line) or (B) 100 IU/mL IFN-γ (dashed line) with or without 1.5 mM NB-DNJ, 2 mM castanospermine, or 160 μM kifunensine. They were analyzed 48 hours later. The surface expression of HLA-DR molecules was determined by flow cytometry with a specific antibody.
Chemical complementation with kifunensine of the response of the patients’ SV40 fibroblasts to IFN-γ. SV40 fibroblasts from a healthy control (WT/WT), 2 patients with recessive complete IFN-γR2 deficiency with detectable surface expression of a nonfunctional IFN-γR2 (RC-R2, 382-387dup/382-387dup and RC-R2-T168N/T168N), a patient with recessive partial IFN-γR2 deficiency (RP-R2, R114C/R114C), patient P1 with the S124F mutation (RP-R2, S124F/S124F), patient P3 with the G141R mutation (RP-R2, G141R/G141R), and a patient with recessive complete IFN-γR2 deficiency and no expression of the receptor at the cell surface (RC-R2, 278delAG/278delAG) were incubated for 72 hours in complete culture medium without (gray filled) stimulation, or with (A) 2.4 × 104 IU/mL IFN-γ (bold line) or (B) 100 IU/mL IFN-γ (dashed line) with or without 1.5 mM NB-DNJ, 2 mM castanospermine, or 160 μM kifunensine. They were analyzed 48 hours later. The surface expression of HLA-DR molecules was determined by flow cytometry with a specific antibody.
Discussion
We report here 3 unrelated MSMD patients with partial, as opposed to complete, AR IFN-γR2 deficiency, and homozygous for 2 novel IFNGR2 mutations, S124F and G141R. The patients’ cells display impaired, but not abolished, responses to IFN-γ. The S124F and G141R mutations affect a residue located in the extracellular domain38-41 and impair but do not abolish the cell surface expression of IFN-γR2. We also found that the 4 (S124F, R114C, G141R, and G227R) hypomorphic IFNGR2 missense alleles encoded misfolded proteins that were abnormally N-glycosylated and largely retained in the ER but did not activate the UPR. Our results also suggest that the high MW of S124F, R114C, G141R, and G227R IFN-γR2 expressed on the cell surface, reflecting excessive N-glycosylation due to a lack of glucose trimming, probably accounts at least in part for the impaired cellular responses to IFN-γ, perhaps more so than the diminished amounts of receptors. This somewhat contrasts with the situation for 382-387dup IFN-γR2 molecules, which had an even higher MW at the cell surface yet were nonfunctional. In addition, as previously described for mutation 382-387dup,25 even though the R114C, S124F, and G141R mutations are not gain of glycosylation, the lack of IFN-γ response was complemented with kifunensine, a modifier of N-glycosylation. These results highlight the important role of glycans in protein folding and quality control in the ER, particularly for IFN-γR2, as previously shown for the 382-387dup in-frame mutation.25 Kifunensine was the only inhibitor tested that complemented all patients’ cells following stimulation with IFN-γ. NB-DNJ and castanospermine also rescued P3 cells. The precise mechanism of action of these modifiers of glycosylation is unclear, but they affect glucosidases I and II, ER-mannosidase I, Golgi mannosidase I, and other enzymes,25,34-37 possibly explaining their different impacts in our experiments. In addition, the mutations may have subtly different structural impacts, in terms of folding and glycosylation of IFN-γR2. Because the three-dimensional structure of IFN-γR2 is unknown, we cannot predict the conformational changes induced by the mutations. Finally, it has been previously shown that some modifiers of N-glycosylation are more potent than others and that not all misfolded mutations are rescued by the same drugs.25 At any rate, our results suggest that protein misfolding with abnormal glycosylation may be disease causing, even for hypomorphic alleles, particularly for proteins undergoing trafficking through the ER. This is apparently at least a general mechanism for IFNGR2 morbid alleles, as all small in-frame mutations encoding surface-expressed receptors detected thus far, whether null (gain-of-glysosylation T168N and misfolding 382-387dup) or hypomorphic (R114C, S124F, and G141R; hypothetically G227R), were rescued with inhibitors of glycosylation.
IFN-γR2 deficiency is one of the rarest genetic etiologies of MSMD, with only 1 known patient with AD and partial IFN-γR2 deficiency, 12 patients with AR and complete deficiency,19-21,23,25,26 and 5 patients with AR and partial IFN-γR2 deficiency,22,27,33 including the 3 patients reported here. The clinical presentation of complete IFN-γR2 deficiency resembles that of complete IFN-γR1 deficiency.32 The disease manifests in early childhood, with severe infections that are often fatal. The most commonly encountered microbial pathogens include Mycobacterium bovis BCG, Mycobacterium avium, and Mycobacterium fortuitum.19,21,23,25 With this poor prognosis, hematopoietic stem cell transplantation (HSCT) is the only curative (but high-risk) option available for these patients.20,32,42-47 The first patient with partial IFN-γR2 deficiency presented with relatively mild infections caused by M bovis BCG and Mycobacterium abscessus.22 Treatment with recombinant IFN-γ was beneficial.22 Partial IFN-γR1 and IFN-γR2 deficiencies generally predispose the patient to curable infections at various ages. Antibiotics, supplemented with IFN-γ treatment, are likely to be effective, and HSCT is therefore not indicated. Despite the treatment of the patients described here with antimycobacterial drugs, they suffered recurrent episodes of MSMD at various ages. However, IFN-γ treatment has recently been initiated in P1, and the clinical status of the patients is improving. Partial IFN-γR1 and IFN-γR2 deficiencies are generally associated with a less severe clinical phenotype than complete deficiencies.22,48,49 Moreover, as modifiers of glycosylation have not been tested in clinical trials, at least at the concentrations used in our assay, IFN-γ treatment (in addition to antimycobacterial drugs) appears to be the best approach for patients with partial deficiencies.22,48 Overall, our findings neatly highlight the importance of early and precise molecular diagnosis of MSMD, to determine the most appropriate treatment available (IFN-γ vs HSCT); this approach may benefit children with other severe infectious diseases.50,51 They also highlight the importance of in-depth investigation of the genetic basis of infectious diseases as an approach to discover novel potential treatments (eg, inhibitors of glycosylation rescuing misfoding mutant proteins).23-25
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the family members for agreeing to participate in this study. The authors also thank all members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions, especially Capucine Picard, Janet Markle, and Minji Byun for critical reading and Tatiana Kochetkov, Hye Kyung Lim, Yelena Nemirovskaya, Tiffany Nivare, and Eric Anderson for technical and secretarial assistance. The authors also thank Masao Kobayashi (Hiroshima University) for providing the ELA2 plasmids and the Flow Cytometry and Bio-Imaging Resource Centers from The Rockefeller University for their important assistance.
This work was supported by grants from the European Research Council (ERC-2010-AdG-268777), Institut National de la Santé et de la Recherche Médicale, University Paris Descartes, French National Agency for Research (ANR), the EU-grant HOMITB (grant HEALTH-F3-2008-200732), the Bill and Melinda Gates Foundation, the St. Giles Foundation, the Jeffrey Modell Foundation,and Talecris Biotherapeutics, Rockefeller University Center for Clinical and Translational Science grant 8UL1TR000043 from the National Center for Research Resources and the National Center for Advancing Sciences (NCATS), the Rockefeller University, and the National Institute of Allergy and Infectious Diseases (grant 1R01AI089970). R.M.-B. is supported by the EMBO Long Term Fellowship program. X.-F.K. is supported by the Stony Wold-Herbert Fund, Choh-Hao Li Memorial Fund Scholar award, and the Shanghai Educational Development Foundation, Y.I. was supported by the AXA Research Fund. V.L.B. was supported by the Stony Wold-Herbert Fund, and A.Y.K. was supported by the Fondation Médicale Medische Stichting Mathilde E. Horlait-Dapsens.
Authorship
Contribution: M.M.-V., S.B.-D., G.V., J.B., and J.-L.C. designed research; M.M.-V., R.M.-B., D.B., X.-F.K., M.M., Q.B.V., and J.B. performed experiments; L.B.-G., C.T., G.A., S.E.-P., M.Y.-N., F.E.-R., and N.K. contributed to the clinical diagnosis of patients; Q.B.V., B.B., Y.I., N.R.-A., S.O., A.Y.K., V.L.B., J.L.F., M.M., and L.A. contributed vital new reagents or analytical tools; M.M.-V., G.V., J.B., S.B.-D., and J.-L.C. edited the paper; and M.M.-V., S.B.-D., and J.-L.C. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jean-Laurent Casanova or Stéphanie Boisson-Dupuis, St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; e-mail: jean-laurent.casanova@rockefeller.edu or stbo603@rockefeller.edu.
References
Author notes
J.B., G.V., J.-L.C., and S.B.-D. contributed equally to this work.