Childhood acute lymphoblastic leukemia (ALL) is a heterogeneous disease including multiple subtypes, defined on the basis of cell lineage and chromosome anomalies. Previous genome-wide association (GWA) studies report that several ARID5B and IKZF1 single nucleotide polymorphisms (SNPs) are associated with the incidence of ALL. However, high resolution melting analysis (HRM) supplies a faster and more convenient technique to detect SNPs. We applied HRM to detect the SNPs in ARID5B and IKZF1 gene.
Seventy-nine pediatric ALL patients and 80 healthy controls were enrolled. Polymorphic variants of IKZF1 (rs6964823, rs4132601, and rs6944602) and ARID5B (rs7073837, rs10740055, and rs7089424) were detected by HRM method. The SNPs were further analyzed the association with childhood ALL.
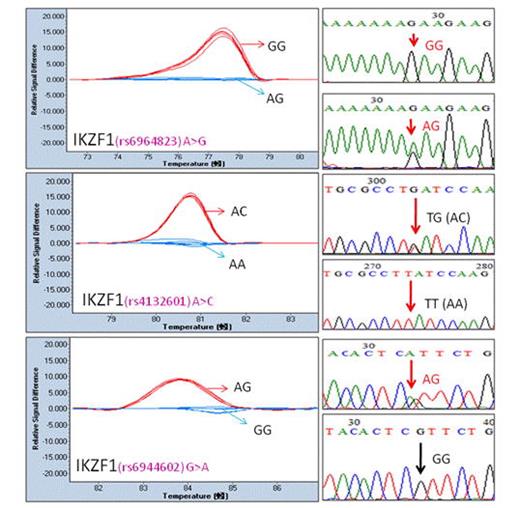
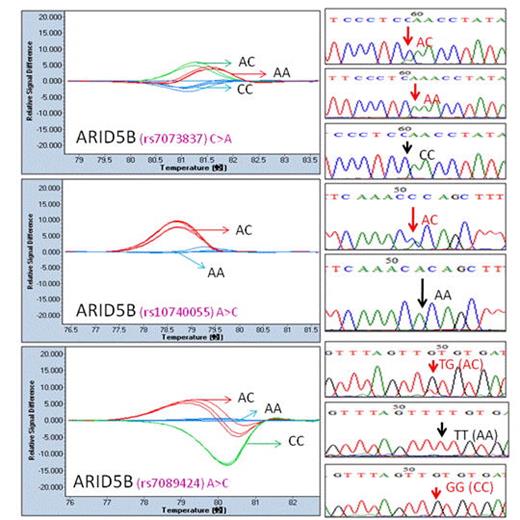
We evaluated 6 mutations by PCR amplicons (81 -176 bp) using the 96-well Light Cycler system. The primer detail was listed in Table 1. The melting curve data showed the difference in normalized temperature and we easily and accurately extended application of HRM analysis to genotyping of IKZF1 (Figure 1) and ARID5B (Figure 2) gene variants.
The HRM primer sequence for IKZF1 and ARID5B gene
| SNP (location) . | Primer sequences . | SNP sequence . | DNA fragment size (bp) . |
|---|---|---|---|
| IKZF1 | |||
| rs6964823 | F : 5'-ATTGCTGTTTTGCCTAATGCTCTGCGGGATTAAAAAAAAA-3' | A>G | 81 |
| R : 5'-CAATGCTCCTAGAGAGACGAATGGTCTTGTTCTTCTTCTT-3' | |||
| IKZF1 | |||
| rs4132601 | F : 5'-CCAATGCACTGAGTGAAGGA-3' | A>C | 176 |
| R : 5'-GGGATGGATGGAAAAGAACA-3' | |||
| IKZF1 | |||
| rs6944602 | F : 5'-AGCTTTCTTACTCATAAAACCGAGA-3' | G>A | 100 |
| R : 5'-CAGGTGGGCAAGTCTGTCTA-3' | |||
| ARID5B | |||
| rs7073837 | F : 5'-CCAGAATGCACACAGTCTCC-3' | C>A | 125 |
| R : 5'-GTGGTTGGGGAAGAACAAAA-3' | |||
| ARID5B | |||
| rs10740055 | F : 5'-GAAAGGCCTAGTCATCCCACT-3' | A>C | 122 |
| R : 5'-AACCACTATGCACTTATCGGAGA-3' | |||
| ARID5B | |||
| rs7089424 | F : 5'-GGAGGTGGAGTTGGCTTTCT-3' | A>C | 126 |
| R : 5'-GCTTTTGCCCTCACTATTGC-3' |
| SNP (location) . | Primer sequences . | SNP sequence . | DNA fragment size (bp) . |
|---|---|---|---|
| IKZF1 | |||
| rs6964823 | F : 5'-ATTGCTGTTTTGCCTAATGCTCTGCGGGATTAAAAAAAAA-3' | A>G | 81 |
| R : 5'-CAATGCTCCTAGAGAGACGAATGGTCTTGTTCTTCTTCTT-3' | |||
| IKZF1 | |||
| rs4132601 | F : 5'-CCAATGCACTGAGTGAAGGA-3' | A>C | 176 |
| R : 5'-GGGATGGATGGAAAAGAACA-3' | |||
| IKZF1 | |||
| rs6944602 | F : 5'-AGCTTTCTTACTCATAAAACCGAGA-3' | G>A | 100 |
| R : 5'-CAGGTGGGCAAGTCTGTCTA-3' | |||
| ARID5B | |||
| rs7073837 | F : 5'-CCAGAATGCACACAGTCTCC-3' | C>A | 125 |
| R : 5'-GTGGTTGGGGAAGAACAAAA-3' | |||
| ARID5B | |||
| rs10740055 | F : 5'-GAAAGGCCTAGTCATCCCACT-3' | A>C | 122 |
| R : 5'-AACCACTATGCACTTATCGGAGA-3' | |||
| ARID5B | |||
| rs7089424 | F : 5'-GGAGGTGGAGTTGGCTTTCT-3' | A>C | 126 |
| R : 5'-GCTTTTGCCCTCACTATTGC-3' |
SNPs in IKZF1 detected by HRM. (A) rs6964823; (B) rs4132601; (C) rs6944602.
SNPs in ARID5B detected by HRM. (A) rs7073837; (B) rs10740055; (C) rs7089424.
The genotype distribution and the allele frequencies of IKZF1 (rs6964823, rs4132601, and rs6944602) and ARID5B (rs7073837, rs10740055, and rs7089424) gene polymorphisms among cases and controls and their association with the risk of childhood ALL were shown in Table 2. The distribution of genotype rs7073837 in ARID5B was significantly different between the ALL group and the control group (P = 0.046), while the distributions for IKZF1 (rs6964823, rs4132601, and rs6944602) and ARID5B (rs10740055, and rs7089424) were not significantly different. In addition, we analyzed the association for those SNPs with B lineage ALL and found rs7073837 in ARID5B conferred a higher risk for B lineage ALL (odds ratio, OR = 1.70, 95% confidence interval, CI = 1.01-2.87, P = 0.049).
Relationship between SNP genotype and risk of childhood ALL in this study
| Genotypes . | Control (n=80) . | Cases (n=79) . | p-value . | OR (95% Cl) . | B-ALL (n=45) . | p-value . | OR (95% Cl) . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | |||||
| IKZF1 | ||||||||||
| rs6964823 A>G | 0.449 | 0.431 | ||||||||
| AA | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| AG | 23 | 28.8 | 21 | 26.6 | 1.0 | 16 | 35.6 | 1.0 | ||
| GG | 57 | 71.3 | 58 | 73.4 | 1.11 (0.56-2.23) | 29 | 64.4 | 0.73 (0.34-1.59) | ||
| G allele | 137 | 85.6 | 137 | 86.7 | 0.871 | 1.11 (0.58-2.07) | 74 | 82.2 | 0.474 | 0.78 (0.39-1.56) |
| rs4132601 A>C | 0.556 | 0.244 | ||||||||
| AA | 61 | 76.3 | 60 | 75.9 | 1.0 | 39 | 86.7 | 1.0 | ||
| AC | 19 | 23.8 | 19 | 24.1 | 1.02 (0.49-2.11) | 6 | 13.3 | 0.49 (0.18-1.35) | ||
| CC | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| C allele | 19 | 11.9 | 19 | 12 | 1.000 | 1.01 (0.52-1.10) | 6 | 6.7 | 0.272 | 0.53 (0.20-1.38) |
| rs6944602 G>A | 0.89 | 0.159 | ||||||||
| GG | 75 | 93.8 | 68 | 86.1 | 1.0 | 45 | 100 | |||
| AG | 5 | 6.3 | 11 | 13.9 | 2.43 (0.80-7.34) | 0 | 0 | |||
| AA | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| A allele | 5 | 3.1 | 11 | 7 | 0.132 | 2.32 (0.79-6.84) | 0 | 0 | 0.163 | |
| ARID5B | ||||||||||
| rs7073837 C>A | 0.046* | 0.002* | ||||||||
| CC | 24 | 30 | 25 | 31.6 | 1.0 | 14 | 31.1 | 1.0 | ||
| AC | 37 | 46.3 | 23 | 29.1 | 0.60 (0.28-1.28) | 8 | 17.8 | 0.37 (0.14-1.02) | ||
| AA | 19 | 23.8 | 31 | 39.2 | 1.57 (0.70-3.49) | 23 | 51.1 | 2.08 (0.85-5.09) | ||
| A allele | 75 | 46.9 | 85 | 53.8 | 0.220 | 1.32 (0.85-2.05) | 54 | 60 | 0.049* | 1.70 (1.01-2.87)* |
| rs10740055 A>C | 0.399 | 0.086 | ||||||||
| AA | 46 | 57.5 | 48 | 60.8 | 1.0 | 33 | 73.3 | 1.0 | ||
| AC | 34 | 42.5 | 31 | 39.2 | 0.87 (0.46-1.65) | 12 | 26.7 | 0.49 (0.22-1.09) | ||
| CC | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| C allele | 34 | 21.3 | 31 | 19.6 | 0.781 | 0.91 (0.52-1.56) | 12 | 13.3 | 0.13 | 0.57 (0.28-1.17) |
| rs7089424 A>C | 0.608 | 0.762 | ||||||||
| AA | 36 | 45 | 31 | 39.2 | 1.0 | 20 | 44.4 | 1.0 | ||
| AC | 26 | 32.5 | 25 | 31.6 | 1.12 (0.54-2.32) | 17 | 37.8 | 1.18 (0.52-2.67) | ||
| CC | 18 | 22.5 | 23 | 29.1 | 1.48 (0.68-3.24) | 8 | 17.8 | 0.80 (0.30-2.17) | ||
| °@ | ||||||||||
| C allele | 62 | 38.8 | 71 | 44.9 | 0.306 | 1.29 (0.83-2.02) | 33 | 36.7 | 0.787 | 0.92 (0.54-1.56) |
| Genotypes . | Control (n=80) . | Cases (n=79) . | p-value . | OR (95% Cl) . | B-ALL (n=45) . | p-value . | OR (95% Cl) . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| n . | % . | n . | % . | n . | % . | |||||
| IKZF1 | ||||||||||
| rs6964823 A>G | 0.449 | 0.431 | ||||||||
| AA | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| AG | 23 | 28.8 | 21 | 26.6 | 1.0 | 16 | 35.6 | 1.0 | ||
| GG | 57 | 71.3 | 58 | 73.4 | 1.11 (0.56-2.23) | 29 | 64.4 | 0.73 (0.34-1.59) | ||
| G allele | 137 | 85.6 | 137 | 86.7 | 0.871 | 1.11 (0.58-2.07) | 74 | 82.2 | 0.474 | 0.78 (0.39-1.56) |
| rs4132601 A>C | 0.556 | 0.244 | ||||||||
| AA | 61 | 76.3 | 60 | 75.9 | 1.0 | 39 | 86.7 | 1.0 | ||
| AC | 19 | 23.8 | 19 | 24.1 | 1.02 (0.49-2.11) | 6 | 13.3 | 0.49 (0.18-1.35) | ||
| CC | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| C allele | 19 | 11.9 | 19 | 12 | 1.000 | 1.01 (0.52-1.10) | 6 | 6.7 | 0.272 | 0.53 (0.20-1.38) |
| rs6944602 G>A | 0.89 | 0.159 | ||||||||
| GG | 75 | 93.8 | 68 | 86.1 | 1.0 | 45 | 100 | |||
| AG | 5 | 6.3 | 11 | 13.9 | 2.43 (0.80-7.34) | 0 | 0 | |||
| AA | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| A allele | 5 | 3.1 | 11 | 7 | 0.132 | 2.32 (0.79-6.84) | 0 | 0 | 0.163 | |
| ARID5B | ||||||||||
| rs7073837 C>A | 0.046* | 0.002* | ||||||||
| CC | 24 | 30 | 25 | 31.6 | 1.0 | 14 | 31.1 | 1.0 | ||
| AC | 37 | 46.3 | 23 | 29.1 | 0.60 (0.28-1.28) | 8 | 17.8 | 0.37 (0.14-1.02) | ||
| AA | 19 | 23.8 | 31 | 39.2 | 1.57 (0.70-3.49) | 23 | 51.1 | 2.08 (0.85-5.09) | ||
| A allele | 75 | 46.9 | 85 | 53.8 | 0.220 | 1.32 (0.85-2.05) | 54 | 60 | 0.049* | 1.70 (1.01-2.87)* |
| rs10740055 A>C | 0.399 | 0.086 | ||||||||
| AA | 46 | 57.5 | 48 | 60.8 | 1.0 | 33 | 73.3 | 1.0 | ||
| AC | 34 | 42.5 | 31 | 39.2 | 0.87 (0.46-1.65) | 12 | 26.7 | 0.49 (0.22-1.09) | ||
| CC | 0 | 0 | 0 | 0 | 0 | 0 | ||||
| C allele | 34 | 21.3 | 31 | 19.6 | 0.781 | 0.91 (0.52-1.56) | 12 | 13.3 | 0.13 | 0.57 (0.28-1.17) |
| rs7089424 A>C | 0.608 | 0.762 | ||||||||
| AA | 36 | 45 | 31 | 39.2 | 1.0 | 20 | 44.4 | 1.0 | ||
| AC | 26 | 32.5 | 25 | 31.6 | 1.12 (0.54-2.32) | 17 | 37.8 | 1.18 (0.52-2.67) | ||
| CC | 18 | 22.5 | 23 | 29.1 | 1.48 (0.68-3.24) | 8 | 17.8 | 0.80 (0.30-2.17) | ||
| °@ | ||||||||||
| C allele | 62 | 38.8 | 71 | 44.9 | 0.306 | 1.29 (0.83-2.02) | 33 | 36.7 | 0.787 | 0.92 (0.54-1.56) |
HRM was a practical method to detect SNPs in ARID5B and IKZF1 gene. We determined the rs7073837 in ARID5B was associated with the risk of childhood B lineage ALL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal