Key Points
In CML, early alterations of the BMP pathway are involved in the survival of hypersensitive LSCs and the expansion of myeloid progenitors.
The leukemic niche provides higher concentrations of BMP2 and BMP4 that fuel a permanent and autonomous pool of leukemic progenitors.
Abstract
Leukemic stem cells in chronic phase chronic myelogenous leukemia (CP-CML) are responsible for disease persistence and eventual drug resistance, most likely because they survive, expand, and are sustained through interactions with their microenvironment. Bone morphogenetic proteins 2 (BMP2) and 4 (BMP4) regulate the fate and proliferation of normal hematopoietic stem cells, as well as interactions with their niche. We show here that the intrinsic expression of members of the BMP response pathway are deregulated in CML cells with differences exhibited in mature (CD34−) and immature (CD34+) compartments. These changes are accompanied by altered functional responses of primitive leukemic cells to BMP2 and BMP4 and strong increases in soluble BMP2 and BMP4 in the CML bone marrow. Using primary cells and a cell line mimicking CP-CML, we found that myeloid progenitor expansion is driven by the exposure of immature cells overexpressing BMP receptor Ib to BMP2 and BMP4. In summary, we demonstrate that deregulation of intracellular BMP signaling in primary CP-CML samples corrupts and amplifies their response to exogenous BMP2 and BMP4, which are abnormally abundant within the tumor microenvironment. These results provide new insights with regard to leukemic stem cell biology and suggest possibilities for the development of novel therapeutic tools specifically targeting the CML niche.
Introduction
Chronic myelogenous leukemia (CML) likely arises from a stem cell (SC) transformation induced by the formation of the BCR-ABL oncogene. Without treatment, the disease evolves to an inexorable fatal blast crisis. Until recently, imatinib has become the gold standard for chronic phase (CP)-CML care.1 However, some discrete Philadelphia-positive (Ph+) leukemic SCs (LSCs) may be insensitive to tyrosine kinase inhibitors (TKIs)2-4 and, therefore, sustain detectable disease for many years. None of the therapeutic agents available to date seem to eradicate the undifferentiated BCR-ABL+ cells that serve as a reservoir for additional oncogenic events leading to disease progression,2,4-6 and they require continued treatment.7,8 Therefore, CML represents a unique model to study LSC biology and to elucidate some of the mechanisms of therapeutic resistance.
Complete elimination of CML clones has rarely been achieved by TKIs because of the development of a variety of cell-intrinsic and cell-extrinsic protective mechanisms. Extrinsic mechanisms are supported by the bone marrow (BM) microenvironment, the so-called “leukemic niche.” Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-β superfamily and regulate hematopoietic SC fate both directly at various stages of SC differentiation9-11 and indirectly through the control of their microenvironment.12,13 These molecules are naturally produced by stromal cells, megakaryocytes, and platelets.12 In humans, BMP2, BMP4, and BMP7 regulate the proliferation, maintenance,14 clonogenicity, and repopulating activities of immature cell populations.9,15 In the absence of erythropoietin, BMP2 induces cell commitment and differentiation toward erythropoiesis,16 whereas BMP4 sustains SC maintenance and megakaryocytopoiesis.12 BMP2 and BMP4 are inhibited by follistatin or Follistatin-related gene (FLRG) binding, which are soluble glycoproteins naturally expressed in the human BM microenvironment.16,17 Follistatin and FLRG are involved in the regulation of immature hematopoietic cell adhesion,18 thus contributing to the regulation of human hematopoiesis.17
Survival of CML LSCs appears to be TKI-independent from the BCR-ABL oncogene,6,19 which strongly suggests the involvement of other signals or of the microenvironment.20 Interestingly, the TGF-β family has been involved in the maintenance of CML LSCs.21 Twist-1, an embryonic transcription factor regulated by BMPs,22,23 may represent a new predictive factor for the effect of TKIs on CML.24 BMP signaling is often altered in numerous cancers and involved in cancer SC properties.25 Here we asked whether BMPs levels in the BM of patients with active CML are altered and whether this is associated with molecular and/or cellular alterations in the responses of LSCs to BMPs.
Methods
Primary cells, cell lines, and culture conditions
Normal BM and CML samples were obtained from healthy donors for allogeneic transplant or patients in the chronic or accelerated phase, all at diagnosis and before treatment. All donors provided written informed consent in accordance with the Declaration of Helsinki. All studies were approved by local ethics committee bylaws (clinical research delegation of the Hospices Civils de Lyon, 3 quai des célestins, BP 2251 69229 Lyon Cedex 02, France). Mononuclear cells were subjected to CD34 immunomagnetic separation (Stemcell Technologies). The purity of the CD34+ cells was checked by flow cytometry and was more than 95% on average. CD34+ cells were seeded at 6 × 105 cells/mL and were cultured in Iscove modified Dulbecco medium (Invitrogen); 15% bovine serum albumin, insulin, and transferrin (Stemcell Technologies); 10 ng/mL interleukin 3 (IL-3), IL6, and IL-11; and 50 ng/mL SC factor (Peprotech). The human TF1 and BM-stromal HS27A cell lines were purchased from American Type Culture Collection. BMP receptor Ib (BMPRIb)-expressing cell lines were obtained by transduction of TF1-green fluorescent protein (GFP) or TF1–breakpoint cluster region (BCR)-Abelson (ABL)-GFP with a murine stem cell virus (MSCV)-based retroviral vector,26 followed by transfection with pCAG– internal ribosome entry site (ires) and pCAG-ires-BMPR1b plasmids, as described in the supplemental Methods. BMP2 and BMP4 (50 ng/mL) and sBMPR1a/b (4 µg/mL; R&D Systems) were added as determined.12
Cell counting and cell viability
Viable cells were counted after trypan blue staining, using a Malassez counting chamber. Cell expansion was represented as the ratio of cell count at day 7 to the number of input cells.
CFC and LTC-IC assays
Colony-forming cell (CFC) and long-term culture-initiating cell (LTC-IC) assays were performed as detailed previously.12 Murine stromal cells MS5 (obtained from Dr Coulombel, INSERM U935, Villejuif, Paris, France) were used as feeders in LTC-IC assays.
Immunohistochemistry, flow cytometry, and cell sorting
BM resin-embedded core biopsy specimen immunohistochemistry was performed after endogene peroxidase inhibition by H2O2, heat–induced antigen retrieval with DAKO-buffer at pH 6 (Dako), antigen labeling overnight with anti-BMP2 or anti-BMP4 antibodies (5 µg/mL), Envision leveling (Dako), and tyramide amplification (Perkin Elmer), followed by streptavidin-phosphatase alkaline (Invitrogen) and Liquid Permanent Red (Dako) revelation.27 For the single cell–derived-CFC formation frequency, cells were directly sorted into U-bottom 96-well plates. Cell staining was performed using standard protocols. The details of the procedure, reagents, and list of antibodies used are provided in the supplemental Methods.
RNA isolation and analysis
Quantitative reverse transcription–polymerase chain reaction (RT-PCR) was performed using standard protocols. The levels for each gene were normalized against the same control transcript for all samples. The details of the procedure, reagents, and list of primers used are provided in the supplemental Methods.
BMP2 and BMP4 quantification
BM plasma was obtained from healthy donors and CP-CML patients at diagnosis, precleared of cellular debris by a quick centrifugation, and processed as described for enzyme-linked immunosorbent assay quantification.12
Statistical analysis
Statistical significance on primary samples was determined using the exact Wilcoxon-Mann-Whitney test. Bivariate analysis of the variance was carried out using the analysis of variance method. Data from the cell line models were analyzed using the paired Student t test. Significant P values are indicated by an asterisk (*P < .05; **P < .005; ***P < .0001). Statistics were performed using R-software version 2.15.2.
Results
Molecular alterations in BMP signaling elements in CML cells
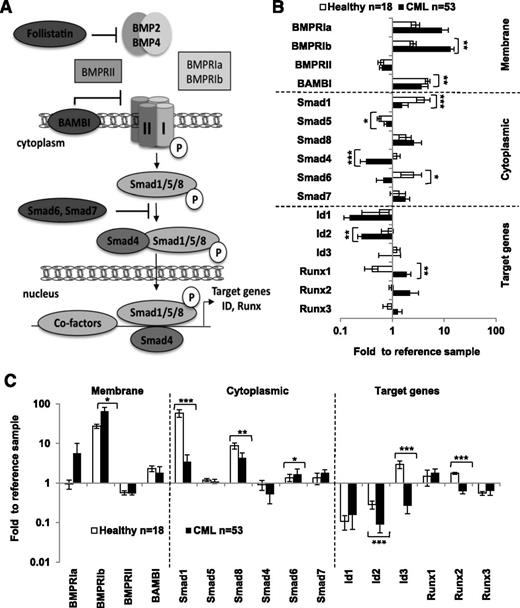
To evaluate whether the endogenous BMP pathway is deregulated in CML, we compared the gene expression profiles of the main elements of the BMP pathway (Figure 1A) in normal cells and CP-CML cells at diagnosis before treatment, using quantitative PCR (qPCR). We found that the expression of several components was deregulated in CML cells (Figure 1B). When compared with normal BM, Smad1 (P < .0001), Smad4 (P < .0001), Smad6 (P = .0062), and Id2 (P = .0025) were significantly downregulated, whereas BMPRIb (P = .0005), Smad5 (P = .0455), and Runx1 (P = .00175) were significantly upregulated. We then investigated whether these changes also affect immature CD34+ CML cells. Although Smad1 (P < .0001), Smad8 (P = .001), and the target genes Id2 (P < .0001), Id3 (P < .0001), and Runx2 (P < .0001) were significantly downregulated, BMPRIb (P = .017) and Smad6 (P = .03) were upregulated (Figure 1C). These results indicate that the BMP pathway is altered in immature CD34+ CML cells at all levels of the pathway. We performed a paired analysis in CD34+ and CD34− cells purified from the same CP-CML samples at diagnosis to compare gene expression between immature and mature cells of the same patient and calculated a ratio for each individual RNA expression. Almost 60% of the BMP-signaling components tested appeared to be differentially expressed between the 2 compartments (supplemental Figure 1). One of the most conserved alterations in both the CD34+ and CD34− cell populations, when compared with their healthy counterpart, was the high levels of BMPRIa and BMPRIb transcripts, together with the very low expression of Smad1. Our results indicate that a major change occurs in the BMP signaling, including an overexpression of type 1 receptors (BMPRIs), a downregulation of Smad-signaling elements, and a striking inversion of target gene expression. These data show that the BMP pathway is affected at all levels in CML.
Expression of BMP signaling elements in primary CML cells. (A) Representative scheme of the BMP signaling pathway that displays the main components of the signaling cascade including extracellular (soluble molecules), membrane, cytoplasm, and nuclear-located responding elements, as well as some BMP target genes. Gene expression (by qPCR) of BMP signaling elements in healthy donor (open bars; n = 18) and CP-CML samples (closed bars; n = 53) at diagnosis for (B) the total number of mononuclear cells or (C) CD34+ immuno-selected hematopoietic cells. Results are expressed as fold change vs the reference value obtained for each gene, using the same CD34− healthy donor sample. *P < .05, **P < .005, and ***P < .0001 indicate differences between the gene expression levels in CML donor samples compared with healthy donor samples.
Expression of BMP signaling elements in primary CML cells. (A) Representative scheme of the BMP signaling pathway that displays the main components of the signaling cascade including extracellular (soluble molecules), membrane, cytoplasm, and nuclear-located responding elements, as well as some BMP target genes. Gene expression (by qPCR) of BMP signaling elements in healthy donor (open bars; n = 18) and CP-CML samples (closed bars; n = 53) at diagnosis for (B) the total number of mononuclear cells or (C) CD34+ immuno-selected hematopoietic cells. Results are expressed as fold change vs the reference value obtained for each gene, using the same CD34− healthy donor sample. *P < .05, **P < .005, and ***P < .0001 indicate differences between the gene expression levels in CML donor samples compared with healthy donor samples.
Deregulation of BMPRIb at the cell surface differentiates normal from immature leukemic cells
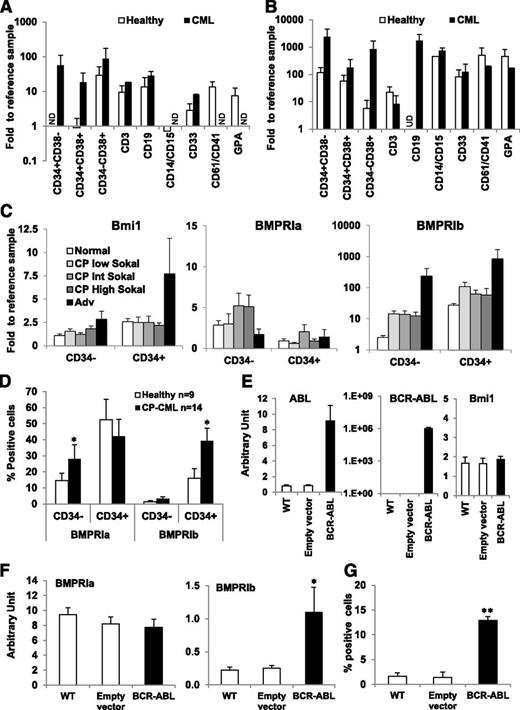
We analyzed gene expression levels in the following sorted cell subpopulations obtained from healthy-donor BM samples (n = 4) or CP-CML cells at diagnosis (n = 3): immature primitive cells (C34+CD38−), immature progenitors (C34+CD38+), lineage-restricted progenitors (C34−CD38+), and cells differentiated as monocytes/granulocytes (CD14+/CD15+), myeloid (CD33+), megakaryocytes (CD61+/CD41+), erythroid (Glycophorin A-GPA+), and T (CD3+) and B (CD19+) lymphocytes. We focused on BMPRIs as mediators of the exogenous effects of BMPs, encoded by one of the most significantly perturbed genes at diagnosis. The higher expression of BMPRIa (Figure 2A) and of BMPRIb (Figure 2B) was present in almost all of the CML subpopulations, indicating that changes in the BMP pathway are a hallmark of leukemic subpopulations. We also analyzed their expression in cells from patients at diagnosis within the 3 established patient groups, based on their Sokal score (low, intermediate, or high), and at advanced stages (accelerated phase, blast crisis). BMPRIb expression progressively increased with disease phase conversely to that of BMPRIa (Figure 2C). However, although the increase of Bmi1 was only observed in late stages of the disease, according to previous data,28 BMPRIb was highly upregulated in CP samples (>10-fold) as early as at diagnosis when compared with normal samples, and again, another 10 times more elevated, during the advanced phases. These data suggest that the expression of BMPRIb is modified early during the course of CML.
BMPRIb is specifically deregulated in primary immature CD34+CML cells. Comparative expression of BMPRIa (A) and BMPRIb (B) genes in distinct sorted subpopulations obtained from healthy donor BM samples (n = 4; open bars) and CP-CML patient samples at diagnosis (n = 3; closed bars). The subpopulations represent immature primitive cells (C34+CD38−), immature progenitors (C34+CD38+), lineage-restricted progenitors (C34−CD38+), monocytes/granulocytes (CD14/CD15+), mature myeloid cells (CD33+), megakaryocytes (CD61/CD41+), erythroid compartments (glycophorin A-GPA+), and T (CD3+) and B (CD19+) lymphocytes. Results are expressed as fold change vs the reference value for each gene, using the same healthy donor sample. *P < .05, **P < .005, and ***P < .0001 indicate differences between the gene expression levels in CML compared with healthy samples. (C) Comparative expression of Bmi1, BMPRIa, and BMPRIb genes in CD34+ and CD34− cells obtained from healthy donors (n = 14; open bars) and CML patient samples either in chronic phase (CP) at diagnosis (n = 53; gray bars) or in advanced (Adv) phase at diagnosis (n = 4; closed bars). Results from patient samples in CP at diagnosis are subdivided between low (<0.8), intermediate (0.8–1.2), and high (> 1.2) Sokal scores (different tone gray bars). (D) Flow cytometry analysis of BMPRIa and BMPRIb cell surface expression in CD34+ and CD34− cells obtained from healthy donors (n = 9; open bars) and CP-CML samples at diagnosis (n = 14; closed bars), using a FACSCalibur cell analyzer (Becton Dickinson). Results are expressed as the percentage of receptor-expressing cells. *P < .05 indicates differences between healthy and CML samples. Comparative expression of (E) ABL, BCR-ABL and Bmi1 genes or (F) BMPRIa and BMPRIb genes by qPCR in parental TF1 cells transduced either with an empty vector (non-BCR-ABL; open bars) or with a vector containing BCR-ABL expressing sequence (closed bars). Results represent the mean value ± standard error of the mean (SEM) of 7 (E) or 14 experiments (F). *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells. (G) Flow cytometry analysis of BMPRIb receptor cell surface expression on either parental TF1 cells transduced with an empty vector (non-BCR-ABL; open bars) or with a vector containing BCR-ABL expressing sequence (closed bars) and, as indicated, performed on viable cells using a FACSCalibur cell analyzer (Becton Dickinson). Results represent the mean value ± SEM of 8 experiments. **P < .005 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells.
BMPRIb is specifically deregulated in primary immature CD34+CML cells. Comparative expression of BMPRIa (A) and BMPRIb (B) genes in distinct sorted subpopulations obtained from healthy donor BM samples (n = 4; open bars) and CP-CML patient samples at diagnosis (n = 3; closed bars). The subpopulations represent immature primitive cells (C34+CD38−), immature progenitors (C34+CD38+), lineage-restricted progenitors (C34−CD38+), monocytes/granulocytes (CD14/CD15+), mature myeloid cells (CD33+), megakaryocytes (CD61/CD41+), erythroid compartments (glycophorin A-GPA+), and T (CD3+) and B (CD19+) lymphocytes. Results are expressed as fold change vs the reference value for each gene, using the same healthy donor sample. *P < .05, **P < .005, and ***P < .0001 indicate differences between the gene expression levels in CML compared with healthy samples. (C) Comparative expression of Bmi1, BMPRIa, and BMPRIb genes in CD34+ and CD34− cells obtained from healthy donors (n = 14; open bars) and CML patient samples either in chronic phase (CP) at diagnosis (n = 53; gray bars) or in advanced (Adv) phase at diagnosis (n = 4; closed bars). Results from patient samples in CP at diagnosis are subdivided between low (<0.8), intermediate (0.8–1.2), and high (> 1.2) Sokal scores (different tone gray bars). (D) Flow cytometry analysis of BMPRIa and BMPRIb cell surface expression in CD34+ and CD34− cells obtained from healthy donors (n = 9; open bars) and CP-CML samples at diagnosis (n = 14; closed bars), using a FACSCalibur cell analyzer (Becton Dickinson). Results are expressed as the percentage of receptor-expressing cells. *P < .05 indicates differences between healthy and CML samples. Comparative expression of (E) ABL, BCR-ABL and Bmi1 genes or (F) BMPRIa and BMPRIb genes by qPCR in parental TF1 cells transduced either with an empty vector (non-BCR-ABL; open bars) or with a vector containing BCR-ABL expressing sequence (closed bars). Results represent the mean value ± standard error of the mean (SEM) of 7 (E) or 14 experiments (F). *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells. (G) Flow cytometry analysis of BMPRIb receptor cell surface expression on either parental TF1 cells transduced with an empty vector (non-BCR-ABL; open bars) or with a vector containing BCR-ABL expressing sequence (closed bars) and, as indicated, performed on viable cells using a FACSCalibur cell analyzer (Becton Dickinson). Results represent the mean value ± SEM of 8 experiments. **P < .005 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells.
We analyzed BMPRI cell surface expression by flow cytometry. Although the increased expression of BMPRIa reaches statistical significance in mature CD34− cells, BMPRIb was significantly overexpressed in immature CD34+ CML cells (P = .037 and P = .04, respectively) (Figure 2D; supplemental Figure 2). We analyzed the effect of the introduction of BCR-ABL in the human cell line TF1, used as a model of immature CD34+ cells, which displays clongenic ability similar to human BM CD34+ cells and is able to differentiate into myeloid lineages.29 When compared with a wild-type cell line or when transduced with an empty vector, BCR-ABL-transduced TF1 cells (TF1-BCR-ABL) increased their transcriptional levels of BCR-ABL and ABL, as expected (Figure 2E), but not levels of Bmi1, the deregulation of which more likely reflects a secondary event28 (Figure 2C). Although no difference was observed between transduced and untransduced cells regarding the BMPRIa expression, the transcriptional level of BMPRIb increased in TF1-BCR-ABL cells (P = .014; Figure 2F). This was confirmed at the protein level (P = .0004; Figure 2G). These data indicate that the TF1-BCR-ABL model reproduces the deregulation observed in CP-CML patients regarding the BMPRI expression patterns, which are directly driven by the oncogene BCR-ABL. Data in both primary cells and the TF1-model strongly suggest a change in the regulation of LSC by the BMP pathway.
Excess BMP2 and BMP4 within the tumoral environment contributes to early increases in BMPRIb expression in immature CML cells
We investigated whether soluble BMPs involved in the regulation of normal hematopoietic SCs (HSCs) and progenitors12,16,17 were present in the CML tumor environment. Using an enzyme-linked immunosorbent assay, we measured the levels of BMP4 and BMP2 present in BM plasma obtained from healthy donors and CP-CML patients at diagnosis, separated into 2 groups on the basis of their number of circulating platelets (elevated, >400 G/L; normal, ≤400 G/L) as a potential source of BMPs12 (Figure 3A). We observed a significantly higher level of BMP2 (P = .0001) and BMP4 (P = .0008) in patients with normal platelet counts and a further increase in some patients with a deregulated platelet count; however, this increase was not significant. Unlike BMP4, BMP2 transcripts are elevated in normal CD34+ blood cells and low in CD34− cells from healthy donors (Figure 3B). In CML blood, lower levels of BMP2 and BMP4 transcripts were detected in CD34+ and CD34− cells isolated from the same samples. Conversely, primary leukemic stromal cells30,31 displayed higher levels of BMP4 transcripts and BMP2 transcripts to a lesser extent (Figure 3C). We used BM biopsies to identify the in situ source of soluble BMP2 and BMP4, using antibody staining. We observed an increase in BMP2 staining in mature polynuclear cells and an increase in BMP4 staining in endothelial sinusoid cells (Figure 3D). We also detected higher levels of BMP2 transcripts in CP-CML-sorted BM-myeloid (CD33+) and BM-megakaryocytic (CD61+) cells, confirming the probable but partial contribution of the megakaryocytic cells to BMP2 levels, together with polynuclear cells (supplemental Figure 3A). Our data indicate that the leukemic microenvironment is the main source of soluble BMP2 and BMP4 involved in the maintenance and expansion of LSCs that do not produce these cytokines in an autocrine fashion in the CP.
High levels of BMP2 and BMP4 produced by the CML microenvironment contributes to BMPRIb-induced expression. (A) Enzyme-linked immunosorbent assay quantification of BMP4 and BMP2 in BM plasma obtained from healthy donors and CP-CML samples at diagnosis. Data are categorized according to circulating platelet number measured for CML patients as either normal (≤400 G/L) or abnormally elevated (>400 G/L). Results expressed in picograms per milliliter represent the mean value ± SEM of the indicated number of analyzed samples. *P < .005 and ***P < .0001 indicate differences between the levels of BMP2 and BMP4 in CP-CML samples at diagnosis compared with healthy samples. Comparative expression of BMP2 and BMP4 genes in (B) CD34+ and CD34− cells obtained from healthy (n = 19; open bars) and CML (n = 41; closed bars) blood samples, *P < .0001 indicates differences between the expression of BMP2 and BMP4 genes in CML patient blood samples in CP at diagnosis compared with healthy samples and in (C) stromal cells derived from 3–4 weeks of culture of unmanipulated BM samples from healthy (n = 4; open bars) and at diagnosis CP-CML (n = 6; closed bars) samples. (D) In situ staining of BMP2 and BMP4 and their control antibody performed on sections of BM biopsies from nonhematologic malignancy patients (n = 2; upper panels) and CP-CML patients at diagnosis (n = 3; lower panels). Pictures were captured on a DMR microscope using PL Fluotar objective (Leica) at a magnification of ×40/1.00–0.50 oil. The following cells are indicated in the picture: (1) polynuclear neutrophils, (2) immature granular neutrophils (promyelocytes/myelocytes), (3) megakaryocytes, (4) erythroblasts, and (5) endothelial cells of the sinusoid. (E) Flow cytometry analysis of BMPRIb receptor cell surface expression on parental TF1 cells either transduced with an empty vector (non–BCR-ABL; open bars) or transduced with a vector containing BCR-ABL expressing sequence (closed bars) after chronic exposure for 4 weeks to either BMP2 or BMP4 (50 ng/mL). Results represent the mean value ± SEM of 8 experiments. *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL–transduced TF1 cells. (F) Selected CD34+ cells isolated from CP-CML samples were incubated in serum-free medium (6 × 105/mL) for 7 days in the presence of BMP4 or BMP2 (50 ng/mL), with or without coculture on a stroma composed of HS27A cells seeded at 2 × 104 cells/mL 1 day before the experiment. Comparative expression of the BMPRIb gene was then performed on CP-CML CD34+cells. Results represent the mean value ± SEM of 3 experiments.
High levels of BMP2 and BMP4 produced by the CML microenvironment contributes to BMPRIb-induced expression. (A) Enzyme-linked immunosorbent assay quantification of BMP4 and BMP2 in BM plasma obtained from healthy donors and CP-CML samples at diagnosis. Data are categorized according to circulating platelet number measured for CML patients as either normal (≤400 G/L) or abnormally elevated (>400 G/L). Results expressed in picograms per milliliter represent the mean value ± SEM of the indicated number of analyzed samples. *P < .005 and ***P < .0001 indicate differences between the levels of BMP2 and BMP4 in CP-CML samples at diagnosis compared with healthy samples. Comparative expression of BMP2 and BMP4 genes in (B) CD34+ and CD34− cells obtained from healthy (n = 19; open bars) and CML (n = 41; closed bars) blood samples, *P < .0001 indicates differences between the expression of BMP2 and BMP4 genes in CML patient blood samples in CP at diagnosis compared with healthy samples and in (C) stromal cells derived from 3–4 weeks of culture of unmanipulated BM samples from healthy (n = 4; open bars) and at diagnosis CP-CML (n = 6; closed bars) samples. (D) In situ staining of BMP2 and BMP4 and their control antibody performed on sections of BM biopsies from nonhematologic malignancy patients (n = 2; upper panels) and CP-CML patients at diagnosis (n = 3; lower panels). Pictures were captured on a DMR microscope using PL Fluotar objective (Leica) at a magnification of ×40/1.00–0.50 oil. The following cells are indicated in the picture: (1) polynuclear neutrophils, (2) immature granular neutrophils (promyelocytes/myelocytes), (3) megakaryocytes, (4) erythroblasts, and (5) endothelial cells of the sinusoid. (E) Flow cytometry analysis of BMPRIb receptor cell surface expression on parental TF1 cells either transduced with an empty vector (non–BCR-ABL; open bars) or transduced with a vector containing BCR-ABL expressing sequence (closed bars) after chronic exposure for 4 weeks to either BMP2 or BMP4 (50 ng/mL). Results represent the mean value ± SEM of 8 experiments. *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL–transduced TF1 cells. (F) Selected CD34+ cells isolated from CP-CML samples were incubated in serum-free medium (6 × 105/mL) for 7 days in the presence of BMP4 or BMP2 (50 ng/mL), with or without coculture on a stroma composed of HS27A cells seeded at 2 × 104 cells/mL 1 day before the experiment. Comparative expression of the BMPRIb gene was then performed on CP-CML CD34+cells. Results represent the mean value ± SEM of 3 experiments.
Therefore, in the leukemic context, CD34+ cells are exposed to higher concentrations of BMP2 and BMP4. We analyzed the effect of such a chronic exposure on the expression of the BMPRIb by treating TF1 cell lines with BMP2 or BMP4 for 4 weeks. Under these conditions, in the presence of the oncogene BCR-ABL, the levels of BMPRIb appeared to be maintained by BMP4 and significantly increased by BMP2 treatment (P = .0268), whereas a slight but not significant effect was observed in nontransduced TF1 cells (Figure 3D). Similar results were obtained using primary CD34+ CP-CML cells that displayed an amplified BMPRIb expression when cocultured with the immortalized human BM-stroma cell line HS27A,32 which overproduces BMP2 and BMP4 (supplemental Figure 3B; Figure 3F). Together, our results show that immature leukemic cells not only display intrinsic alterations as a result of the BCR-ABL oncogene but also increased and sustained high levels of BMPRIb expression regulated by their niche.
BMP2 and BMP4 expand immature CD34+ cells from CP-CML patients
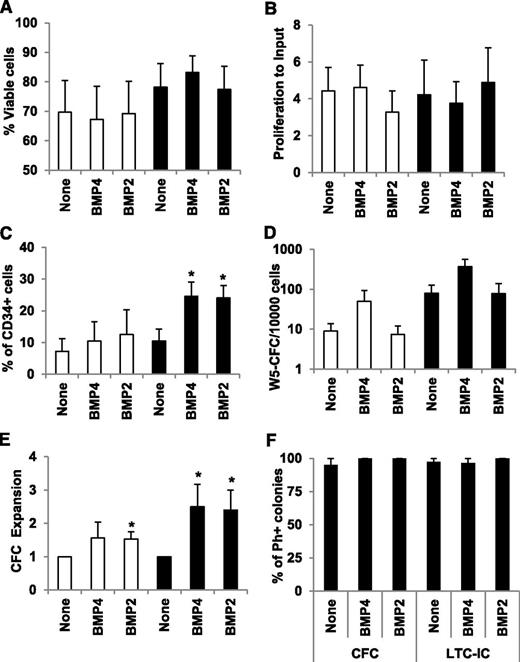
Deregulations in the BMP pathway in immature leukemic cells, together with a significant increase in available soluble BMPs within the leukemic environment, predict a perturbed response of LSCs to BMPs. To investigate this hypothesis, we compared the biological response of CD34+ immature cells isolated from healthy BM donors or CP-CML patients at diagnosis to soluble BMP4 and BMP2. CD34+ cells were incubated for 7 days in serum-free medium supplemented with BMP2 or BMP4 (50 ng/mL) and analyzed for cell viability, proliferation, and CD34 cell surface expression. We observed no differences in cell viability (Figure 4A) and proliferation (Figure 4B) compared with untreated cells (none), which suggests a lack of toxic effects of BMP2 or BMP4 in primitive cells. The number of CD34-expressing cells increased in the CML samples in the presence of BMP4 (P = .03) and BMP2 (P = .026), but not in healthy donors (Figure 4C). This increase in CD34+ cells suggests either higher survival rates/maintenance or the amplification of LSCs, as shown for BMP4 in normal HSC.12,15 We then examined the quantities of stem and progenitor cells within the BMP-treated CD34+ cells, using LTC-IC and CFC assays. BMP2 had no effect, but LTC-IC numbers increased with BMP4 by 5.6-fold (P = .06) in healthy cells and, similarly, by 4.7-fold in leukemic cells (Figure 4D). However, 58% (7/12) of healthy CD34+ cells were expanded for their LTC-IC content by BMP4; this frequency decreased to 34% (3/9) in CP-CML samples. Surprisingly, although fewer CML-CD34+ cells responded to BMP4, when they did, it was more pronounced, as indicated by the large number of LTC-IC colonies we detected (data not shown). The progenitor compartment (CFC) was increased by both BMP4 and BMP2 treatment in healthy (1.6-fold and 1.5-fold [P = .024], respectively) and leukemic (2.5-fold [P = .033] and 2.4-fold [P = .022], respectively) samples (Figure 4E). To establish whether the maintenance or amplification of CFC or LTC-IC by BMPs affect remaining residual healthy or leukemic cells, we harvested individual colonies and analyzed them by qPCR for their expression of the normal Abelson (ABL) gene and the BCR-ABL oncogene. Colonies derived from healthy samples expressed ABL, but not BCR-ABL (data not shown). In the presence of BMP2 (5/5) and BMP4 (21/22), colonies detected in LTC-IC assays from patient samples were truly leukemic, as more than 95% of them expressed BCR-ABL (Figure 4F). All single CFC colonies (100%) derived from treated CD34+ cells expressed BCR-ABL. These results indicate that BMP2 and BMP4 specifically affect the leukemic primitive cell compartment. In summary, these results indicate that changes in the expression of multiple BMP signaling factors correlate with a deregulated and amplified biological response in leukemic CD34+ cells after exposure to soluble BMPs.
Effects of BMP2 and BMP4 treatments on leukemic progenitor amplification. Selected CD34+ cells isolated from healthy donor (open bars) or CP-CML (closed bars) samples were incubated in serum-free medium (6 × 105/mL) for 7 days in the presence of BMP2 or BMP4 (50 ng/mL). *P < .05 indicate differences between healthy and CML samples. Cell viability (A) and proliferation (B) were evaluated using trypan blue staining and cell counting. Results are expressed as the percentage of viable cells or the proliferation ratio, which is the ratio of cell counts to the number of input cells. The mean ± SEM of 10 experiments is presented. (C) Phenotypic analysis of CD34, the hematopoietic progenitor marker, was performed using flow cytometry on a FACSCalibur cell analyzer (Becton Dickinson) and gated for viable cells. Results are presented as the mean ± SEM of 5 experiments for healthy donors and 8 experiments for CML patients. (D) SC content of treated cells was analyzed by the LTC-IC coculture assay, and results are presented as the mean value ± SEM for 12 or 9 experiments for healthy donors and CML patients, respectively. Results are presented as the total LTC-IC–derived colonies after 5 weeks of coculture per 1 × 104 seeded cells. (E) The progenitor content of treated cells was analyzed by the clonogenic CFC assay. Results are expressed as the ratio of treated to untreated cells and represent the mean value ± SEM of 12 or 19 experiments for healthy donors and CML patients, respectively. (F) The number of leukemic colonies in either CFC- or LTC-IC–derived colonies was assessed by picking individual colonies to quantify expression of ABL, BCR-ABL, and the reference genes BGUS and TBP by qPCR. Leukemic positive colonies were determined by simultaneous detection of all genes, whereas normal negative colonies were stated as ABL-positive but BCR-ABL–negative. In all cases, internal gene controls were positive; otherwise, those samples would have been removed. Results are expressed as the percentage of positive BCR-ABL colonies of the total assayed colonies of 3 experiments for healthy donors and CML patients.
Effects of BMP2 and BMP4 treatments on leukemic progenitor amplification. Selected CD34+ cells isolated from healthy donor (open bars) or CP-CML (closed bars) samples were incubated in serum-free medium (6 × 105/mL) for 7 days in the presence of BMP2 or BMP4 (50 ng/mL). *P < .05 indicate differences between healthy and CML samples. Cell viability (A) and proliferation (B) were evaluated using trypan blue staining and cell counting. Results are expressed as the percentage of viable cells or the proliferation ratio, which is the ratio of cell counts to the number of input cells. The mean ± SEM of 10 experiments is presented. (C) Phenotypic analysis of CD34, the hematopoietic progenitor marker, was performed using flow cytometry on a FACSCalibur cell analyzer (Becton Dickinson) and gated for viable cells. Results are presented as the mean ± SEM of 5 experiments for healthy donors and 8 experiments for CML patients. (D) SC content of treated cells was analyzed by the LTC-IC coculture assay, and results are presented as the mean value ± SEM for 12 or 9 experiments for healthy donors and CML patients, respectively. Results are presented as the total LTC-IC–derived colonies after 5 weeks of coculture per 1 × 104 seeded cells. (E) The progenitor content of treated cells was analyzed by the clonogenic CFC assay. Results are expressed as the ratio of treated to untreated cells and represent the mean value ± SEM of 12 or 19 experiments for healthy donors and CML patients, respectively. (F) The number of leukemic colonies in either CFC- or LTC-IC–derived colonies was assessed by picking individual colonies to quantify expression of ABL, BCR-ABL, and the reference genes BGUS and TBP by qPCR. Leukemic positive colonies were determined by simultaneous detection of all genes, whereas normal negative colonies were stated as ABL-positive but BCR-ABL–negative. In all cases, internal gene controls were positive; otherwise, those samples would have been removed. Results are expressed as the percentage of positive BCR-ABL colonies of the total assayed colonies of 3 experiments for healthy donors and CML patients.
BMP2 and BMP4 induce the specific amplification of committed myeloid progenitors
To confirm the effect of BMP2 and BMP4 on leukemic CD34+ progenitors, we analyzed their activities on the different categories of clonogenic cells (supplemental Figure 4A). The most significant effects were observed in the granulo-monocytic compartment, with a 9.6-fold increase in late leukemic colony forming unit (CFU)–granulocyte macrophage (GM) induced by BMP4 treatment (P = .045). BMP2 exposure led to a 13-fold increase in early granulo-monocytic progenitors (P = .05; Figure 5A). No difference was observed in the number of erythroid or CFU–granulocyte erythrocyte macrophage megakaryocyte mixed colonies (not shown). These data strongly support the notion that BMP2 and BMP4 are involved in the amplification of leukemic myeloid progenitors.
BMP2 and BMP4 amplify leukemic myeloid progenitors. Selected CD34+ cells (6 × 105/mL) were incubated in serum-free medium for 7 days in the presence of either BMP2 or BMP4 (50 ng/mL). The progenitor content of the treated cells was analyzed using the CFC assay. (A) We scored them as indicated as early or late erythroid-E and granulo-monocytic-GM (as defined in supplemental Figure 4). Results represent the mean value ± SEM of 12 or 15 experiments for healthy donors and CML patients, respectively, and are expressed as ratio of treated to untreated cells (none). (B) CFC values obtained from CD34+ cells treated in the same experiments with BMP2 or BMP4 and isolated from 12 or 18 different healthy or CP-CML patients at diagnosis, respectively, are expressed as a percentage of the total number of colonies obtained and are presented as pie charts. P < .05 indicates differences between treated and untreated cells. (C) Sorted CD34+CD38− cells from healthy and CML samples were plated at 1 cell/well in serum-free medium in the presence of 50 ng/mL BMP2 or BMP4. After 7 days, methylcellulose was added to the wells, and single cell–derived colonies were scored 2 weeks later. We scored all colonies (total CFC) as well as CFU-GM content. Results are expressed as percentage of wells that give rise to colonies and represent the mean value ± SEM of 5 experiments for healthy donors and CML patients.
BMP2 and BMP4 amplify leukemic myeloid progenitors. Selected CD34+ cells (6 × 105/mL) were incubated in serum-free medium for 7 days in the presence of either BMP2 or BMP4 (50 ng/mL). The progenitor content of the treated cells was analyzed using the CFC assay. (A) We scored them as indicated as early or late erythroid-E and granulo-monocytic-GM (as defined in supplemental Figure 4). Results represent the mean value ± SEM of 12 or 15 experiments for healthy donors and CML patients, respectively, and are expressed as ratio of treated to untreated cells (none). (B) CFC values obtained from CD34+ cells treated in the same experiments with BMP2 or BMP4 and isolated from 12 or 18 different healthy or CP-CML patients at diagnosis, respectively, are expressed as a percentage of the total number of colonies obtained and are presented as pie charts. P < .05 indicates differences between treated and untreated cells. (C) Sorted CD34+CD38− cells from healthy and CML samples were plated at 1 cell/well in serum-free medium in the presence of 50 ng/mL BMP2 or BMP4. After 7 days, methylcellulose was added to the wells, and single cell–derived colonies were scored 2 weeks later. We scored all colonies (total CFC) as well as CFU-GM content. Results are expressed as percentage of wells that give rise to colonies and represent the mean value ± SEM of 5 experiments for healthy donors and CML patients.
We compared the effects of BMP2 and BMP4 in the same experiments on CD34+-CML cells obtained from 18 patients. The absolute number of colonies, obtained in each condition from the same initial number of purified CD34+ CML cells, was used to normalize the number of colonies obtained for the erythroid and granulo-monocytic categories, and values were plotted as the percentage of progenitor subtype. Compared with untreated CML cells (54%), those treated with BMP4 or BMP2 amplified more granulo-monocytic progenitors (61% and 65%, respectively) at the expense of erythroid progenitors (Figure 5B). The balance between granulo-monocytic and erythroid colonies thus significantly shifted toward a more granulo-monocytic phenotype with either BMP2 (P = .01) or BMP4 (P = .02) in a leukemic context but remained unchanged for healthy cells. To evaluate whether BMP2 and BMP4 induced LSC commitment toward the myeloid lineage, we sorted CD34+CD38− cells from CP-CML patients at diagnosis (n = 5) and from healthy donors (n = 5). The cells were plated at 1 cell/well in serum-free conditions with or without BMP2 or BMP4. After 7 days, methylcellulose was added to the wells, and single cell–derived colonies were scored 2 weeks later. Consistent with the bulk culture results, less than 2% of normal single cells gave rise to colonies (Figure 5C), whereas the CML samples exhibited a strong increase in cloning efficiency (10%–16%). Only the leukemic CD34+CD38− cells responded to BMP2 and BMP4, with a slight increase in total and CFU-GM colonies by BMP4 and no effect in the presence of BMP2 (Figure 5C; supplemental Figure 4B). These data suggest that only BMP4 has a very weak effect on single-cell proliferation, and the clonogenic ability of the LSC while BMP2 is likely to contribute to the expansion of more differentiated myeloid progenitors.
BMPRIb drives primitive leukemic myeloid progenitor amplification
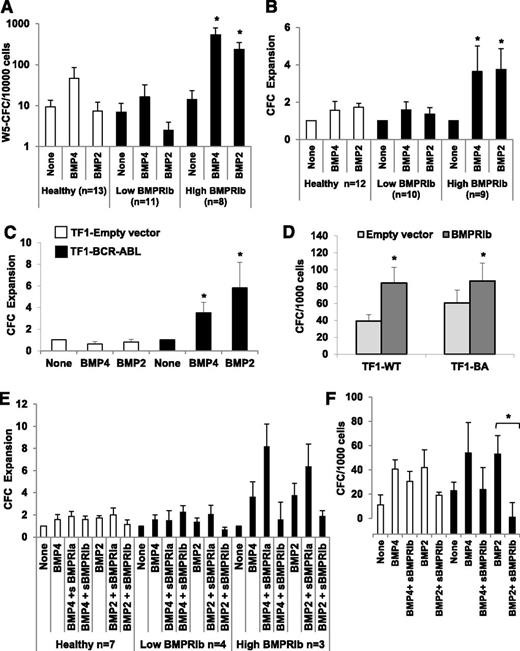
To characterize the direct involvement of the BMP pathway in the amplification of leukemic myeloid progenitors, we analyzed the effect of BMP2 and BMP4 treatments on the CFC and LTC-IC output of primary CD34+ cells. Cells were isolated from healthy donors or CP-CML patients but divided into 2 groups on the basis of the level of BMPRIb qPCR expression. The BMPRIb-low group expressed transcript levels similar to normal samples (<10 arbitrary units), contrary to the BMPRIb-high group, which had transcript levels increased by 10-fold. In contrast to the BMPRIb-low patients, the BMPRIb-high group of samples strongly responded to BMP4 or BMP2 exposure, as measured by a major increase in LTC-IC (107-fold [P = .0018] and 15-fold [P = .003], respectively) (Figure 6A) and CFC (4-fold [P = .05] and 4-fold [P = .008], respectively) (Figure 6B). These data suggest that although BMP4 has a very weak effect and BMP2 has no effect at all on cell fate decisions, in the overall CP-CML analysis of LSCs, BMP2 and BMP4 both appear to induce a very strong expansion of LSCs overexpressing BMPRIb that correlates to CFC amplification. Using the TF1-BCR-ABL model, we reproduced the CFC expansion after BMP4 or BMP2 chronic exposure to a similar extent as measured in primary cells, with a 3.5-fold and 6-fold CFC increase in the presence of BMP4 or BMP2, respectively (Figure 6C), and no effect with control cells (TF1-Empty vector). The introduction of BMPRIb was by itself sufficient to increase the CFC number by 2.1-fold (P = .003), whereas BCR-ABL alone led to only a 1.5-fold increase (P = .05) (Figure 6D). Introducing BMPRIb into TF1-BA cells did not further expand CFCs. Last, we treated primary CD34+ cells taken at diagnosis from CP-CML patients from BMPRIb-low or BMPRIb-high groups with soluble forms of BMPRIa (sBMPRIa) or BMPRIb (sBMPRIb), using previously determined optimal conditions,12 to outcompete BMP binding to endogenous membrane-bound receptors. The BMP2- and BMP4-mediated CFC increases were reduced in the presence of sBMPRIb and inversely increased when incubated with sBMPRIa (Figure 6E). Similarly, a significant (P = .042) inhibition by of BMP2-driven CFC expansion was observed (Figure 6F) by treating TF1 cells transduced or not with BCR-ABL for 48 hours with sBMPRIb. These data confirm that changes in BMPRIb expression are associated with an alteration of the BMP2 and BMP4 response involved in the amplification of the leukemic myeloid stem and progenitor compartment in CP-CML patients.
BMPRIb receptor mediates BMP2 and BMP4 effects on LSC and myeloid progenitors. After qPCR analysis of BMPRIb expression, CP-CML samples were divided as low and high BMPRIb-expressing CML samples. CD34+ cells were then isolated from healthy donors (open bars) or both groups of CP-CML samples (closed bars) and incubated at 6 × 105/mL in serum-free medium for 7 days in the presence of BMP2 or BMP4 (50 ng/mL). (A) The SC content of the treated cells was analyzed using the LTC-IC cocultured assay; results represent the mean value ± SEM of the indicated number of samples. Results are presented as the total LTC-IC-derived week 5 colonies per 1 × 104 seeded cells. (B) The progenitor content of treated cells was analyzed using the CFC assay. Results are expressed as ratio of treated to untreated cells and represent the mean value ± SEM of the indicated number of samples. (C) BCR-ABL-transduced (closed bars) or empty vector-transduced (open bars) TF1 cells were continuously treated for 4 weeks by 50 ng/mL BMP2 or BMP4 and then assayed by CFC assay for their progenitor content. Results are expressed as the ratio of treated to untreated cells and represent the mean value ± SEM of 5 experiments. (D) Parental TF1 cells (TF1-wild-type) or TF1-BCR-ABL (TF1-BA) cells were transfected with a control empty (gray bars) or a BMPRIb-encoding (dark gray bars) vector. The effect on CFC output was analyzed using the CFC assay. Results are expressed as the total CFC colonies per 1 × 103 seeded cells and represent the mean value ± SEM of 7 or 3 experiments for TF1-wild-type and TF1-BCR-ABL, respectively. (E) The progenitor content of CD34+ cells isolated from healthy donors (open bars) or CP-CML (closed bars) samples. The cells were divided as low- and high-BMPRIb-expressing CML-samples and were treated for 7 days in serum-free medium by BMP2 or BMP4 (50 ng/mL) in the presence or absence of soluble BMPRIa or BMPRIb receptor (4 μg/mL). Results are expressed as ratio of treated to untreated cells and represent the mean value ± SEM of the indicated number of experiments. (F) The progenitor content of parental TF1 cells (TF1-wild-type; open bars) or TF1-BCR-ABL cells (closed bars) was analyzed by CFC assay after 48 hours of treatment by BMP2 or BMP4 (50 ng/mL), with or without soluble BMPRIb receptor (4 μg/mL). Results are expressed as the total CFC colonies per 1 × 103 seeded cells and represent the mean value ± SEM of 5 experiments. *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells.
BMPRIb receptor mediates BMP2 and BMP4 effects on LSC and myeloid progenitors. After qPCR analysis of BMPRIb expression, CP-CML samples were divided as low and high BMPRIb-expressing CML samples. CD34+ cells were then isolated from healthy donors (open bars) or both groups of CP-CML samples (closed bars) and incubated at 6 × 105/mL in serum-free medium for 7 days in the presence of BMP2 or BMP4 (50 ng/mL). (A) The SC content of the treated cells was analyzed using the LTC-IC cocultured assay; results represent the mean value ± SEM of the indicated number of samples. Results are presented as the total LTC-IC-derived week 5 colonies per 1 × 104 seeded cells. (B) The progenitor content of treated cells was analyzed using the CFC assay. Results are expressed as ratio of treated to untreated cells and represent the mean value ± SEM of the indicated number of samples. (C) BCR-ABL-transduced (closed bars) or empty vector-transduced (open bars) TF1 cells were continuously treated for 4 weeks by 50 ng/mL BMP2 or BMP4 and then assayed by CFC assay for their progenitor content. Results are expressed as the ratio of treated to untreated cells and represent the mean value ± SEM of 5 experiments. (D) Parental TF1 cells (TF1-wild-type) or TF1-BCR-ABL (TF1-BA) cells were transfected with a control empty (gray bars) or a BMPRIb-encoding (dark gray bars) vector. The effect on CFC output was analyzed using the CFC assay. Results are expressed as the total CFC colonies per 1 × 103 seeded cells and represent the mean value ± SEM of 7 or 3 experiments for TF1-wild-type and TF1-BCR-ABL, respectively. (E) The progenitor content of CD34+ cells isolated from healthy donors (open bars) or CP-CML (closed bars) samples. The cells were divided as low- and high-BMPRIb-expressing CML-samples and were treated for 7 days in serum-free medium by BMP2 or BMP4 (50 ng/mL) in the presence or absence of soluble BMPRIa or BMPRIb receptor (4 μg/mL). Results are expressed as ratio of treated to untreated cells and represent the mean value ± SEM of the indicated number of experiments. (F) The progenitor content of parental TF1 cells (TF1-wild-type; open bars) or TF1-BCR-ABL cells (closed bars) was analyzed by CFC assay after 48 hours of treatment by BMP2 or BMP4 (50 ng/mL), with or without soluble BMPRIb receptor (4 μg/mL). Results are expressed as the total CFC colonies per 1 × 103 seeded cells and represent the mean value ± SEM of 5 experiments. *P < .05 indicates differences between parental TF1 cells transduced with an empty vector and BCR-ABL-transduced TF1 cells.
Discussion
In various cancers including leukemia, the SC niche is often deregulated without being transformed per se,33,34 but its contribution to the maintenance, survival, and resistance of LSCs has only started to be deciphered. Here, we report a significant increase in the availability of soluble BMP2 and BMP4 in the BM of CP-CML patients at diagnosis, combined with an oversensitivity of LSCs to these molecules. In line with a recent in vivo study that demonstrated that CML-associated changes in the microenvironment confer a selective growth advantage to LSC,35 we revealed for the first time the crucial role of alterations in the BMP pathway during the early stages of CML.
By analyzing more than 70 samples from CP-CML patients at diagnosis, we demonstrated that the expression of a multitude of elements in the BMP pathway is altered in almost all CML subpopulations, including the SC/progenitor compartment, indicating a pathologic deregulation of this pathway, while not reflecting the prevalence of a specific subpopulation. Some changes become more striking during the course of disease progression, such as BMPRIb overexpression. Our results, together with the fact that we restricted our analysis to samples obtained from patients that only display the t(9;22) translocation without additional clonal abnormalities, suggest that the BMP signaling alterations represent an early event in the transformation process.
We demonstrated that alterations in the BMP pathway do not prevent leukemic cells from responding to exogenous BMP2 and BMP4. These data were confirmed using a model of BCR-ABL-positive immature CD34+ cells reproducing the main features observed in primary immature CP-CML cells. Using this model, we further showed that overexpression of BCR-ABL increased BMPRIb surface expression. The erythroid–myeloid progenitor balance changed in favor of the granulo-monocytic lineage when treating CML-CD34+ cells with BMP2 and BMP4. In addition, BMP4-expanded LTC-ICs were all genotypically Ph+, suggesting that the biological response of CD34+ cells to BMP2 and BMP4 is specifically altered in CML. It then suggests that BMP4-mediated amplification and maintenance of LSCs and BMP2-dependent expansion of myeloid progenitors are related to a hypersensitivity to exogenous BMP2 and BMP4 signaling mediated by BMPRIb overexpression. This would further explain how LSCs, in contrast to normal HSCs, respond to soluble BMPs, thus contributing to the vast increase in the myeloid compartment observed in CML.
Soluble BMPs, naturally provided by the BM niche,31 have been implicated in myelofibrotic processes.36 In CML, changes in the SC niche such as secondary myelofibrosis37 are often correlated with disease progression. Myelofibrosis is characterized by an increased production in collagens, proteins known to sequester BMPs and control the gradient of these molecules in the microenvironment. In addition, TKIs induce BMP2 expression by mesenchymal SCs.38 Furthermore, BMP4 increases hematopoietic progenitor adhesion to the stroma and regulates HSC behavior.12,17,18 Therefore, CML evolution and TKI treatment may contribute to a local increase in BMPs in the LSC niche. This also explains why both normal and leukemic SCs, as well as BM alterations observed in patients, are amplified throughout disease progression and may fuel a permanent and autonomous pool of leukemic progenitors.
In summary, we have demonstrated the existence of molecular and functional alterations in the BMP pathway in CML cells, as well as alterations in the quantities of soluble BMPs present in the tumoral niche itself. These alterations are involved in the CP of the disease through their role in the survival of LSCs, as well as in the expansion of myeloid progenitors (Figure 7). The analysis of the BMP pathway may therefore represent an interesting prognostic tool, allowing the design of drugs directly targeting the reservoir of LSCs.
Proposed model for BMP pathway alteration effects on CML LSC survival and myeloid progenitor expansion.
Proposed model for BMP pathway alteration effects on CML LSC survival and myeloid progenitor expansion.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank S. Morisset for statistics (Centre Hospitalier Lyon Sud, Pierre-Benite, France), S. Joly and I. Durand (Cytometry Facility, Cancer Research Center of Lyon, France) for their excellent technical assistance, and Elsevier for providing cell representation items.
This study was funded by Novartis, the patient association “Regarde un jour le monde” grants (to F.E.N. and V.M.-S.), and the Institut National de la Santé et de la Recherche Médicale, Ligue Nationale contre le Cancer (Saone et Loire), “Cent pour cent la vie” and Clara-INCA grants (to V.M.-S.). B.L is the recipient of a fellowship from the French government. K.S. was funded by grants from Novartis, Bristol-Myers Squibb, and the patients associations “Pense à Moelle,” “Guillaume Espoir,” and “Arhepan.”
Authorship
Contribution: B.L, S.J., K.S, B.K., S.R., M.F. and T.V. performed experiments and data analysis; S.S. provided tools, contributed to discussions, and proofread the manuscript; F.E.N. followed-up the patients, provided patient samples, analyzed the data, and contributed to writing the manuscript; and V.M.-S. designed the study, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Véronique Maguer-Satta, Cancer Research Center of Lyon-CRCL, U1052-UMR5286, 28 rue Laennec, 69373 Lyon Cedex 08, France; e-mail: veronique.maguer-satta@lyon.unicancer.fr; and Franck E. Nicolini, Hematology Department 1G, Pavillon Marcel Bérard, Centre Hospitalier Lyon-Sud, 165 Chemin du grand Revoyet, 69495 Pierre Bénite Cedex France; e-mail: franck-emmanuel.nicolini@chu-lyon.fr.
References
Author notes
B.L. and S.J. contributed equally to this study.