Key Points
Amino acid substitution at peptide-binding residues of the HLA class I molecule is associated with graft-versus-host disease and mortality.
Avoidance of donor-recipient combinations that result in amino acid substitution at peptide-binding residues may improve transplant outcomes.
Abstract
HLA disparity has a negative impact on the outcomes of hematopoietic cell transplantation (HCT). We studied the independent impact of amino acid substitution (AAS) at peptide-binding positions 9, 99, 116, and 156, and killer immunoglobulin-like receptor binding position 77 of HLA-A, B, or C, on the risks for grade 3-4 acute graft-versus-host disease (GVHD), chronic GVHD, treatment-related mortality (TRM), relapse, and overall survival. In multivariate analysis, a mismatch at HLA-C position 116 was associated with increased risk for severe acute GVHD (hazard ratio [HR] = 1.45, 95% confidence interval [CI] = 1.15-1.82, P = .0016). Mismatch at HLA-C position 99 was associated with increased transplant-related mortality (HR = 1.37, 95% CI = 1.1-1.69, P = .0038). Mismatch at HLA-B position 9 was associated with increased chronic GVHD (HR = 2.28, 95% CI = 1.36-3.82, P = .0018). No AAS were significantly associated with outcome at HLA-A. Specific AAS pair combinations with a frequency >30 were tested for association with HCT outcomes. Cysteine to tyrosine substitution at position 99 of HLA-C was associated with increased TRM (HR = 1.78, 95% = CI 1.27-2.51, P = .0009). These results demonstrate that donor-recipient mismatch for certain peptide-binding residues of the HLA class I molecule is associated with increased risk for acute and chronic GVHD and death.
Introduction
The majority of allogeneic hematopoietic cell transplantation (HCT) procedures use adult volunteer unrelated donors, and robust evidence supports the adverse impact of donor-recipient HLA disparity on important HCT outcomes.1-7 In the largest analysis to date, Lee et al3 reported that high-resolution DNA matching at HLA-A, -B, -C, and -DRB1 (8/8 match) resulted in optimal outcomes, whereas single antigen- or allele-level mismatch was associated with increased hazard for acute graft-versus-host disease (GVHD) and an ∼10% reduction in 1-year survival; multiple mismatches further compounded this risk. Unfortunately, an 8/8 match cannot be found for all patients in need of transplantation. The National Marrow Donor Program (NMDP) estimates that ∼30% of Caucasian and up to 70% of minority patients will not find an 8/8 match.
Insights into the relationship between the nature and position of HLA mismatch and its functional consequences are needed to mitigate risk for severe acute GVHD and mortality. Studies have come to divergent conclusions regarding the impact of mismatch at individual HLA loci.3,8 Investigators have attempted to estimate allogenicity of individual HLA class I or II donor-recipient mismatches.9-11 Others have attempted to identify specific nonpermissive donor-recipient allele combinations or specific amino acid substitutions (AAS) associated with increased risk for severe acute GVHD and treatment-related mortality (TRM),12-15 primary malignancy relapse,16 or 100-day mortality post-HCT.17
We proposed that AAS at peptide-binding pockets or killer immunoglobulin-like receptor (KIR) binding domains would have greater impact on GVHD and mortality. We anticipated that AAS at peptide-binding sites would alter antigen presentation and therefore confer greater risk for serious acute GVHD compared with AAS at other sites. In a large analysis facilitated by the Center for International Blood and Marrow Transplant Research, we aimed to: (1) establish the impact of AAS at peptide-binding positions 9, 99, 116, and 156, and KIR binding position 77,18,19 of the HLA class I molecule on HCT outcome; (2) determine whether this effect is restricted to particular HLA class I loci; and (3) examine the impact of specific AAS residue pairs on HCT outcome.
Patients and methods
Study population
The study population included adult and pediatric patients who underwent a myeloablative or reduced intensity/nonmyeloablative first unrelated bone marrow or peripheral blood stem cell HCT for AML, ALL, CML, or MDS between 1988 and 2009. Patients and donors had complete high-resolution typing for HLA-A, B, C, and DRB1 validated through the ongoing NMDP retrospective high-resolution typing program.20 Donor-recipient pairs were categorized as fully high resolution matched for HLA-A, B, C, and DRB1 (8/8) or had a single mismatch at HLA-A, B, or C (7/8). The 7/8 class I mismatched pairs were categorized according to the presence or absence of AAS at peptide-binding sites (positions 9, 99, 116, or 156) and KIR binding position 77. Position 80 was not included in this analysis, because it is in strong linkage disequilibrium with position 77 of HLA-C. Pairs with 2 or more mismatched class I loci were excluded. Available DPB1 high-resolution typing information was used in secondary analyses, given previously reported association of HLA-DP mismatch and acute GVHD3,21 ; HLA-DQ mismatch was not considered, given insufficient evidence of such an effect. All participants provided informed consent for inclusion of clinical data and biospecimens. Research was approved and conducted under supervision of the NMDP Institutional Review Board. This study was conducted in accordance with the Declaration of Helsinki.
Outcomes
Overall survival (OS) summarized time from HCT to death from any cause. TRM included death in continuous remission from primary disease; events were summarized by the cumulative incidence estimate with relapse as a competing risk. Severe acute GVHD included development of grade III-IV acute GVHD according to consensus grading.22 Chronic GVHD was reported as the cumulative incidence of limited or extensive chronic GVHD.23 Primary malignancy relapse was summarized by cumulative incidence estimate, with death as a competing risk.
Variables tested
The main effect tested was the presence of AAS at positions 9, 77, 99, 116, and 156 among 7/8 matched donor-recipient pairs on HCT outcomes. The frequency of donor-recipient allele mismatch combinations, as well as discrepant amino acid pairs, was summarized. Those AAS pairs with frequency of >30 were tested for association with HCT outcomes. In secondary analyses, we examined the impact of specific combinations of AAS at positions 9, 77, 99, 116, and 156, and also the impact of the sum of AAS on HCT outcomes. Analyses examining the relationship between AAS and outcomes of acute or chronic GVHD only considered donor-recipient mismatching in the GVH vector.
Additional patient, disease, and transplantation variables were considered in all multivariate models. Patient variables at the time of HCT included: age, race/ethnicity, and Karnofsky performance status (KPS). Disease variables included: disease, disease stage at time of HCT, and time from diagnosis to HCT. Transplant variables included: stem cell source, donor age, year of HCT, donor-recipient gender match, cytomegalovirus (CMV) serostatus, and ABO matching, conditioning regimen intensity, GVHD prophylaxis, use of in vivo T-cell depletion (ATG or Campath), and transplantation center.
Statistical methods
Descriptive statistics were used to summarize the characteristics of the data set. The χ-square test was used to compare discrete factors between groups. The Kruskal-Wallis test was used to compare continuous factors between groups. Probabilities for OS and disease-free survival were calculated using the Kaplan-Meier estimator with variance estimated by Greenwood's formula. Comparison of survival curves was made using the log-rank test. Relapse and TRM were calculated as cumulative incidence using a linear approximation to estimate the variance.
Multivariate analyses were performed using the proportional hazards model. All clinical variables were tested for the affirmation of the proportional hazards assumption, and factors violating the proportional hazards assumption were adjusted through stratification. A stepwise model building approach was used for the primary outcomes. Interactions among the significant AAS variables and clinical variables were tested. Multivariate analysis for the amino acid mismatches was performed by treating the AAS at each position as an independent variable. A stepwise model selection procedure was adopted to identify AAS sites most associated with the outcomes, controlling for other mismatches present. Adjusted cumulative incidence curves were also generated to illustrate major study findings.24
In the primary analysis, 7/8 mismatched donor-recipient pairs with AAS at the residue of interest (separately tested 9, 77, 99, 116, and 156) were compared with 7/8 pairs without AAS at the residue of interest. In all analyses, the comparator group without AAS of interest did not include 8/8 pairs. The 8/8 pairs were used only in multivariate models to estimate the impact of non-HLA variables on outcome. Separate models were also constructed to examine the impact of AAS at these residues when limited to particular HLA class I locus (HLA-A, B, C) mismatches. For selecting the adjusted covariates, a significance level of 0.05 was used. The frequency of discrepant donor-recipient allele and amino acid residue combinations was summarized. For amino acid residue combinations with a frequency >30 (assumed minimum frequency for this analysis), we examined association with HCT outcomes. To account for multiple testing, based on the Bonferroni criterion, a significance level of 0.01 was used for testing the 5 AAS mismatches, and the significance level of 0.00125 was used for testing specific residue pairs. Given the extent of missing high-resolution typing data for DPB1 (42% of sample), we included the DPB1 match variable in the above multivariate models as a secondary approach. In additional analyses, we examined the relationship between combinations of AAS of interest as well as the sum of total AAS and HCT outcomes. SAS software version 9.2 (SAS Institute, Cary, NC) was used in all analyses.
Results
Included patients
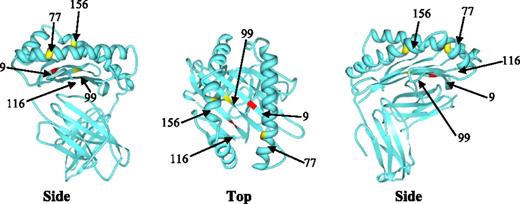
There were 1713 pairs mismatched for HLA-A, -B, or -C at peptide-binding residue 9, 99, 116, or 156, or KIR binding residue 77, either alone or in combination, and 318 pairs mismatched at one HLA class I molecule without these AAS of interest. Patient characteristics are summarized in Table 1. The observed frequency of AAS at residues 9, 77, 99, 116, and 156 alone or in combination are presented in Table 2. The physical location of these amino acid residues in the HLA class I molecule is presented in Figure 1.
Characteristics of recipients receiving first transplants for AML, ALL, CML, or MDS that are high-resolution typed for HLA-A, -B, -C, and -DRB1 and are 8/8 allele-matched or 7/8 allele-matched with single mismatch at Class I locus
| . | 8/8 Matched . | 7/8 with Study AAS . | 7/8 without Study AAS . | P . |
|---|---|---|---|---|
| N (%) | N (%) | N (%) | ||
| Number of patients | 5282 | 1713 | 318 | |
| Number of centers | 166 | 155 | 93 | |
| Recipient age, median (range), y | 41 (<1-74) | 36 (<1-74) | 37 (<1-68) | <.0001 |
| Age at transplant, y | ||||
| 0-9 | 377 (7) | 166 (10) | 33 (10) | <.0001 |
| 10-19 | 523 (10) | 234 (14) | 40 (13) | |
| 20-29 | 744 (14) | 257 (15) | 50 (16) | |
| 30-39 | 870 (16) | 303 (18) | 48 (15) | |
| 40-49 | 1118 (21) | 352 (21) | 78 (25) | |
| 50 and older | 1650 (31) | 401 (23) | 69 (22) | |
| Male sex | 3001 (57) | 940 (55) | 170 (53) | .22 |
| Karnofsky prior to transplant >90 | 3425 (70) | 1141 (71) | 206 (69) | .79 |
| Disease at transplant | ||||
| AML | 2122 (40) | 677 (40) | 112 (35) | <.0001 |
| ALL | 1038 (20) | 395 (23) | 90 (28) | |
| CML | 1146 (22) | 380 (22) | 71 (22) | |
| MDS | 976 (18) | 261 (15) | 45 (14) | |
| Disease status at transplant | ||||
| Early | 3713 (70) | 1186 (69) | 219 (69) | .43 |
| Intermediate | 267 (5) | 104 (6) | 14 (4) | |
| Advanced | 1302 (25) | 423 (25) | 85 (27) | |
| Graft type | ||||
| Bone marrow | 2902 (55) | 1052 (61) | 199 (63) | <.0001 |
| PBSC | 2380 (45) | 661 (39) | 119 (37) | |
| Conditioning regimen | ||||
| Myeloablative | 4082 (77) | 1412 (82) | 268 (84) | <.0001 |
| Reduced intensity | 880 (17) | 229 (13) | 36 (11) | |
| Nonmyeloablative | 320 (6) | 72 (4) | 14 (4) | |
| In vivo T-cell depletion (ATG or Campath) | ||||
| Yes | 1266 (24) | 492 (29) | 75 (24) | .0003 |
| No | 4016 (76) | 1221 (71) | 243 (76) | |
| GVHD prophylaxis | ||||
| FK506 ± MTX ± other | 2617 (50) | 730 (43) | 133 (42) | <.0001 |
| CSA ± MTX ± other | 2303 (44) | 813 (47) | 162 (51) | |
| T-cell depletion | 159 (3) | 90 (5) | 16 (5) | |
| Other | 203 (4) | 80 (5) | 7 (2) | |
| Interval from diagnosis to TX – CML, mo | 13 (1-296) | 15 (2-309) | 12 (2-112) | .13 |
| Interval from diagnosis to TX – MDS, mo | 8 (<1-325) | 9 (<1-342) | 9 (<1-283) | .40 |
| HLA matching for HLA-A, -B, -C and -DRB1 | ||||
| 8/8 allele matched | 5282 (100) | 0 | 0 | <.0001 |
| Single MM at HLA-A | 0 | 631 (37) | 67 (21) | |
| Single MM at HLA-B | 0 | 226 (13) | 91 (29) | |
| Single MM at HLA-C | 0 | 856 (50) | 160 (50) | |
| Donor/recipient sex match | ||||
| Male/male | 2136 (40) | 573 (33) | 116 (36) | <.0001 |
| Male/female | 1349 (26) | 411 (24) | 80 (25) | |
| Female/male | 865 (16) | 367 (21) | 54 (17) | |
| Female/female | 932 (18) | 362 (21) | 68 (21) | |
| Donor/recipient CMV match | ||||
| Negative/negative | 1688 (32) | 526 (31) | 96 (30) | .0009 |
| Negative/positive | 1678 (32) | 485 (28) | 97 (31) | |
| Positive/negative | 696 (13) | 253 (15) | 49 (15) | |
| Positive/positive | 952 (18) | 382 (22) | 65 (20) | |
| Unknown donor or recipient CMV status | 268 (5) | 67 (4) | 11 (3) | |
| Donor age, median (range), y | 34 (18-61) | 36 (19-61) | 37 (19-58) | <.0001 |
| 18-29 | 1763 (33) | 436 (25) | 78 (25) | <.0001 |
| 30-39 | 1928 (37) | 660 (39) | 99 (31) | |
| 40-49 | 1279 (24) | 476 (28) | 100 (31) | |
| 50 and older | 312 (6) | 141 (8) | 41 (13) | |
| Year of transplant | ||||
| 1988 | 11 (<1) | 2 (<1) | 0 | <.0001 |
| 1989 | 37 (1) | 11 (1) | 2 (1) | |
| 1990 | 48 (1) | 16 (1) | 3 (1) | |
| 1991 | 73 (1) | 20 (1) | 7 (2) | |
| 1992 | 91 (2) | 37 (2) | 6 (2) | |
| 1993 | 94 (2) | 44 (3) | 10 (3) | |
| 1994 | 147 (3) | 54 (3) | 6 (2) | |
| 1995 | 143 (3) | 75 (4) | 17 (5) | |
| 1996 | 159 (3) | 64 (4) | 15 (5) | |
| 1997 | 193 (4) | 69 (4) | 14 (4) | |
| 1998 | 184 (3) | 74 (4) | 19 (6) | |
| 1999 | 210 (4) | 96 (6) | 23 (7) | |
| 2000 | 260 (5) | 99 (6) | 26 (8) | |
| 2001 | 255 (5) | 119 (7) | 26 (8) | |
| 2002 | 261 (5) | 74 (4) | 12 (4) | |
| 2003 | 321 (6) | 124 (7) | 19 (6) | |
| 2004 | 466 (9) | 148 (9) | 34 (11) | |
| 2005 | 562 (11) | 161 (9) | 24 (8) | |
| 2006 | 639 (12) | 153 (9) | 23 (7) | |
| 2007 | 630 (12) | 147 (9) | 18 (6) | |
| 2008 | 309 (6) | 93 (5) | 8 (3) | |
| 2009 | 189 (4) | 33 (2) | 6 (2) | |
| Median follow-up of recipients, mo (range) | 67 (3-264) | 74 (3-240) | 96 (3-234) | .0003* |
| . | 8/8 Matched . | 7/8 with Study AAS . | 7/8 without Study AAS . | P . |
|---|---|---|---|---|
| N (%) | N (%) | N (%) | ||
| Number of patients | 5282 | 1713 | 318 | |
| Number of centers | 166 | 155 | 93 | |
| Recipient age, median (range), y | 41 (<1-74) | 36 (<1-74) | 37 (<1-68) | <.0001 |
| Age at transplant, y | ||||
| 0-9 | 377 (7) | 166 (10) | 33 (10) | <.0001 |
| 10-19 | 523 (10) | 234 (14) | 40 (13) | |
| 20-29 | 744 (14) | 257 (15) | 50 (16) | |
| 30-39 | 870 (16) | 303 (18) | 48 (15) | |
| 40-49 | 1118 (21) | 352 (21) | 78 (25) | |
| 50 and older | 1650 (31) | 401 (23) | 69 (22) | |
| Male sex | 3001 (57) | 940 (55) | 170 (53) | .22 |
| Karnofsky prior to transplant >90 | 3425 (70) | 1141 (71) | 206 (69) | .79 |
| Disease at transplant | ||||
| AML | 2122 (40) | 677 (40) | 112 (35) | <.0001 |
| ALL | 1038 (20) | 395 (23) | 90 (28) | |
| CML | 1146 (22) | 380 (22) | 71 (22) | |
| MDS | 976 (18) | 261 (15) | 45 (14) | |
| Disease status at transplant | ||||
| Early | 3713 (70) | 1186 (69) | 219 (69) | .43 |
| Intermediate | 267 (5) | 104 (6) | 14 (4) | |
| Advanced | 1302 (25) | 423 (25) | 85 (27) | |
| Graft type | ||||
| Bone marrow | 2902 (55) | 1052 (61) | 199 (63) | <.0001 |
| PBSC | 2380 (45) | 661 (39) | 119 (37) | |
| Conditioning regimen | ||||
| Myeloablative | 4082 (77) | 1412 (82) | 268 (84) | <.0001 |
| Reduced intensity | 880 (17) | 229 (13) | 36 (11) | |
| Nonmyeloablative | 320 (6) | 72 (4) | 14 (4) | |
| In vivo T-cell depletion (ATG or Campath) | ||||
| Yes | 1266 (24) | 492 (29) | 75 (24) | .0003 |
| No | 4016 (76) | 1221 (71) | 243 (76) | |
| GVHD prophylaxis | ||||
| FK506 ± MTX ± other | 2617 (50) | 730 (43) | 133 (42) | <.0001 |
| CSA ± MTX ± other | 2303 (44) | 813 (47) | 162 (51) | |
| T-cell depletion | 159 (3) | 90 (5) | 16 (5) | |
| Other | 203 (4) | 80 (5) | 7 (2) | |
| Interval from diagnosis to TX – CML, mo | 13 (1-296) | 15 (2-309) | 12 (2-112) | .13 |
| Interval from diagnosis to TX – MDS, mo | 8 (<1-325) | 9 (<1-342) | 9 (<1-283) | .40 |
| HLA matching for HLA-A, -B, -C and -DRB1 | ||||
| 8/8 allele matched | 5282 (100) | 0 | 0 | <.0001 |
| Single MM at HLA-A | 0 | 631 (37) | 67 (21) | |
| Single MM at HLA-B | 0 | 226 (13) | 91 (29) | |
| Single MM at HLA-C | 0 | 856 (50) | 160 (50) | |
| Donor/recipient sex match | ||||
| Male/male | 2136 (40) | 573 (33) | 116 (36) | <.0001 |
| Male/female | 1349 (26) | 411 (24) | 80 (25) | |
| Female/male | 865 (16) | 367 (21) | 54 (17) | |
| Female/female | 932 (18) | 362 (21) | 68 (21) | |
| Donor/recipient CMV match | ||||
| Negative/negative | 1688 (32) | 526 (31) | 96 (30) | .0009 |
| Negative/positive | 1678 (32) | 485 (28) | 97 (31) | |
| Positive/negative | 696 (13) | 253 (15) | 49 (15) | |
| Positive/positive | 952 (18) | 382 (22) | 65 (20) | |
| Unknown donor or recipient CMV status | 268 (5) | 67 (4) | 11 (3) | |
| Donor age, median (range), y | 34 (18-61) | 36 (19-61) | 37 (19-58) | <.0001 |
| 18-29 | 1763 (33) | 436 (25) | 78 (25) | <.0001 |
| 30-39 | 1928 (37) | 660 (39) | 99 (31) | |
| 40-49 | 1279 (24) | 476 (28) | 100 (31) | |
| 50 and older | 312 (6) | 141 (8) | 41 (13) | |
| Year of transplant | ||||
| 1988 | 11 (<1) | 2 (<1) | 0 | <.0001 |
| 1989 | 37 (1) | 11 (1) | 2 (1) | |
| 1990 | 48 (1) | 16 (1) | 3 (1) | |
| 1991 | 73 (1) | 20 (1) | 7 (2) | |
| 1992 | 91 (2) | 37 (2) | 6 (2) | |
| 1993 | 94 (2) | 44 (3) | 10 (3) | |
| 1994 | 147 (3) | 54 (3) | 6 (2) | |
| 1995 | 143 (3) | 75 (4) | 17 (5) | |
| 1996 | 159 (3) | 64 (4) | 15 (5) | |
| 1997 | 193 (4) | 69 (4) | 14 (4) | |
| 1998 | 184 (3) | 74 (4) | 19 (6) | |
| 1999 | 210 (4) | 96 (6) | 23 (7) | |
| 2000 | 260 (5) | 99 (6) | 26 (8) | |
| 2001 | 255 (5) | 119 (7) | 26 (8) | |
| 2002 | 261 (5) | 74 (4) | 12 (4) | |
| 2003 | 321 (6) | 124 (7) | 19 (6) | |
| 2004 | 466 (9) | 148 (9) | 34 (11) | |
| 2005 | 562 (11) | 161 (9) | 24 (8) | |
| 2006 | 639 (12) | 153 (9) | 23 (7) | |
| 2007 | 630 (12) | 147 (9) | 18 (6) | |
| 2008 | 309 (6) | 93 (5) | 8 (3) | |
| 2009 | 189 (4) | 33 (2) | 6 (2) | |
| Median follow-up of recipients, mo (range) | 67 (3-264) | 74 (3-240) | 96 (3-234) | .0003* |
ALL, acute lymphoblastic leukemia; AML, acute myelogenous leukemia; CML, chronic myelogenous leukemia; CSA, cyclosporine; FK506, tacrolimus; MDS, myelodysplastic syndrome; MTX, methotrexate; PBSC, peripheral blood mobilized stem cell.
Log-rank P value.
Frequency of AAS at residues of interest in peptide-binding region of HLA molecule
| Single AAS . | Frequency . | % . | Total AAS . | Frequency . | % . |
|---|---|---|---|---|---|
| 9 | 59 | 15 | Zero | 318 | 16 |
| 77 | 70 | 18 | One | 400 | 20 |
| 99 | 42 | 11 | Two | 403 | 20 |
| 116 | 137 | 34 | Three | 404 | 20 |
| 156 | 92 | 23 | Four | 230 | 11 |
| Five | 276 | 14 |
| Single AAS . | Frequency . | % . | Total AAS . | Frequency . | % . |
|---|---|---|---|---|---|
| 9 | 59 | 15 | Zero | 318 | 16 |
| 77 | 70 | 18 | One | 400 | 20 |
| 99 | 42 | 11 | Two | 403 | 20 |
| 116 | 137 | 34 | Three | 404 | 20 |
| 156 | 92 | 23 | Four | 230 | 11 |
| Five | 276 | 14 |
Single AAS, observed frequency of single AAS of interest in isolation; total AAS, observed frequency of total number of AAS.
Position of studied amino acid residues within the class I HLA molecule.
Impact of type and number of AAS on outcome, irrespective of HLA-locus
In the primary analysis, only AAS at position 116 was associated with a significant increase in grade III-IV acute GVHD (hazard ratio [HR] = 1.3, 95% confidence interval [CI] = 1.1-1.6, P = .0012) after adjustment for significant covariates, including recipient age. Similar findings were observed (HR = 1.3, 95% CI = 1.1-1.5, P = .0006) when DPB1 mismatch was considered. No other significant association was detected between the AAS studied (9, 77, 99, 116, 156) and clinical outcomes.
In a separate analysis, we examined the impact of specific AAS in combination as well as the total sum of AAS. The sum of AAS was associated with increasing hazard for TRM, with a sum of 5 AAS (HR = 1.7, 95% CI = 1.2-2.5, P = .006). Additionally, the sum of AAS among 7/8 matched pairs was associated with grade III-IV acute GVHD (P < .0001). This effect was significant among those with mismatch at HLA-C (P = .0003), but not those with a mismatch at HLA-A (P = .03) or -B (P = .09). We could not demonstrate a consistent association of combinations of AAS and any of the studied HCT outcomes.
AAS restricted to each class I HLA locus
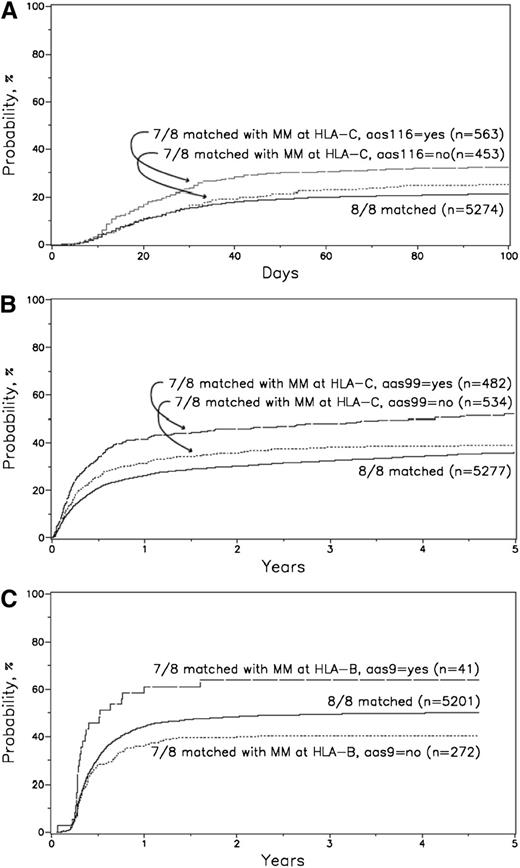
When restricting AAS comparisons within each class I HLA locus, most significant findings were among HLA-C mismatches (Table 3). AAS at position 116 was associated with significantly increased risk for grade III-IV acute GVHD (HR = 1.45, 95% CI = 1.15-1.82, P = .0016), adjusting for disease stage, stem cell source, KPS, recipient age, donor-recipient sex match, disease, GVHD prophylaxis, conditioning regimen, in vivo T-cell depletion, and year of transplant. AAS at position 116 was associated with worse OS (HR = 1.2, 95% CI = 1.01-1.4, P = .03); however, this did not reach our prespecified significance level. AAS at position 99 was associated with significantly worse TRM (HR = 1.37, 95% = CI 1.1-1.69, P = .0038), adjusting for CMV match, donor age, disease category, disease stage, donor race, recipient age, graft type, KPS, GVHD prophylaxis, conditioning regimen, interval from diagnosis to transplant, and year of transplant. These findings were not changed when the DPB1 match variable was included in multivariate analysis. Adjusted cumulative incidence plots are presented in Figure 2.
Results of HLA class I restricted multivariate analyses
| HLA class I locus . | AAS considered . | Outcome . | N . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| HLA-A | AAS 116 absent | OS | 433 | 1.00 | — | |
| HLA-A | AAS 116 present | OS | 265 | 0.96 | (0.78-1.18) | .70 |
| HLA-B | AAS 116 absent | OS | 153 | 1.00 | — | |
| HLA-B | AAS 116 present | OS | 164 | 0.98 | (0.72-1.32) | .88 |
| HLA-C | AAS 116 absent | OS | 453 | 1.00 | — | |
| HLA-C | AAS 116 present | OS | 563 | 1.20 | (1.01-1.41) | .03 |
| HLA-A | AAS 99 absent | TRM | 612 | 1.00 | — | |
| HLA-A | AAS 99 present | TRM | 85 | 0.86 | (0.58-1.30) | .48 |
| HLA-B | AAS 99 absent | TRM | 308 | 1.00 | — | |
| HLA-B | AAS 99 present | TRM | 9 | 0.53 | (0.13-2.24) | .39 |
| HLA-C | AAS 99 absent | TRM | 534 | 1.00 | — | |
| HLA-C | AAS 99 present | TRM | 482 | 1.37 | (1.11-1.69) | .0038 |
| HLA-A | AAS 116 absent | Grades III-IV acute GVHD | 450 | 1.00 | — | |
| HLA-A | AAS 116 present | Grades III-IV acute GVHD | 247 | 1.18 | (0.90-1.56) | .23 |
| HLA-B | AAS 116 absent | Grades III-IV acute GVHD | 159 | 1.00 | — | |
| HLA-B | AAS 116 present | Grades III-IV acute GVHD | 157 | 1.20 | (0.82-1.75) | .35 |
| HLA-C | AAS 116 absent | Grades III-IV acute GVHD | 484 | 1.00 | — | |
| HLA-C | AAS 116 present | Grades III-IV acute GVHD | 531 | 1.45 | (1.15-1.82) | .0016 |
| HLA-A | AAS 9 absent | Chronic GVHD | 291 | 1.00 | — | |
| HLA-A | AAS 9 present | Chronic GVHD | 374 | 1.18 | (0.91-1.53) | .20 |
| HLA-B | AAS 9 absent | Chronic GVHD | 269 | 1.00 | — | |
| HLA-B | AAS 9 present | Chronic GVHD | 40 | 2.28 | (1.36-3.82) | .0018 |
| HLA-C | AAS 9 absent | Chronic GVHD | 434 | 1.00 | — | |
| HLA-C | AAS 9 present | Chronic GVHD | 552 | 1.12 | (0.91-1.39) | .28 |
| HLA class I locus . | AAS considered . | Outcome . | N . | HR . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| HLA-A | AAS 116 absent | OS | 433 | 1.00 | — | |
| HLA-A | AAS 116 present | OS | 265 | 0.96 | (0.78-1.18) | .70 |
| HLA-B | AAS 116 absent | OS | 153 | 1.00 | — | |
| HLA-B | AAS 116 present | OS | 164 | 0.98 | (0.72-1.32) | .88 |
| HLA-C | AAS 116 absent | OS | 453 | 1.00 | — | |
| HLA-C | AAS 116 present | OS | 563 | 1.20 | (1.01-1.41) | .03 |
| HLA-A | AAS 99 absent | TRM | 612 | 1.00 | — | |
| HLA-A | AAS 99 present | TRM | 85 | 0.86 | (0.58-1.30) | .48 |
| HLA-B | AAS 99 absent | TRM | 308 | 1.00 | — | |
| HLA-B | AAS 99 present | TRM | 9 | 0.53 | (0.13-2.24) | .39 |
| HLA-C | AAS 99 absent | TRM | 534 | 1.00 | — | |
| HLA-C | AAS 99 present | TRM | 482 | 1.37 | (1.11-1.69) | .0038 |
| HLA-A | AAS 116 absent | Grades III-IV acute GVHD | 450 | 1.00 | — | |
| HLA-A | AAS 116 present | Grades III-IV acute GVHD | 247 | 1.18 | (0.90-1.56) | .23 |
| HLA-B | AAS 116 absent | Grades III-IV acute GVHD | 159 | 1.00 | — | |
| HLA-B | AAS 116 present | Grades III-IV acute GVHD | 157 | 1.20 | (0.82-1.75) | .35 |
| HLA-C | AAS 116 absent | Grades III-IV acute GVHD | 484 | 1.00 | — | |
| HLA-C | AAS 116 present | Grades III-IV acute GVHD | 531 | 1.45 | (1.15-1.82) | .0016 |
| HLA-A | AAS 9 absent | Chronic GVHD | 291 | 1.00 | — | |
| HLA-A | AAS 9 present | Chronic GVHD | 374 | 1.18 | (0.91-1.53) | .20 |
| HLA-B | AAS 9 absent | Chronic GVHD | 269 | 1.00 | — | |
| HLA-B | AAS 9 present | Chronic GVHD | 40 | 2.28 | (1.36-3.82) | .0018 |
| HLA-C | AAS 9 absent | Chronic GVHD | 434 | 1.00 | — | |
| HLA-C | AAS 9 present | Chronic GVHD | 552 | 1.12 | (0.91-1.39) | .28 |
Adjusted cumulative incidence of grade III-IV acute GVHD for 8/8 matched vs 7/8 HLA-C with AAS 116 vs 7/8 HLA-C without AAS 116. (A) Adjusted cumulative incidence of grade III-IV acute GVHD at 100 days: (1) 8/8 = 21%; (2) 7/8 without HLA-C AAS 116 = 25%; and (3) 7/8 with HLA-C AAS 116 = 32%. P value for comparisons: (2 vs 1) P = .071; (3 vs 1) P < .0001; (3 vs 2) P = .0088. Adjusted variables: disease stage, graft type, Karnofsky score, patient age, sex match, disease, GVHD prophylaxis, conditioning regimen, in vivo T-cell depletion, year of transplant. (B) Adjusted cumulative incidence of TRM for 8/8 matched vs 7/8 HLA-C with AAS 99 vs 7/8 HLA-C without AAS 99. Adjusted cumulative incidence of TRM for 8/8 matched vs 7/8 HLA-C with AAS 99 vs 7/8 HLA-C without AAS 99 at 1 year: (1) 8/8 = 26%; (2) 7/8 without HLA-C AAS 99 = 31%; and (3) 7/8 with HLA-C AAS 99 = 42%. P value for comparisons: (2 vs 1) P = .012; (3 vs 1) P < .0001; (3 vs 2) P = .00035. Adjusted variables: CMV match, disease, disease stage, donor age, donor race, patient age, graft type, KPS, GVHD prophylaxis, conditioning regimen, interval from diagnosis to transplant, year of transplant. (C) Adjusted cumulative incidence of chronic GVHD for 8/8 matched vs 7/8 HLA-B with AAS 9 vs 7/8 HLA-B without AAS 9. Adjusted cumulative incidence of chronic GVHD for 8/8 matched vs 7/8 HLA-B with AAS 9 vs 7/8 HLA-B without AAS 9 at 2 years: (1) 8/8 = 48%; (2) 7/8 without HLA-B AAS 9 = 40%; and (3) 7/8 with HLA-B AAS 9 = 64%. P value for comparisons: (2 vs 1) P = .0056; (3 vs 1) P = .023; (3 vs 2) P = .0012. Adjusted variables: in vivo T-cell depletion, patient age, sex match, disease, graft type, GVHD prophylaxis, CMV match, conditioning regimen, year of transplant.
Adjusted cumulative incidence of grade III-IV acute GVHD for 8/8 matched vs 7/8 HLA-C with AAS 116 vs 7/8 HLA-C without AAS 116. (A) Adjusted cumulative incidence of grade III-IV acute GVHD at 100 days: (1) 8/8 = 21%; (2) 7/8 without HLA-C AAS 116 = 25%; and (3) 7/8 with HLA-C AAS 116 = 32%. P value for comparisons: (2 vs 1) P = .071; (3 vs 1) P < .0001; (3 vs 2) P = .0088. Adjusted variables: disease stage, graft type, Karnofsky score, patient age, sex match, disease, GVHD prophylaxis, conditioning regimen, in vivo T-cell depletion, year of transplant. (B) Adjusted cumulative incidence of TRM for 8/8 matched vs 7/8 HLA-C with AAS 99 vs 7/8 HLA-C without AAS 99. Adjusted cumulative incidence of TRM for 8/8 matched vs 7/8 HLA-C with AAS 99 vs 7/8 HLA-C without AAS 99 at 1 year: (1) 8/8 = 26%; (2) 7/8 without HLA-C AAS 99 = 31%; and (3) 7/8 with HLA-C AAS 99 = 42%. P value for comparisons: (2 vs 1) P = .012; (3 vs 1) P < .0001; (3 vs 2) P = .00035. Adjusted variables: CMV match, disease, disease stage, donor age, donor race, patient age, graft type, KPS, GVHD prophylaxis, conditioning regimen, interval from diagnosis to transplant, year of transplant. (C) Adjusted cumulative incidence of chronic GVHD for 8/8 matched vs 7/8 HLA-B with AAS 9 vs 7/8 HLA-B without AAS 9. Adjusted cumulative incidence of chronic GVHD for 8/8 matched vs 7/8 HLA-B with AAS 9 vs 7/8 HLA-B without AAS 9 at 2 years: (1) 8/8 = 48%; (2) 7/8 without HLA-B AAS 9 = 40%; and (3) 7/8 with HLA-B AAS 9 = 64%. P value for comparisons: (2 vs 1) P = .0056; (3 vs 1) P = .023; (3 vs 2) P = .0012. Adjusted variables: in vivo T-cell depletion, patient age, sex match, disease, graft type, GVHD prophylaxis, CMV match, conditioning regimen, year of transplant.
Further analyses were performed to discern the relative importance of AAS 99 and 116 at HLA-C on grade III-IV acute GHVD and TRM. Among pairs with HLA-C mismatch, AAS 99 alone occurred in 120, AAS 116 alone in 201, and AAS 99+116 together in 362. In the subset analysis restricted to only pairs with HLA-C mismatch, AAS 116 increased risk for grade III-IV acute GVHD (HR = 1.46, 95% CI = 1.03-2.06, P = .03), whereas AAS 99 did not (HR = 0.96, 95% CI =0.61-1.5, P = .85). In multivariate analysis for TRM, both AAS 99 and 116 increased hazard and exerted an additive effect in combination (HR = 1.88, 95% CI = 1.33-2.66, P = .0004).
Among those with mismatch at HLA-B, AAS at position 9 was associated with significantly increased risk for chronic GVHD (HR = 2.28, 95% = CI 1.36-3.82, P = .0018), adjusting for in vivo T-cell depletion, recipient age, donor-recipient sex match, disease category, graft type, GVHD prophylaxis, CMV match, conditioning regimen, and year of transplant. These findings were not changed when the DPB1 match variable was included. Adjusted cumulative incidence plots are presented in Figure 2.
No significant association was found between any of the other studied AAS and relapse. In addition, no significant associations were detected among those with HLA-A mismatches.
AAS pair frequency and outcome
In multivariate analysis, cysteine (C) to tyrosine (Y) substitution at position 99 at HLA-C was associated with increased TRM (HR = 1.78, 95% CI = 1.27-2.51, P = .0009) adjusted for significant covariates, including disease status and recipient age. This finding was not observed when DPB1 match was included in the model. Otherwise, we did not detect significant relationships between the tested specific AAS pairs and any of the studied HCT outcomes. We were not able to confirm the previously reported findings per Kawase et al12,16 regarding acute GVHD or malignancy relapse, although the number of pairs mismatched in these particular combinations was limited. Comparison of the Japan Marrow Donor Program and CIBMTR/NMDP data are presented in supplemental Table 1 (available on the Blood Web site), and donor-recipient allele combinations resulting in AAS significantly associated with adverse outcomes in our analysis are presented in supplemental Table 2.
Discussion
Enormous diversity exists in the human HLA system, and thus many potential HCT candidates will not find a suitably matched unrelated donor. However, current knowledge does not readily help identify the donor that poses minimal risk for acute GVHD and mortality. Structural studies have characterized the peptide-binding groove of the HLA class I molecule25,26 and have highlighted the importance of peptide-binding pockets in binding and presentation of specific peptides.27,28 Emerging clinical outcome data suggests that AAS at key peptide-binding residues of the HLA class I molecule may result in increased risk for severe acute GVHD12 and mortality.17 In a large analysis, strengthened by a priori hypothesis and rigorous statistical methods focused on the impact of AAS among 7/8 mismatched unrelated donor-recipient pairs, we aimed to determine the impact of AAS at peptide-binding positions 9, 99, 116, and 156 and KIR binding position 77 of HLA-A, -B, or -C on HCT outcomes.
The major finding from this study is that AAS at position 116 among HLA-C mismatches is associated with significantly increased risk for severe acute GVHD. This finding advances our understanding beyond a prior report from Ferrara et al,14 as we have demonstrated that AAS at 116 confers increased risk for severe acute GVHD above the risk imposed by other mismatches and that this effect is limited to the HLA-C locus. These clinical findings recapitulate prior structural biologic and functional studies highlighting the importance of position 116 in the class I HLA molecule. The 116 residue of the class I HLA molecule has been demonstrated to form the floor of the peptide-binding F pocket and interact with bound peptide.28-30 AAS at position 116 affects the steric conformation of the F pocket,29 altering peptide binding and allorecognition.31,32 Thus, our HCT outcome data support the functional implications of AAS at this key residue.
We have also identified that AAS at position 99 and 116 among 7/8 pairs mismatched at HLA-C are associated with significantly increased risk for TRM. In our analysis of specific amino acid pairs and HCT outcomes, we demonstrated that the cysteine (C) to tyrosine (Y) substitution at position 99 at HLA-C was associated with increased TRM. We could not detect significant association between other amino acid pairs and outcome. AAS previously reported by Kawase et al12,16 as important for severe acute GVHD and malignancy relapse, respectively, were not substantiated by our analysis. The frequency of these AAS pairs is generally lower in the NMDP/CIBMTR data set (supplemental Table 1). A notable exception is asparagine-serine at position 77 of HLA-C; whereas the observed frequency is greater in our sample, we could not demonstrate the previously reported increased risk for severe acute GVHD. The effects reported by Kawase et al12,16 may be more apparent in a more homogenous population.
A novel finding of our analysis was the association of AAS at position 9 among 7/8 pairs mismatched at HLA-B with risk for chronic GVHD. Providing potential mechanistic support for this finding, previous biological studies have demonstrated that AAS at position 9 alters peptide binding and allorecognition.33-35 In addition, although this association has not been previously reported at the AAS level, prior clinical outcome data have suggested the association of class I HLA,36,37 or specifically HLA-B allelic mismatch,4 with risk for chronic GVHD. In contrast, other investigators have not detected an impact of HLA-B allelic disparity on risk for chronic GVHD development following HCT.3,11,13
We acknowledge the following limitations of our analysis. Low numbers of observations in certain subgroups may limit the power to detect significant differences; for example, HLA-C mismatch and AAS 116 have greater representation in this sample than mismatch at other HLA class I loci and other AAS positions. The overall diversity of this sample limited our analysis to only a subset of AAS pairs of sufficient frequency. Our analysis is limited to only adult volunteer unrelated donor transplantation, and we cannot comment on alternative donor sources. Low numbers of observed events precluded study of the impact of AAS on engraftment failure. Finally, we acknowledge that differences in patient, disease, and transplantation characteristics may impact our results. The observed differences suggest that clinicians take patient factors such as age and disease into account when electing to use mismatched donors and modify transplant practices in this setting. We have tried to address these concerns through the conduct of multivariate analyses controlling for any significant covariates, but this may not fully account for such factors.
In summary, these data support that AAS at key peptide-binding residues of the HLA class I molecule is associated with severe acute GVHD and death. Donor-recipient pairs with such AAS should be avoided to optimize HCT outcomes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The CIBMTR is supported by Public Health Service Grant/Cooperative Agreement U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung, and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID); Grant/Cooperative Agreement 5U10HL069294 from NHLBI and NCI; contract HHSH250201200016C with Health Resources and Services Administration; 2 grants, N00014-12-1-0142 and N00014-13-1-0039, from the Office of Naval Research; and grants from Actinium Pharmaceuticals, Allos Therapeutics, Inc., Amgen, Inc., Anonymous donation to the Medical College of Wisconsin, Ariad, Be the Match Foundation, Blue Cross and Blue Shield Association, Celgene Corporation, Fred Hutchinson Cancer Research Center, Fresenius-Biotech North America, Inc., Gamida Cell Teva Joint Venture Ltd., Genentech, Inc.,Gentium SpA, Genzyme Corporation, GlaxoSmithKline, Health Research, Inc. Roswell Park Cancer Institute, HistoGenetics, Inc., Incyte Corporation, Jeff Gordon Children’s Foundation, Kiadis Pharma, The Leukemia & Lymphoma Society, Medac GmbH, The Medical College of Wisconsin, Merck & Co., Inc., Millennium: The Takeda Oncology Co., Milliman USA, Inc., Miltenyi Biotec, Inc., National Marrow Donor Program, Onyx Pharmaceuticals, Optum Healthcare Solutions, Inc., Osiris Therapeutics, Inc., Otsuka America Pharmaceutical, Inc., PerkinElmer, Inc., Remedy Informatics, Sanofi US, Seattle Genetics, Sigma-τ Pharmaceuticals, Soligenix, Inc., St. Baldrick’s Foundation, StemCyte, A Global Cord Blood Therapeutics Co., Stemsoft Software, Inc., Swedish Orphan Biovitrum, Tarix Pharmaceuticals, TerumoBCT, Teva Neuroscience, Inc., THERAKOS, Inc., University of Minnesota, University of Utah, and Wellpoint, Inc. The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration, or any other agency of the U.S. Government.
Authorship
Contribution: J. Pidala, S.J.L., M.H., T.W., C.A., and S.R.S. designed and performed research, analyzed and interpreted data, and wrote the manuscript; and M.A., M. Battiwalla, L.A.B.-L., M. Bitan, M.F.V., M.G., A.A.J., M.M., S.R.M., S.G.E.M., M.O., J. Palmer, V.K.P., V.R., O.R., W.S., S.S., K.R.S., M.S., E.T., E.V.T., and A.E.W. analyzed and interpreted data and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joseph Pidala, Blood and Marrow Transplantation, Moffitt Cancer Center, 12902 Magnolia Dr, Tampa, FL 33612; e-mail: joseph.pidala@moffitt.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal