Key Points
The authors developed a novel method for isolating tumor membrane antigens.
LCP1 is functionally relevant to leukemia chemokine induced migration.
Abstract
Membrane antigens are critical to the pathogenesis of chronic lymphocytic leukemia (CLL) as they facilitate microenvironment homing, proliferation, and survival. Targeting the CLL membrane and associated signaling patterns is a current focus of therapeutic development. Many tumor membrane targets are simultaneously targeted by humoral immunity, thus forming recognizable immunoglobulin responses. We sought to use this immune response to identify novel membrane-associated targets for CLL. Using a novel strategy, we interrogated CLL membrane-specific autologous immunoglobulin G reactivity. Our analysis unveiled lymphocyte cytosolic protein 1 (LCP1), a lymphocyte-specific target that is highly expressed in CLL. LCP1 plays a critical role in B-cell biology by crosslinking F-actin filaments, thereby solidifying cytoskeletal structures and providing a scaffold for critical signaling pathways. Small interfering RNA knockdown of LCP1 blocked migration toward CXCL12 in transwell assays and to bone marrow in an in vivo xenotransplant model, confirming a role for LCP1 in leukemia migration. Furthermore, we demonstrate that the Bruton’s tyrosine kinase inhibitor ibrutinib or the PI3K inhibitor idelalisib block B-cell receptor induced activation of LCP1. Our data demonstrate a novel strategy to identify cancer membrane target antigens using humoral anti-tumor immunity. In addition, we identify LCP1 as a membrane-associated target in CLL with confirmed pathogenic significance. This clinical trial was registered at clinicaltrials.gov; study ID number: OSU-0025 OSU-0156.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common type of adult leukemia and is currently incurable. CLL develops through accumulation of malignant B lymphocytes that circulate in the blood but are continuously supported by microenvironments within the bone marrow, lymph nodes, and spleen.1 The most common therapeutic strategies coordinate chemotherapy with monoclonal antibody therapies that target specific membrane antigens, such as CD20.2,3 Recently, immunomodulatory agents (iMiDs) have elicited promising clinical trial results by enhancing anti-tumor immunity in CLL patients. Lenalidomide, an iMiD currently under investigation, was shown to bolster humoral autoimmunity against known CLL-specific membrane antigens.4 The significance of this autoimmune signature is unknown; however, humoral immunity has previously pinpointed a number of relevant tumor targets including p53, Her2/neu, WT1, MUC1, NY-ESO-1, and, in CLL, ROR1.5-8 In terms of CLL therapy, ROR1 is currently under development as both a monoclonal antibody target and chimeric antigen receptor epitope–based therapeutic.9 Curiously, proteomic identification of ROR1 was serendipitous with the identification of autoantibody responses against the epitope in CLL patients.4,8
In CLL, oncogenic signaling derived from multicellular interactions within the specialized niche environment drive the proliferation of an otherwise quiescent leukemic clone.10 In this respect, receptors, ligands, and other membrane-associated proteins represent potential therapeutic targets, as they are the initiators of intracellular signaling, microenvironment homing, proliferation, and survival. Bruton’s tyrosine kinase (BTK) and phosphatidylinositol-3 kinase (PI3K) are two relevant examples of critically important membrane-integrated proteins. BTK and PI3K are essential components of the B-cell receptor (BCR) signalosome, a pathway that is heavily relied on by malignant B cells.11 The recent clinical success of specific BTK and PI3K inhibitors demonstrates that our understanding of membrane-derived CLL signaling is sufficient to attack its molecular core.11,12 Unfortunately, the BCR pathway represents a singular input in a system defined by multiple heterogenic signaling patterns. Therefore, identifying novel membrane-associated targets and their associated role in CLL biology is a current focus of therapeutic development.
A comparative molecular approach has successfully identified signaling pathways commandeered by CLL.13 However, simultaneous reports identified humoral anti-tumor autoimmunity to the same molecular components, implying that autoimmune signatures may be an alternative and efficient means of identification.4,8 Under this premise, we developed a novel strategy by which we interrogate purified CLL membrane extracts with autologous serum to identify immunoreactive membrane antigens. Mass spectroscopic identification showed specific immunoglobulin G (IgG) reactivity against l-plastin (lymphocyte cytosolic protein 1) (LCP1), a membrane-integral component of CLL pathology.
Our data confirm that CLL cells constitutively express LCP1 at high levels, consistent with a previous report.14 We also show that LCP1 protein is exported within exosome structures from CLL cells. Our data demonstrate elevated serum IgG reactivity against LCP1 in CLL patients that surpasses the immunoglobulin response generated against potent vaccine antigens. Functional knockdown of LCP1 impairs migration of CLL cells toward the powerful microenvironment chemokine CXCL12. In vivo characterization showed that LCP1 knockdown impaired bone marrow migration in a xenotransplant leukemia model. Furthermore, therapeutically relevant BTK inhibitor ibrutinib or PI3K inhibitor idelalisib blocked LCP1 activation downstream of BCR ligation. Overall, we use a novel methodology to identify and validate a membrane-associated target in CLL with multifaceted pathogenic potential.
Materials and methods
Patient populations and cell lines
Sera and peripheral blood mononuclear cells were obtained from normal donors or patients with CLL by RosetteSep and density gradient isolation (StemCell Technologies, Vancouver, BC, Canada) to >95% and purity as confirmed by fluorescence-activated cell sorter analysis. As defined by the International Workshop on Chronic Lymphocytic Leukemia 2008 criteria, all subjects provided written, informed consent for their blood to be used for research under two institutional review board–approved protocols in accordance with the Declaration of Helsinki. Blood was collected at The Ohio State University Wexner Medical Center (Columbus, OH). Peripheral blood mononuclear cells were used fresh or stored in 1 mL aliquots at −140°C, and sera were stored in aliquots at −80°C until used. The 697 acute lymphoblastic cell line and the MEC1 CLL cell line were obtained from DSMZ (Braunschwieg, Germany).
Murine xenotransplant leukemia model
Eight-week-old CB17/SCID mice (The Jackson Laboratory, Bar Harbor, ME) were housed in microisolator cages under controlled light cycle, temperature, and humidity and were given a standard pelleted rodent chow and filter-purified water ad libitum. All animal procedures were performed in accordance with Federal and Institutional Animal Care and Use Committee requirements with formal prospective protocol review. Mice were engrafted with 1 × 107 live 697, 697-Empty Vector, or 697-LCP1 short hairpin RNA (shRNA) cells by tail vein injection. After an 18-day engraftment period, mice were humanely killed, and femoral, tibial, and sternal bone marrows were isolated for flow cytometric and histopathologic analysis. Bone marrow analysis was performed using a fluorochrome-labeled mAb against human CD19 (Becton Dickinson, San Jose, CA). FC500 flow cytometric data were analyzed with Kaluza software (Beckman Coulter, Indianapolis, IN) on a minimum of 100 000 collected events. Histopathologic analysis confirmed the distribution of leukemic cells within the bone marrow.
Serological identification of autologous membrane antigens
Membrane isolates were made from freshly purified RosetteSep (StemCell Technologies)-isolated primary CLL cells using the cell surface protein isolation kit (Pierce, Rockford, IL) according to manufacturer instructions. In brief, cells were resuspended in biotinylation buffer for 30 minutes and quenched. Lysates were then made from cell pellets, and biotinylated proteins were extracted using nutravidin column processing. Three wash steps were performed, followed by elution with DTT-containing lysis buffer. Coomassie gel analysis of lysates, wash steps, and final membrane fractions were conducted to ensure purity. Purified membrane isolates were loaded onto sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and were electrophoretically separated according to conventional methods.
Immunoblot
Experiments were conducted using conventional methodology previously described.15 Blotting was conducted using anti-CD52 (alemtuzumab), CD20 (rituximab), HSP70, CD63 (SBI Biosciences, Mountain View, CA), LCP1 #ls-c26109 (LifeSpan Biosciences, Seattle, WA), Erk, pErk (Cell Signaling Technologies, Danvers, MA), BRG1 #sc-17796, α-tubulin #sc-8035, and β-actin #sc-1616 (Santa Cruz Biotechnology, Santa Cruz, CA). Western blotting for pSer5-LCP1 was conducted using rabbit polyclonal antibody, a kind gift from Dr Eric Brown. In circumstances where human IgG was interrogated by immunoblot, anti-human IgG-HRP #2040-05 (Southern Biotech, Birmingham, AL) was used.
Enzyme-linked immunosorbent assay (ELISA)
Ninety-six-well high-binding plates (Corning, Tewksbury, MA) were coated with purified recombinant LCP1 protein (OriGene, Rockville, MD) or tetanus toxin (Sigma-Aldrich, St. Louis, MO) at 1 μg/mL in 50 mmol/L sodium carbonate buffer (pH 9.6) overnight at 4°C. After blocking for 2 hours at room temperature with PBS/1%BSA, wells were filled with 1:25 diluted CLL patient serum or healthy donor serum in blocking solution and were incubated overnight at 4°C. To detect autoantibody, plates were washed three times with PBS/0.1% Tween-20, and anti-human IgG-HRP was added at 1:1000 in blocking buffer. After a three-wash step, reactivity was measured using tetramethyl benzidine substrate (KPL, Gaithersburg, MD) according to the manufacturer’s instructions.
BCR signaling inhibitor treatment
Purified ibrutinib and idealisib chemical inhibitors were obtained from Selekchem Inc. Idelalisib treatment was conducted with continuous exposure to 1 µM idelalisib throughout stimulation. Ibrutinib was used as a pretreatment with 30-minute exposure to 1 µM compound followed by two successive washes to remove unbound free drug. Immediately following washes, CLL cells were stimulated with plate-bound anti-IgM (MP-Biomedicals cat#55073) plated at 10 μg/mL and then washed for 15 minutes.
Exosome purification
Exosome purification was conducted using ExoQuick-TC (SBI Biosciences) according to manufacturer’s specifications. Briefly, primary CLL or RosetteSep isolated (StemCell Technologies) healthy B cells were cultured in RPMI/FBS for 24 hours at 37°C/5% CO2 with or without CpG stimulation. ExoQuick-TC reagent was added to cell-depleted, 24-hour, primary cell culture supernatant and was incubated at 4°C for 12 hours. Exosomes were then centrifugally pelleted at 13 000g for 30 minutes, resuspended in radio-immunoprecipitation assay lysis buffer, and assayed for protein content via immunoblot.
Confocal immunofluorescent microscopy
Cells were centrifugally concentrated on microscope slides using a Cytospin3 centrifuge (Thermo, Asheville, NC) and stained as previously described.16 Cells were then fixed in PBS/2% paraformaldehyde. Slides were incubated in blocking solution (4% bovine serum albumin in PBS) and stained for LCP1 (LifeSpan Biosciences) by incubating with the primary antibodies overnight at 4°C, followed by incubation with secondary antibody conjugated to Alexa fluor 488 (Invitrogen, Carlsbad, CA). Nuclei were stained blue with DAPI (Vector Laboratories, Burlingame, CA). Images were taken using a ×60 objective and ×4 digital zoom with Olympus Fluoview 1000 Laser Scanning Confocal microscope at the Ohio State University Campus Microscopy and Imaging Facility.
Transwell migration assays
Six-hour migration assays were conducted using 8 µm pore size 24-well transwell plates (Corning). One hundred microliters of Suspension of cell line culture (MEC1 or 697) were plated at 1 × 106 in the upper chamber and were allowed to migrate toward 200 ng/mL CXCL-12 containing RPMI for 6 hours at 37°C. Transwell inserts were then removed, and migrated cells were counted using a Coulter FC500 flow cytometer. Fold migration was calculated vs unstimulated baseline migration.
Results
Identifying autoreactive membrane-associated antigens expressed on leukemia cells
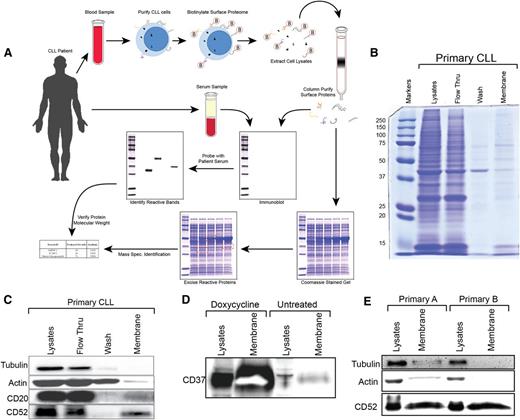
We sought specifically to elucidate novel leukemia membrane antigens using CLL patient IgG immunity. CLL cells were collected from peripheral blood by centrifugal separation and B-cell Rosette-Sep isolation (Figure 1A). The purity of CLL cell isolates was greater than 95%. Following biotinylation of the surface proteins, whole cell lysates were separated from membrane-associated proteins on a streptavidin column. Membrane lysates were then simultaneously analyzed on twin polyacrylamide gels. One gel was transferred to nitrocellulose and probed with autologous serum to identify IgG autoreactive proteins specific to the leukemic membrane. Highly immunoreactive bands were excised from the twin gel after coomassie staining, and mass spectroscopic analysis was used to generate a list of candidate target genes for further validation.
Schema and validation for isolation and identification of autoreactive CLL surface proteins. Panel A alone depicts a schematic representation of an idealized antigen-identification process. Primary CLL cells were surface labeled with biotin. Lysates were loaded onto streptavidin columns, and biotinylated membrane proteins were collected. Cell surface proteome aliquots were run on two identical sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels. One gel was transferred to nitrocellulose and probed with autologous serum at a dilution of 1:100, probed with anti-human IgG-HRP, washed, and developed by autoradiography. Any autoreactive bands were isolated from the coomassie stained gel and subjected to mass spectroscopic protein identification. (B) A coomassie stained gel shows relative protein recovery at each step in the isolation of membrane proteins from freshly purified primary CLL B cells. (C) Immunoblot analysis of cytosolic proteins β-actin and α-tubulin as well as membrane proteins CD20 and CD52 demonstrates enrichment of membrane protein isolation from primary CLL cells. (D) MEC1 CLL cells containing a doxycycline-inducible CD37 construct were utilized to demonstrate the fidelity of cellular membrane isolation. (E) Membrane fractions from primary CLL B cells isolated from two different patients on two different days to ensure repeatable enrichment of membrane proteins.
Schema and validation for isolation and identification of autoreactive CLL surface proteins. Panel A alone depicts a schematic representation of an idealized antigen-identification process. Primary CLL cells were surface labeled with biotin. Lysates were loaded onto streptavidin columns, and biotinylated membrane proteins were collected. Cell surface proteome aliquots were run on two identical sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels. One gel was transferred to nitrocellulose and probed with autologous serum at a dilution of 1:100, probed with anti-human IgG-HRP, washed, and developed by autoradiography. Any autoreactive bands were isolated from the coomassie stained gel and subjected to mass spectroscopic protein identification. (B) A coomassie stained gel shows relative protein recovery at each step in the isolation of membrane proteins from freshly purified primary CLL B cells. (C) Immunoblot analysis of cytosolic proteins β-actin and α-tubulin as well as membrane proteins CD20 and CD52 demonstrates enrichment of membrane protein isolation from primary CLL cells. (D) MEC1 CLL cells containing a doxycycline-inducible CD37 construct were utilized to demonstrate the fidelity of cellular membrane isolation. (E) Membrane fractions from primary CLL B cells isolated from two different patients on two different days to ensure repeatable enrichment of membrane proteins.
To ensure that our membrane extraction was robust, repeatable, and of sufficient purity, we conducted a preliminary isolation using primary CLL cells. Coomassie staining showed that only a small fraction of proteins were retained in the membrane enrichment (Figure 1B). To verify that these proteins were specifically membrane-associated, we conducted a simultaneous immunoblot for the membrane antigens CD52 and CD20, as well as the highly expressed cytoplasmic proteins α-tubulin and β-actin. We found that membrane fractions from primary CLL cells were enriched for specific CLL membrane antigens (Figure 1C).
To verify the fidelity of membrane extraction, we used the MEC1 CLL cell line harboring a doxycycline-inducible CD37 vector. As expected, doxycycline induction of the CD37 membrane antigen produced high levels of protein expression that were specifically enriched in our membrane isolates (Figure 1D).
As a final validation of our procedure, we sought to confirm repeatability using multiple primary CLL donors isolated on separate days using identical methodology. Immunoblot analysis confirmed that membrane fractions were enriched for CD52 expression and were virtually devoid of cytoplasmic proteins α-tubulin and β-actin (Figure 1E).
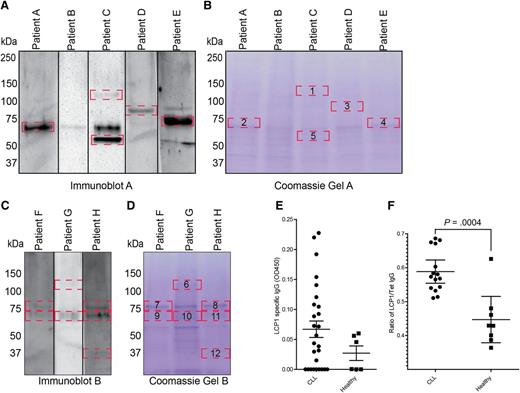
Mass spectroscopy of autoimmune IgG responses identifies LCP1 as a highly reactive CLL membrane-associated protein in multiple donors
Using autologous IgG responses, we screened autoimmune membrane antigens from 8 patients with CLL (supplemental Table 1, available on the Blood Web site). Our analysis unveiled a number of IgG autoreactive membrane antigens, which were further excised from a coomassie stained gel and subjected to mass spectrometric identification (Figure 2A-D). Our results identified a number of candidate proteins, including LCP1, which appeared in multiple instances at a consistent size of 71 kDa (Table 1). Oncomine (Compendia Bioscience, Ann Arbor, MI) database microarray analysis of healthy and CLL B cells showed a significant elevation of LCP1 mRNA (P < .0001) within CLL cells, thus corroborating our identification of a membrane-specific IgG response (supplemental Figure 1).17
Autoreactive membrane associated proteins were identified in a subset of eight CLL patients. (A-D) Immunoblot analysis of autoreactive membrane associated proteins demonstrated immunoreactive bands (delineated by red boxes) (A,C). The identified autoreactive bands were subsequently excised from the coomassie-stained twin gels (B,D) and analyzed by mass spectroscopy to obtain preliminary protein identifications (Table 1). (E) ELISA analysis of LCP1-specific IgG in 33 previously untreated CLL patients and 6 healthy donor controls. (F) ELISA was used to compare autoimmune IgG responsiveness to LCP1 relative to the known vaccine antigen, tetanus toxin (Tet). All CLL patients assayed by ELISA had not received any previous intravenous immunoglobulin therapy. Statistical analysis was performed by Student t test; Error bars indicate standard error of the mean (P = .0004).
Autoreactive membrane associated proteins were identified in a subset of eight CLL patients. (A-D) Immunoblot analysis of autoreactive membrane associated proteins demonstrated immunoreactive bands (delineated by red boxes) (A,C). The identified autoreactive bands were subsequently excised from the coomassie-stained twin gels (B,D) and analyzed by mass spectroscopy to obtain preliminary protein identifications (Table 1). (E) ELISA analysis of LCP1-specific IgG in 33 previously untreated CLL patients and 6 healthy donor controls. (F) ELISA was used to compare autoimmune IgG responsiveness to LCP1 relative to the known vaccine antigen, tetanus toxin (Tet). All CLL patients assayed by ELISA had not received any previous intravenous immunoglobulin therapy. Statistical analysis was performed by Student t test; Error bars indicate standard error of the mean (P = .0004).
Autoreacive membrane-associated antigens identified by mass spectrometry
| Coomassie gel . | Band ID . | CLL patient . | Protein ID . | Predicted Mw (kD) . | GenBank . |
|---|---|---|---|---|---|
| Gel A | 1 | C | hnRNP U | 90 | 32358 |
| ICAM 3 | 60 | 32628 | |||
| Platelet Glycoprotein lib | 71 | 35520 | |||
| l-Plastin (LCPl) | 71 | 62898171 | |||
| 2 | A | Unnamed Prot. | 71 | 189054552 | |
| hnRNP R | 71 | 5031755 | |||
| Unnamed Prot. | 71 | 35218 | |||
| 3 | D | None Identified | — | — | |
| 4 | E | l-Plastin (LCP1) | 71 | 62898171 | |
| Unnamed Prot. | 66 | 1890541782 | |||
| Unnamed Prot. | 71 | 189054552 | |||
| 5 | C | Trans. Upreg Nuclear Prot. | 51 | 4607892 | |
| Unnamed Prot. | 66 | 189054178 | |||
| T-complex Prot. | 57 | 58761484 | |||
| Gel B | 6 | G | Hypothetical Prot. | 78 | 52545896 |
| hnRNP U | 90 | 32358 | |||
| Splicing Factor 3A | 89 | 5032087 | |||
| Matrin 3 | 95 | 21626466 | |||
| 7 | F | Unnamed Prot. | 71 | 18905552 | |
| Unnamed Prot. | 64 | 194373695 | |||
| 8 | H | Lamin B1 | 67 | 5031755 | |
| Unnamed Prot. | 61 | 32356 | |||
| 9 | F | ICAM 2 | 31 | 32624 | |
| Vimentin | 54 | 340219 | |||
| ATP synthase | 60 | 4757810 | |||
| Coronin-like Prot. | 52 | 1002923 | |||
| Nucleosome Assy Prot. | 46 | 4758756 | |||
| Vimentin | 54 | 47115317 | |||
| 10 | G | hnRNP H | 49 | 5031753 | |
| Unnamed Prot. | 51 | 221041944 | |||
| Polyprimidine tract Prot. | 60 | 4506243 | |||
| ICAM 2 | 31 | 32624 | |||
| 11 | H | Calreticulin Precursor | 48 | 4757900 | |
| PDCD4 | 36 | 223673990 | |||
| Lyn | 59 | 4505055 | |||
| Transmembrane Prot. | 24 | 148233642 | |||
| 12 | H | Unnamed Prot. | 27 | 32532 | |
| Elongation Factor 1 | 25 | 4503477 | |||
| Histone H1.2 | 21 | 4885375 |
| Coomassie gel . | Band ID . | CLL patient . | Protein ID . | Predicted Mw (kD) . | GenBank . |
|---|---|---|---|---|---|
| Gel A | 1 | C | hnRNP U | 90 | 32358 |
| ICAM 3 | 60 | 32628 | |||
| Platelet Glycoprotein lib | 71 | 35520 | |||
| l-Plastin (LCPl) | 71 | 62898171 | |||
| 2 | A | Unnamed Prot. | 71 | 189054552 | |
| hnRNP R | 71 | 5031755 | |||
| Unnamed Prot. | 71 | 35218 | |||
| 3 | D | None Identified | — | — | |
| 4 | E | l-Plastin (LCP1) | 71 | 62898171 | |
| Unnamed Prot. | 66 | 1890541782 | |||
| Unnamed Prot. | 71 | 189054552 | |||
| 5 | C | Trans. Upreg Nuclear Prot. | 51 | 4607892 | |
| Unnamed Prot. | 66 | 189054178 | |||
| T-complex Prot. | 57 | 58761484 | |||
| Gel B | 6 | G | Hypothetical Prot. | 78 | 52545896 |
| hnRNP U | 90 | 32358 | |||
| Splicing Factor 3A | 89 | 5032087 | |||
| Matrin 3 | 95 | 21626466 | |||
| 7 | F | Unnamed Prot. | 71 | 18905552 | |
| Unnamed Prot. | 64 | 194373695 | |||
| 8 | H | Lamin B1 | 67 | 5031755 | |
| Unnamed Prot. | 61 | 32356 | |||
| 9 | F | ICAM 2 | 31 | 32624 | |
| Vimentin | 54 | 340219 | |||
| ATP synthase | 60 | 4757810 | |||
| Coronin-like Prot. | 52 | 1002923 | |||
| Nucleosome Assy Prot. | 46 | 4758756 | |||
| Vimentin | 54 | 47115317 | |||
| 10 | G | hnRNP H | 49 | 5031753 | |
| Unnamed Prot. | 51 | 221041944 | |||
| Polyprimidine tract Prot. | 60 | 4506243 | |||
| ICAM 2 | 31 | 32624 | |||
| 11 | H | Calreticulin Precursor | 48 | 4757900 | |
| PDCD4 | 36 | 223673990 | |||
| Lyn | 59 | 4505055 | |||
| Transmembrane Prot. | 24 | 148233642 | |||
| 12 | H | Unnamed Prot. | 27 | 32532 | |
| Elongation Factor 1 | 25 | 4503477 | |||
| Histone H1.2 | 21 | 4885375 |
Unconfirmed peptide identities were obtained from 11 of the 12 bands; l-plastin (AKA: LCP1) was identified twice at the correct size in two different donors. Gel, patient, and band identifiers can be referenced to Figure 1A-D.
To confirm LCP1-specific humoral immunity in CLL patients, we screened leukemic and healthy donor serum samples against purified LCP1 via immunoblot. Fifteen of 22 CLL patients and 5 of 8 healthy donors displayed anti-LCP1 IgG responses (supplemental Figure 2A). To quantify LCP1 IgG in CLL patients, we screened previously untreated CLL patients via ELISA, which showed LCP1-specific IgG responses in 18 of 33 patients (Figure 2E). It has previously been noted that autoimmune responses to cancer-specific antigens such as MUC1 and cyclin B1 can be identified in healthy donor populations; therefore, we decided to further characterize humoral immune function in CLL patients.18-20 Although the majority of CLL patients maintained detectable LCP1-specific IgG, only 6 of 12 were able to mount an identifiable IgG response to the vaccinated tetanus antigen. Notably, 4 of the 6 responders had recently received intravenous allogeneic immunoglobulin as part of their ongoing therapy and, therefore, may be demonstrating adopted humoral immunity (supplemental Figure 2B). As expected, IgG specific immunoblot reactivity to tetanus toxin was identified in 7 of 7 healthy donors.
Our results imply that CLL patients possess the ability to form anti-LCP1 IgG despite the lack of IgG-based immunity to strong vaccine antigens. To confirm these results, we conducted ELISA analyses on serum IgG from a separate CLL patient cohort, none of whom had received previous intravenous allogeneic immunoglobulin therapy. The data clearly demonstrated a significant increase in LCP1 IgG responsiveness (P = .0004) compared with tetanus toxin in the CLL patient cohort, thereby indicating that LCP1 autoimmunity is maintained in spite of profound immune dysfunction (Figure 2F).
LCP1 protein identified in the nucleus, cytoplasm, and exosomes of both CLL cells and healthy B cells
To determine if LCP1 is expressed in healthy tissues and cancers other than CLL, the Oncomine microarray database was probed to generate a comprehensive expression profile for LCP1 mRNA in cancer cell lines (supplemental Figure 3A). We found that LCP1 mRNA was highly expressed in leukemia and lymphoma cell lines, including 3 known CLL cell lines (MEC1, MEC2, and WaC3). Cell line data were compared with LCP1 mRNA expression in nonhematopoietic and hematopoietic primary tissues. Among primary tumors, we found that LCP1 mRNA expression was restricted to leukemia, lymphoma, and a small subset of prostate cancers as compared with healthy hematopoietic and nonhematopoietic tissues (supplemental Figure 3B).
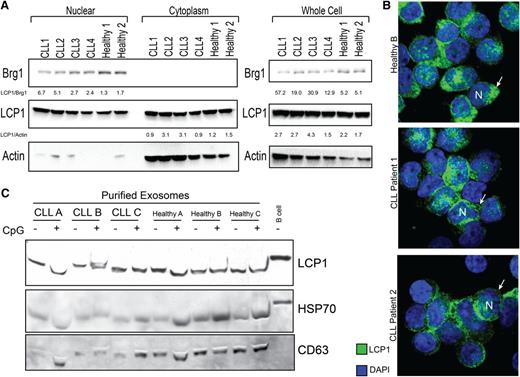
Pathogenic function of tumor antigens can often be inferred by their subcellular localization. To determine whether LCP1 protein expression is nuclear or cytosolic, a subcellular immunoblot analysis was performed. We found that LCP1 protein was highly expressed in both the nuclear and cytosolic fractions of both healthy and malignant B cells (Figure 3A). Our immunoblot results correlated with mRNA expression data, indicating that CLL B cells maintained higher LCP1 levels as compared with healthy B cells. To confirm these results, the localization of LCP1 was verified by confocal microscopy (Figure 3B).
LCP1 protein was identified in the nucleus, cytoplasm, and exosomes of both CLL cells and healthy B cells. (A) Immunoblot analysis of whole cell, nuclear, and cytoplasmic extracts demonstrates that LCP1 protein is present in both subcellular compartments in healthy donor and CLL B cells. Brg1 protein is used as a nuclear loading control, and β-actin is used as a cytoplasmic loading control. Quantitative analysis of band density relative to Actin or Brg1 controls is provided. (B) Confocal immunofluorescent analysis of LCP1 localization in CLL cells and healthy donor B cells confirms evidence that LCP1 is heavily represented in both the nuclear (N) and cytoplasmic (arrow) compartments of malignant and healthy B lymphocytes. (C) Purified exosomes isolated from ex-vivo cultured CLL cells or healthy B cells with or without CpG stimulation were probed for LCP1 by immunoblot. CD63 and HSP70 were used as exosome-specific loading controls.
LCP1 protein was identified in the nucleus, cytoplasm, and exosomes of both CLL cells and healthy B cells. (A) Immunoblot analysis of whole cell, nuclear, and cytoplasmic extracts demonstrates that LCP1 protein is present in both subcellular compartments in healthy donor and CLL B cells. Brg1 protein is used as a nuclear loading control, and β-actin is used as a cytoplasmic loading control. Quantitative analysis of band density relative to Actin or Brg1 controls is provided. (B) Confocal immunofluorescent analysis of LCP1 localization in CLL cells and healthy donor B cells confirms evidence that LCP1 is heavily represented in both the nuclear (N) and cytoplasmic (arrow) compartments of malignant and healthy B lymphocytes. (C) Purified exosomes isolated from ex-vivo cultured CLL cells or healthy B cells with or without CpG stimulation were probed for LCP1 by immunoblot. CD63 and HSP70 were used as exosome-specific loading controls.
Exosomes and microvesicles represent a new area of cancer research. The proteins expressed within these subcellular cancer-derived vesicles can induce adaptive humoral immunity.21,22 These proteins now form a new class of cancer-exosome antigens. To explore the possibility that LCP1 may be packaged into leukemic exosomes, we centrifugally precipitated exosomes from CLL and healthy B cells. We identified LCP1 in exosome fractions from both leukemic and healthy B cells (Figure 3C). Notably, toll-like receptor ligation with CpG did not alter the incorporation of LCP1 into exosome products.
LCP1 function is required for CXCL12-induced CLL cell migration
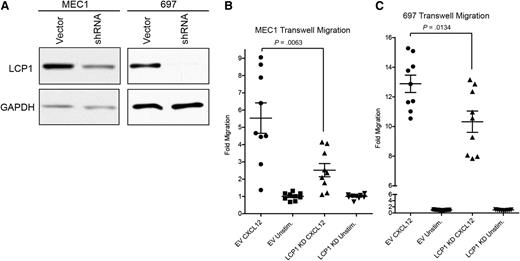
LCP1 is critical to the migratory potential of lymphocytes and certain tumors.23,24 To identify a functional effect of LCP1 on the chemotactic ability of CLL cells, we generated stable shRNA LCP1 knockdown MEC1CLL and 697 (Acute Lymphoblastic Leukemia) cell lines (Figure 4A). The in vitro phenotype, morphology, proliferative rate, and survival of these cell lines were indistinguishable from their parental counterparts. Transwell migration assays confirmed that knockdown of LCP1 significantly reduced leukemic cell migration in response to CXCL12 (Figure 4B). Our results implicate LCP1 as an important factor in CXCL12 responsive migration of leukemic cells.
LCP1 is integral to the CXCL12-driven migration of leukemic cells. (A) Immunoblot analysis of MEC1 or 697 cell lines stably transduced with either an empty vector construct or a shRNA lentiviral knockdown vector to confirm absence of LCP1. GAPDH was used as a loading control. (B) Transwell migration analysis of MEC1 empty vector and MEC1-LCP1 knockdown (KD) in the presence of the chemokine CXCL12 to evaluate chemotactic importance of LCP1. (C) Transwell migration analysis of 697-empty vector and 697-LCP1 knockdown in the presence of chemokine CXCL12 to evaluate importance of LCP1 to cellular chemotaxis. (B-C) Statistical analysis by two tailed Student t test; error bars represent standard error of the mean.
LCP1 is integral to the CXCL12-driven migration of leukemic cells. (A) Immunoblot analysis of MEC1 or 697 cell lines stably transduced with either an empty vector construct or a shRNA lentiviral knockdown vector to confirm absence of LCP1. GAPDH was used as a loading control. (B) Transwell migration analysis of MEC1 empty vector and MEC1-LCP1 knockdown (KD) in the presence of the chemokine CXCL12 to evaluate chemotactic importance of LCP1. (C) Transwell migration analysis of 697-empty vector and 697-LCP1 knockdown in the presence of chemokine CXCL12 to evaluate importance of LCP1 to cellular chemotaxis. (B-C) Statistical analysis by two tailed Student t test; error bars represent standard error of the mean.
Ablation of LCP1 blocks bone marrow migration in vivo
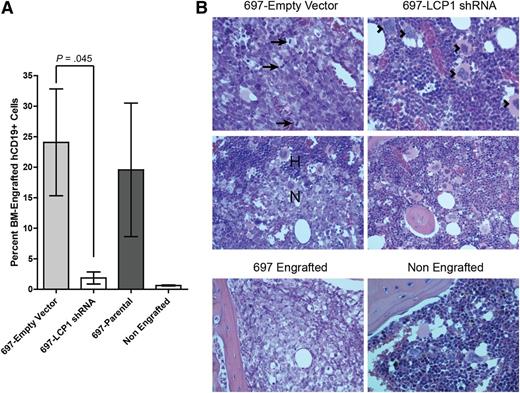
Given these promising in vitro experimental results, we wanted to establish pathologic relevance for LCP1 in vivo. We engrafted CB17/SCID mice with 1 × 107 green fluorescent protein (GFP)-labeled 697-Empty Vector or 697-LCP1 shRNA transfected cells. After 18 days, mice were killed, and bone marrow infiltration of 697 cells was quantified by flow cytometric analysis of GFP and human CD19 (Figure 5A; supplemental Figure 4). Data showed significantly higher levels of GFP+CD19+ bone marrow infiltration in mice engrafted with 697-Empty Vector cells (P = .045). In contrast, mice engrafted with 697-LCP1 shRNA cells displayed no engraftment of GFP+CD19+ cells. Notably, 2 of the mice engrafted with 697-LCP1 shRNA cells showed signs of a small GFP negative CD19+ cell population within the bone marrow, which possibly derived from a subclone that had lost stable expression of the GFP+ shRNA vector and therefore expression of functional LCP1 is assumed (supplemental Figure 4). These data were verified with routine histologic evaluation of femoral and sternal bone marrow. H&E stained sections of the 697-Empty Vector engrafted mice were consistent with fluorescence-activated cell sorter analysis, demonstrating multifocal infiltration and replacement of normal bone marrow with neoplastic lymphocytes (Figure 5B). Infiltrating neoplastic cells demonstrated cytologic criteria of malignancy including high mitotic index, anisokaryosis, anisocytosis, and karyomegaly. In contrast, mice engrafted with 697-LCP1 shRNA cells demonstrated histologic morphology consistent with a nonengrafted control mouse.
LCP1 is critical to bone marrow migration of leukemia in an in vivo xenotransplantation model. (A) CB17/SCID mice were engrafted with 1E7 live 697-parental, 697-Empty Vector, or 697-LCP1 shRNA cells and sacrificed on day 18. Bone marrow infiltration of GFP+human-CD19+ cells was quantified by flow cytometry. Bars depict average bone marrow infiltration of CD19+ cells. Error bars indicate standard error of the mean (B) H&E stained sections of femoral bone marrow from 697-Empty Vector (left), note the bone marrow in this photomicrograph is largely replaced by a monomorphic population of neoplastic round cells containing numerous mitotic figures (arrows). Low-magnification view of the neoplastic cell population (N), which is invading and replacing normal hematopoietic bone marrow (H), as compared with sections from 697-LCP1 shRNA (right), which depict significantly less pathology than the control group. This photomicrograph depicts normal bone marrow in a mouse from this group. Several megakaryocytes (arrowheads) are surrounded by myeloid and erythroid precursor cells. Controls (bottom row) included a mouse engrafted with the parental 697 cell line and an animal that received no xenograft.
LCP1 is critical to bone marrow migration of leukemia in an in vivo xenotransplantation model. (A) CB17/SCID mice were engrafted with 1E7 live 697-parental, 697-Empty Vector, or 697-LCP1 shRNA cells and sacrificed on day 18. Bone marrow infiltration of GFP+human-CD19+ cells was quantified by flow cytometry. Bars depict average bone marrow infiltration of CD19+ cells. Error bars indicate standard error of the mean (B) H&E stained sections of femoral bone marrow from 697-Empty Vector (left), note the bone marrow in this photomicrograph is largely replaced by a monomorphic population of neoplastic round cells containing numerous mitotic figures (arrows). Low-magnification view of the neoplastic cell population (N), which is invading and replacing normal hematopoietic bone marrow (H), as compared with sections from 697-LCP1 shRNA (right), which depict significantly less pathology than the control group. This photomicrograph depicts normal bone marrow in a mouse from this group. Several megakaryocytes (arrowheads) are surrounded by myeloid and erythroid precursor cells. Controls (bottom row) included a mouse engrafted with the parental 697 cell line and an animal that received no xenograft.
Pharmacologic inhibition of BTK or PI3K blocks BCR-induced activation of LCP1
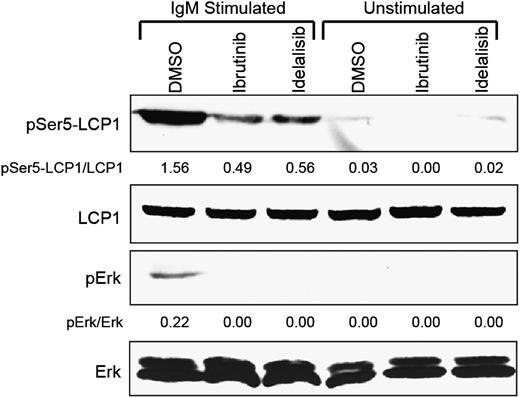
Clinical application of BCR-targeted therapies, such as ibrutinib and idelalisib, in patients with B-cell leukemias and lymphomas has consistently demonstrated early blood lymphocytosis in concert with reduction in lymph node and bone marrow disease.12 This class-specific phenomenon of BCR-targeted agents has been attributed to the downregulation of B-cell integrins, although inhibition of LCP1 may also contribute to inhibiting migratory behavior. To verify this, we treated primary CLL cells ex vivo with ibrutinib, idelalisib, or DMSO control and stimulated the BCR pathway using plate-bound anti-IgM. These studies demonstrate that both of these BCR-targeted inhibitors block phosphorylation of LCP1 at serine-5, a critical residue that drives F-actin binding and cellular migration (Figure 6).25
Ibrutinib and idelalisib block BCR-induced LCP1 serine-5 activation. Immunoblot analysis of freshly obtained primary CLL cells either pretreated with 1 µM ibrutinib or continuously treated with 1 µM idelalisib and stimulated for 15 minutes with plate-bound anti-IgM. Control samples included DMSO-treated cells and matching unstimulated samples. Immunoblots were probed for pSer5-LCP1, total LCP1, pErk, and total Erk. Densitometry measurements indicate intensity relative to total protein controls. Immunoblot is representative of 3 independent experiments performed on different CLL donors.
Ibrutinib and idelalisib block BCR-induced LCP1 serine-5 activation. Immunoblot analysis of freshly obtained primary CLL cells either pretreated with 1 µM ibrutinib or continuously treated with 1 µM idelalisib and stimulated for 15 minutes with plate-bound anti-IgM. Control samples included DMSO-treated cells and matching unstimulated samples. Immunoblots were probed for pSer5-LCP1, total LCP1, pErk, and total Erk. Densitometry measurements indicate intensity relative to total protein controls. Immunoblot is representative of 3 independent experiments performed on different CLL donors.
Discussion
Identification of tumor critical membrane-associated targets represents a therapeutic goal in epithelial and hematopoietic tumors. Herein we describe a novel methodology for the identification and characterization of membrane antigens using preexisting autoimmune IgG responses. We successfully use this methodology in CLL to identify a membrane-associated target, LCP1. Expression of LCP1 is typically limited to the lymphoid lineage.14 However, it is ectopically expressed in many malignant conditions, suggesting that its expression is stimulated accompanying tumorigenesis.23,26,27 We confirm this pathogenic role using an in vivo xenotransplantation model of leukemia. Finally, we demonstrate that BCR-targeted therapies may deactivate LCP1, thereby blocking migratory behavior, a mechanism of action consistent with clinical observations of peripheral lymphocytosis using these inhibitors.
A primary motivation for examining CLL is that these tumor cells are driven to proliferate and survive through microenvironment-driven external stimulation. Moreover, this external stimulation is heterogenic and multifactorial, which implies that many potential membrane-based activating interactions remain to be identified. Previous efforts to identify autoantigens in CLL have been hindered by the prevalence of hypogammaglobulinemia. As a result, only a handful of autoantigens have been identified in CLL. Our methodology for autoantigen identification proved robust and efficient in a notoriously difficult tumor system. Furthermore, our data support the hypothesis that continued antigenic stimulation provides adequate and detectable IgG responses in CLL.
One unanswered question pertains to the mechanism by which CLL patients develop autoimmune signatures for LCP1 while simultaneously suppressing immunity toward relevant external antigens and, as evidenced by our data, vaccines. We have found that both healthy B cells and CLL cells produce LCP1 antigen-laden exosomes. As a result, one possible explanation is that accumulating CLL cells produce excessively high levels of exosomes, thereby driving exosome-antigen presentation.22 Conceivably, a similar situation could occur in healthy donors as a result of a rapidly expanding B-cell population responding to an external pathogen. The continued antigenic stimulation hypothesis serves to explain the stochastic LCP1 autoimmunity found in healthy donors as well as the more frequent and higher level autoimmunity found in CLL patients. Moreover, a well documented membrane-associated cancer antigen, MUC1, has demonstrated similar autoimmune characteristics as a result of virus-induced molecular presentation28-30
We have specifically identified LCP1 as a target in CLL and confirmed that it is functionally relevant in the migration of leukemic cells toward the critical microenvironment homing factor CXCL12. Specific identification of LCP1 within the nucleus and cancer exosomes, however, implies that LCP1 may have additional roles in CLL pathogenesis. Intriguingly, two recent studies identified LCP1 as a molecular signature of highly aggressive IGVH unmutated and 11q23 deletion CLL.14,31 Previous studies of the molecular function of LCP1 indicate that it is critical to the molecular scaffolding process driven by PKC signaling in lymphocytes.32 Moreover, as an actin-bundling protein, LCP1 may be instrumental in the formation or export of tumor microvesicles and exosomes.
Given the robust clinical success of BCR-targeted therapeutics such as ibrutinib, our future studies will be aimed at evaluating further the linkage between LCP1 and the BCR pathway with a specific focus on PKC-β derived signaling. A defined molecular pathogenesis will enhance our understanding of the oncogenic signaling that drives CLL and will help determine the extent to which LCP1 may be a useful new target for therapeutic development.
Portions presented in abstract form at the 54th annual meeting of the American Society for Hematology, Atlanta, GA, December 9, 2012.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Eric Brown for providing the pSer5-LCP1 polyclonal antibody used in this study.
This work was supported by the Leukemia and Lymphoma Society, the D. Warren Brown Foundation, Mr and Mrs Michael Thomas, Harry Mangurian Foundation, the National Cancer Institute (P50 CA140158, K12 CA133250-05, R01 CA177292, P01 CA095426), American Cancer Society 125039-PF-13-246-01-LIB, Pelotonia Undergraduate Fellowship at The Ohio State University, and by scholarship through Choose Ohio First for Bioinformatics.
Authorship
Contribution: J.A.D. conceived the project, planned and organized the research, performed experiments, analyzed data, and cowrote the manuscript; D.L.C. helped plan the research, performed experiments, maintained the mouse colonies, contributed to animal studies, and cowrote the manuscript; B.K.H. ran in vivo experiments; K.A. and M.E.P. analyzed exosomal data; L.A.A., J.M.F., and J.A.J. provided clinical samples; B.B. and A.J.J. provided editorial assistance with the manuscript preparation; and J.C.B. and N.M. conceived the project, supervised the research, reviewed manuscript drafts, obtained funding for the research, and approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John C. Byrd, 455 Wiseman Hall/CCC, 410, West 12th Ave, Columbus, OH 43210; e-mail: john.byrd@osumc.edu; and Natarajan Muthusamy, 455E, OSUCCC, 410, West 12th Ave, Columbus, OH 43210; e-mail: raj.muthusamy@osumc.edu.
References
Author notes
J.A.D. and D.L.C. contributed equally to this study.
Senior authors J.C.B. and N.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal